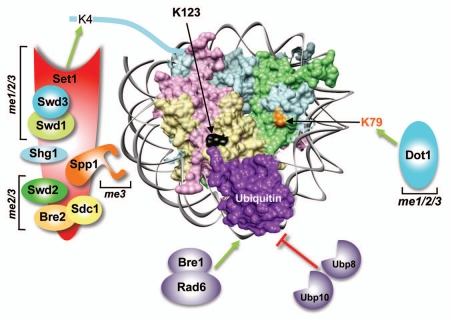
Figure 1.
The trans-histone crosstalk between histone H2B ubiquitination and H3K4/-K79 methylation in budding yeast. A simulated structure for an ubiquitinated nucleosome is shown by placing the C-terminal residue of ubiquitin (PDB: 1UBQ) close to lysine 123 (K123) of H2B present in the yeast nucleosome (PDB 1ID3; H3, pale blue; H4, pale green; H2A, pale pink; H2B, pale yellow). Rad6 (the ubiquitin-conjugating enzyme) and Bre1 (the E3 ligase) conjugate ubiquitin onto K123 in the H2B C-terminal helix. The conjugated ubiquitin is removed by two deubiquitinases, Ubp8 and Ubp10. Histone methyltransferases Set1 and Dot1 catalyze mono- (me1), di- (me2) and trimethylation (me3) of H3 K4 and K79 residues, respectively. Set1 is present in a multi-protein complex (COMPASS) along with seven subunits, which regulate the complex stability, integrity and processive methylation.