Abstract
Chromatin structure is greatly influenced by histone tail post-translational modifications (PTM), which also play a central role in epigenetic processes. Antibodies against modified histone tails are central research reagents in chromatin biology and molecular epigenetics. We applied Celluspots peptide arrays for the specificity analysis of 36 commercial antibodies from different suppliers, which are directed towards modified histone tails. The arrays contained 384 peptides from eight different regions of the N-terminal tails of histones, viz. H3 1–19, 7–26, 16–35 and 26–45, H4 1–19 and 11–30, H2A 1–19 and H2B 1–19, featuring 59 post-translational modifications in many different combinations. Using various controls we document the reliability of the method. Our analysis revealed previously undocumented details in the specificity profiles of the tested antibodies. Most of the antibodies bound well to the PTM they have been raised for, but some failed. In addition, some antibodies showed high cross-reactivity and most antibodies were inhibited by specific additional PTMs close to the primary one. Furthermore, specificity profiles for antibodies directed toward the same modification sometimes were very different. The specificity of antibodies used in epigenetic research is an important issue. We provide a catalog of antibody specificity profiles for 36 widely used commercial histone tail PTM antibodies. Better knowledge about the specificity profiles of antibodies will enable researchers to implement necessary control experiments in biological studies and allow more reliable interpretation of biological experiments using these antibodies.
Key words: histone modification, histone methylation, histone acetylation, histone phosphorylation, chromatin, antibody, specificity, ChIP
Introduction
The regulation of gene expression and chromatin state by epigenetic modifications is an essential process in development, physiology and disease.1–4 Epigenetic modifications comprise the methylation of the DNA and the post-translational modification (PTM) of histone tails including acetylation, phosphorylation or methylation, which can occur at Arg and Lys and lead to monomethylation, dimethylation (symmetric or asymmetric in case of Arg) or trimethylation in case of Lys. Altogether, over 100 different PTMs of histone proteins have been identified so far, many of them with defined and important roles in the regulation of gene expression, chromatin biology, cell cycle regulation, DNA repair and replication.1,2 Different methods were established for the biochemical detection of DNA methylation, including the conversion of unmethylated cytosines to uracils by treatment with sodium bisulfite, the protection of methylated DNA against digestion with restriction enzymes or the selective binding of methylated DNA to antibodies or other methylated DNA binding proteins.5,6 In contrast to this, the locus specific investigation of histone tail PTMs in chromatin completely relies on a single method—the specific interaction of modified histone tails with antibodies.7,8 Antibodies are used for pull-down of DNA bound to chromatin carrying a certain modified histone tail, followed by various downstream analytical techniques including quantitative PCR, binding to oligonucleotide arrays or next generation sequencing. In addition, antibodies are used for direct staining of modified chromatin in fixed cells and for studying of chromatin states. Furthermore, antibodies are essential tools to investigate the binding specificities of histone PTM reading domains and in the specificity analysis of histone modifying enzymes and they are an important component in ELISA based screening assays for inhibitors of histone-modifying enzymes. Consequently, much research in molecular epigenetics and chromatin biology is based on PTM specific antibodies against histone tails, and several companies offer hundreds of different monoclonal or polyclonal antibodies directed against histone PTMs and various academic groups developed their own reagents.
In experimental studies, antibody binding is taken as evidence for the presence or absence of certain PTMs on particular residues of histones. Since this interpretation is totally dependent on the specificity of the antibody, detailed information on the antibody specificity profile is required. Antibodies may show cross-reactivity and bind to secondary sites, giving rise to a signal in the absence of the primary epitope (false positives). In addition, secondary modifications within the binding epitope of the antibody may lead to the loss or reduction of binding, although the primary modification is present (false negatives). In this context, it is relevant that the N-terminal histone tails are amongst the most extensively modified protein regions known. For example, seven residues are currently known to be modified within position 2–11 of the N-terminal tail of H3.
Currently, there is no standardized, cost efficient, accurate and detailed method available for specificity analysis of histone tail antibodies. Quality control of histone PTM antibodies is usually done by testing the interaction with different modified histone tail peptides,9 but there are no established criteria with respect to the number of different peptides to be used and how the results are documented. As a consequence, many important studies have been conducted without providing detailed information on the binding properties of the used antibodies.
Results
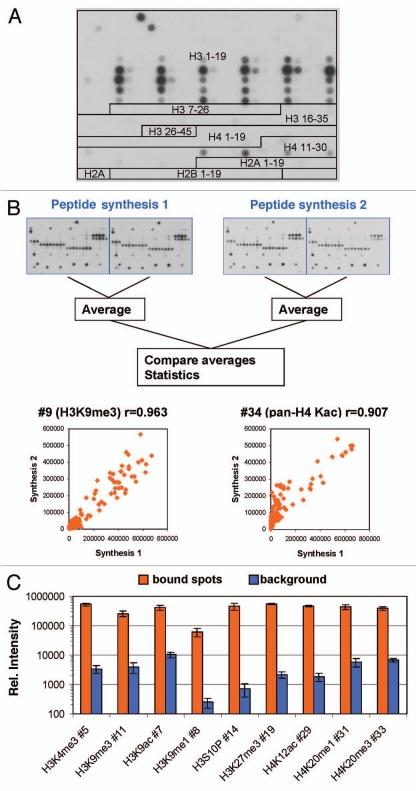
Peptide arrays synthesized on cellulose membranes by the SPOT method were introduced by R. Frank in 1992.10 Since that time, peptide arrays developed into valuable tools11–16 that were applied, for example, to analyze the specificity of peptide modifying enzymes17–21 and the binding specificities of antibodies and proteins including epigenetic reading domains.12,15,16,19,21–25 Here, we used Celluspots peptide arrays comprising 384 peptides to study the specificity of the interaction of 36 different commercial antibodies raised for modified histone tails. In the Celluspots technique, peptides are synthesized on cellulose support as in conventional SPOT synthesis, but after synthesis the cellulose matrix together with the peptides is solubilized and spotted on glass slides.26 Due to miniaturization when compared with conventional SPOT synthesis, Celluspots arrays can be produced at lower costs and assays can be performed using much less reagent. The arrays contained peptides from eight different regions of the N-terminal tails of histones, viz. H3 1–19, 7–26, 16–35 and 26–45, H4 1–19 and 11–30, H2A 1–19 and H2B 1–19 (Fig. 1A and Sup. Table 1), featuring 59 post-translational modifications (most of them identified, some hypothetical) in many different combinations.
Figure 1.
(A) Design of the peptide array used in this study (detailed information on the sequence and modification of each peptide is given in Sup. Table 1). Here the image obtained with antibody #1 is used for illustration. (B) Antibody binding to independently synthesized peptides. Spot intensities were averaged from the two internal repeats of the array and compared between arrays that were synthesized independently. One image obtained with H3K4me3 is chosen for illustration. The scatter plots show a comparison of the intensity of peptide binding in both arrays. The r value refers to the Pearson correlation coefficient of both intensities. (C) Reproducibility of peptide binding intensities between different peptides on the same array. For several antibodies, the binding intensities to all peptides containing the primary epitope (after exclusion of false negatives) were averaged and plotted in log scale (orange columns, the error bars display the standard deviations). As background, binding intensities to the 100 weakest spots were used (blue columns, the error bars display the standard deviations). For antibody numbers cf. Supplemental Table 2.
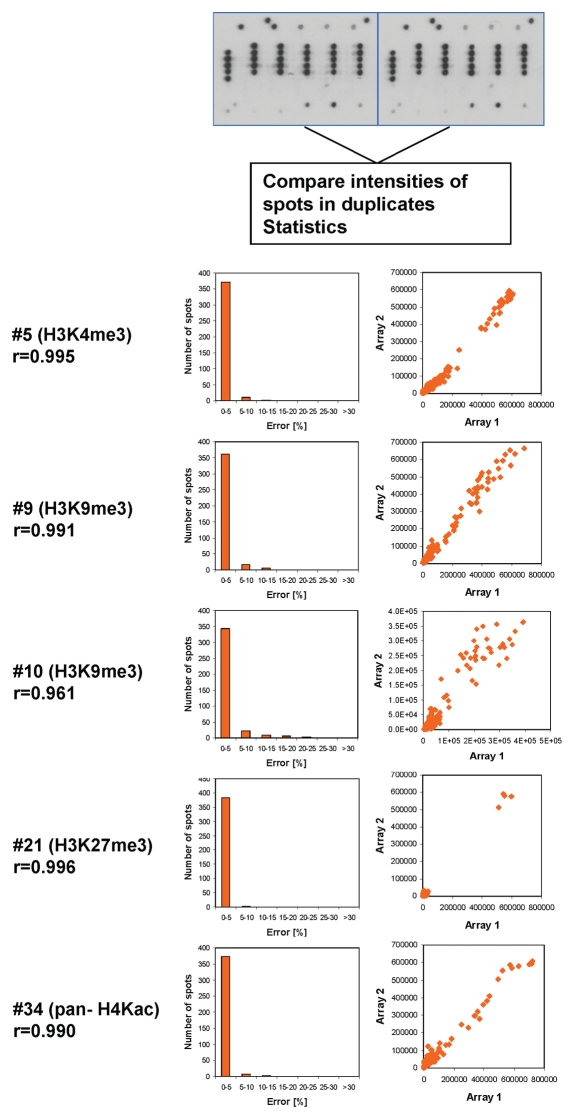
Antibody binding was visualized using a secondary antibody and ECL detection system. To avoid overestimation of cross-reactivity and potential saturation of the detection system, antibodies with high levels of cross-reactivity or high signal intensity were assayed at higher dilutions until the results no longer changed with further dilution. For quality control, each glass slide contained two copies of the array. Binding to these internal duplicates was highly reproducible, which ensures reproducible spotting (Fig. 2). Binding to slides with independently synthesized peptides was found to be very similar as well (Fig. 1B). We compared the binding of antibodies to different peptides containing the primary epitope (by excluding the false negatives, see below) and observed very reproducible binding intensities with standard errors within 5–30% while the difference between bound spots and background was usually more than 100-fold (Fig. 1C). These observations indicated that the synthesis of the peptides was comparable between different peptides and between batches and the signal/noise ratio of the readout is excellent.
Figure 2.
Internal reproducibility of the peptide binding. Spot intensities were quantified on the two internal repeats of the array and compared. One image obtained with H3K4me3 is chosen for illustration. Examples of the results are given for several antibodies. The box plots show the number of pairs of spots within certain error margins obtained after normalization to the maximum binding. The scatter plots show a comparison of the intensity of each peptide in both arrays. The r value refers to the Pearson correlation coefficient of both intensities.
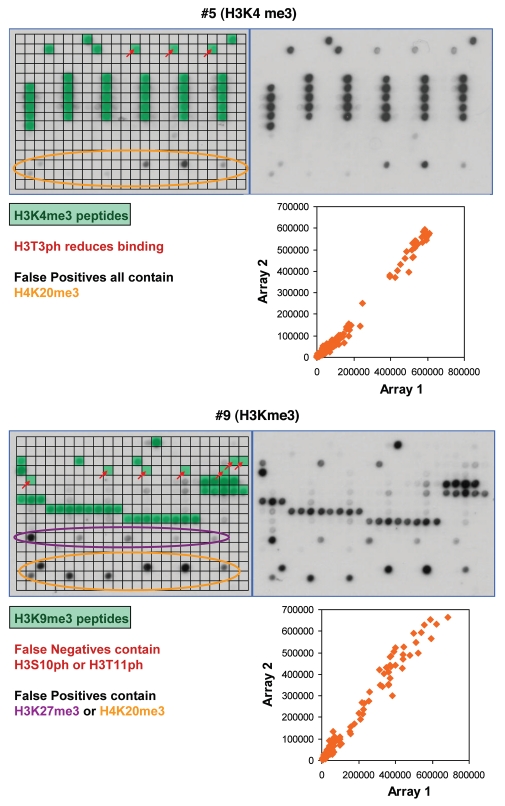
Altogether, we tested the peptide tail binding specificity of 36 antibodies from major suppliers and analyzed the results in detail (Table 1 and Sup. Table 1, examples of the results are shown in Figs. 3–5, all technical details are given in Sup. Table 2, the tables of spot intensities are provided in Sup Table 2). In most cases the antibodies showed highest specificity towards the expected target although some of them failed (like #25), which is critical for previous studies using these reagents. Furthermore, our data revealed important details on the specificity of the antibodies which were not documented before. For example, antibody #5 directed against H3K4me3 showed weak binding to peptides also containing H3T3ph (false negatives) and cross-reactivity at some H4K20me3 spots (false positives) (Fig. 3). Antibody #9 directed against H3K9me3 bound weakly to peptides also containing H3S10ph or H3T11ph and it showed cross-reactivity at H3K27me3 and H4K20me3. The different “pan”-antibodies supposed to bind to a defined set of related modifications were particular critical cases, since all of them showed clear preferences; sometimes the binding spectrum was rather restricted. The importance of an antibody specificity analysis can be further exemplified by the results obtained with four different H3K27me3 antibodies (Fig. 4). While all of them bind to the single modified H3K27me3 peptide with good preference, antibody #19 did not bind to any of the peptides carrying additional modifications like R26me2a, R26me2s (which is hypothetical at present) or S28ph and it showed weak cross-reactivity at H3K27ac. Antibody #20 bound to all H3K27me3 peptides but it showed strong cross-reactivity at H4K20me3 and weak cross-reactivity at H3K4m3 and H3K9me3. Antibody #21 did not show cross-reactivity, but it did not bind to peptides containing S28ph in addition to K27me3. Antibody #22 showed cross-reactivity at H4K20me3 and showed only weak binding to peptides containing S28ph in addition to K27me3. Therefore, studies on H3K27me3 distribution likely will come to different conclusions if these different antibodies are used.
Table 1.
Compilation of the results of the specificity analyses with different antibodies
| No. | Antibody target site | SFT | SFN | SFT/SFN |
| 1 | H3K4me1 | 30 | 3 | 10 |
| 2 | H3K4me2 | 42 | 1.3 | 33 |
| 3 | H3K4me2 | 62 | 3 | 19 |
| 4 | H3K4me3 | 31 | 4 | 9 |
| 5 | H3K4me3 | 32 | 2 | 16 |
| 6 | H3K4me3 | 9 | 3 | 3 |
| 7 | H3K9ac | 44 | 1 | 44 |
| 8 | H3K9me1 | 19 | 16 | 1 |
| 9 | H3K9me3 | 18 | 7 | 2 |
| 10 | H3K9me3 | 44 | 1 | 44 |
| 11 | H3K9me3 | 28 | 3 | 11 |
| 12 | H3K9me3 | 47 | 3 | 15 |
| 13 | H3K9me3 S10ph | 51 | 13 | 4 |
| 14 | H3S10ph | 48 | 1 | 48 |
| 15 | H3S10ph | 46 | 1 | 46 |
| 16 | H3S10ph | 138 | 1 | 138 |
| 17 | H3K14ac | 26 | 28 | 1 |
| 18 | H3K27me2 | 18 | 14 | 1 |
| 19 | H3K27me3 | 34 | 9 | 4 |
| 20 | H3K27me3 | 36 | 10 | 4 |
| 21 | H3K27me3 | 172 | 1 | 172 |
| 22 | H3K27me3 | 74 | 14 | 5 |
| 23 | H3K36me2 | 71 | 44 | 2 |
| 24 | H3K36me3 | 26 | 23 | 1 |
| 25 | H3K36me3 | a | a | a |
| 26 | H3K36ac | 504 | 6 | 85 |
| 27 | H3-Pan-Kme2/me3 | 9b | 19 | 1 |
| 28 | H4R3me2a | 77 | 3 | 31 |
| 29 | H4K12ac | 30 | 4 | 8 |
| 30 | H4K16ac | 179 | 1 | 179 |
| 31 | H4K20me1 | 77 | 6 | 12 |
| 32 | H4K20me2 | 228 | 4 | 61 |
| 33 | H4K20me3 | 155 | 1 | 155 |
| 34 | H4-Pan-Kac | 25c | 4 | 6 |
| 35 | H4-Pan-Kac | 14d | 3 | 5 |
| 36 | H2aK9ac | 121 | 10 | 12 |
In this table, the antibody number (referring to Sup. Table 2), the antibody target site, the specificity factor for binding at the target site (SFT) and for the best non-target site (SFN) and the ratio of both are given. Examples of the results are shown in Supplemental Table 1.
not applicable since no specific binding to target.
calculated for H3K9me3.
calculated for K8ac or K5ac or K12ac.
calculated for K12ac or K5ac or K8ac.
Figure 3.
Examples of antibody specificity analysis. The upper part of each panel shows the image of the array. Peptide spots are annotated on the left copy of the duplicates, color coded as described below the image. On the left side, spots are annotated as follows: all peptides containing the primary PTM are shaded in green, false negatives (or very weakly bound peptides which contain the primary PTM) are highlighted with red arrows. False positives are encircled in orange or violet color. The secondary PTMs present in the false negative, and false positive spots are specified below the pictures. In the lower part a scatter plot of the binding intensities to both repeats is shown.
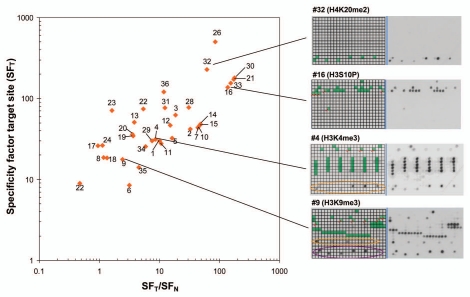
Figure 5.
Summary of antibody specificities. In this graph, the specificity factor for binding at the target site (SFT) is plotted on the y-axis and the ratio of the specificity factors for binding at the target site and the best non-target site (SFT/SFN) on the x-axis. Best antibodies are located in the upper right corner of the graph. Some examples of antibodies are shown on the right side, for coloring cf. Supplemental Table 1.
Figure 4.
Examples of results obtained with different antibodies binding to H3K27me3. Peptide spots are annotated on the left copy of the duplicates, color coded as described next to the image.
To confirm our data, we have performed a dot blot analysis of selected antibodies using purified peptides (Sup. Fig. 2). The H3K9me3 antibody #11 showed weaker binding to H3K9me3/S10ph than to H3K9me3, while antibody #10 bound both peptides equally. Furthermore, H3K9me3 antibody #9 showed cross-reactivity with H4K20me3 and H3K27me3. The H3K27me3 antibody #20 showed cross-reactivity with H3K9me3 and H4K20me3. Finally, the H4K20me3 antibody #33 did not show binding of H3K27me3 or H3K9me3. These results qualitatively agree with the data from the Celluspots arrays indicating that Celluspots arrays are a reliable tool to study the binding specificities of the antibodies.
Discussion
SPOT and Celluspots peptide arrays have been successfully applied in different applications including specificity analysis of peptide modifying enzymes,17,20,21 antibodies and epigenetic reading domains.12,16,21–23,25,27 SPOT peptide synthesis allows a cost efficient synthesis of peptide libraries containing modified and unmodified peptides. Due to miniaturization, many Celluspots arrays can be produced from one SPOT synthesis and only very little reagent is needed for the actual binding reaction. One drawback of the method is that peptides do not undergo purification. Therefore, we conducted several experiments to investigate the reliability of the approach. First, we confirmed the identity of representative peptides synthesized in parallel and cleaved off from the matrix by mass spectrometry (Sup. Fig. 1). Second, we tested the binding of antibodies to peptide arrays synthesized independently and observed very reproducible binding patterns (Fig. 1B). Third, we showed that different peptides containing a particular antibody epitope are bound by the matching antibody with very similar binding intensities although the different peptides were produced in independent synthesis (Fig. 1C). Fourth, we confirmed exemplary binding results using purified peptides (Sup. Fig. 2). From all of these observations, we conclude that Celluspots peptide arrays are reliable tools for screening of antibody specificity.
Summarizing our data (Table 1 and Sup. Table 1), we found that most of the antibodies bound well to the PTM they have been raised for. However, with few exceptions (like antibody #33), our analysis revealed previously unknown details of the binding properties that are important for the interpretation of experimental results. Some antibodies showed lost or reduced interaction with the predicted peptides in the presence of a second modification (false negatives), others showed high cross-reactivity (false positives). Most antibodies were inhibited by some additional PTMs close to the primary one. Therefore, lack of binding of the antibody in a biological sample can not only be due to the absence of the primary PTM, but also to the presence of an inhibiting secondary PTM and it cannot be taken as evidence of the absence of the primary PTM. On the other hand, based on our data, it would be incorrect to assume that a secondary modification near the primary PTM will always prevent binding. The reliability of the identification of the false negatives and false positives in our data set in many instances is confirmed by internal comparison of different antibodies. Often peptides identified as false negatives for one antibody are efficiently bound by other antibodies. Conversely, peptides identified as false positives with one antibody often are not bound by another antibody directed towards the same primary epitope.
To compare the specificities of antibodies, we have calculated the specificity factor for binding at its target site (SFT), which is defined as the ratio of the average binding activities to all spots which contain the target modification of the antibody, divided by the average binding to all other spots. This value integrates the amount and intensity of false positives and false negatives into a single number. In addition, we computed the specificity factor for the best non-target site (SFN), which is a measure for the binding preference to the second best site, and calculated the ratio of these numbers. SFT vs. SFT/SFN plots (Fig. 5) are a convenient way to display the quality of antibodies, where best antibodies will cluster in the upper right corner of the plot. However, it is important to evaluate the suitability of an antibody in the light of the actual application. For example, cross-reactivity on H4 of an antibody targeting H3K9me3 would not be relevant in western blot analysis performed after gel separation of histone proteins because the staining of H3 and H4 can be discriminated on the blot. Similarly, false negatives caused by additional modifications are not of concern for an in vitro histone methyltransferase assay using a recombinant histone protein as substrate because secondary modifications cannot occur. In addition, it may be possible to optimize antibody specificities for particular applications by variations in the buffer composition, which was not tested here.
Materials and Methods
Celluspots arrays spotted on glass slides were prepared as described.26 They are now commercially available from Active Motif (Cat. No. 13001). To confirm the identity of the peptides, analysis representative peptides were synthesized in parallel starting with an acid labile Rink linker and cleaved off from the matrix and subjected to mass spectrometric characterization (Sup. Fig. 1). For antibody binding, the array was blocked by incubation in TTBS buffer (10 mM Tris/HCl pH 7.5, 0.05% Tween-20 and 150 mM NaCl) containing 5% non-fat milk powder at 4°C overnight, then washed three times with TTBS buffer and incubated at room temperature for 1 h with antibody dilutions as recommended by the supplier for western blot staining. Antibodies were dissolved in 2 ml of TTBS buffer. In case of high cross-reactivity or strong signal, antibody concentrations were decreased stepwise until results were stable with further dilution. Final antibody concentrations used are given in Supplemental Table 2. The array was washed three times with TTBS and incubated with horseradish peroxidase conjugated anti-rabbit or anti-mouse antibody (GE Healthcare, 1:2,500) in TTBS for 1 h at room temperature. Finally, the array was washed three times with TTBS and submerged in ECL developing solution (Thermo Fisher Scientific) and the image was captured in X-ray film. To avoid saturation of the ECL detection system and the X-ray film, all images were developed several times with different exposition and only images which were not saturated were used for the analysis. Typical exposure times were 0.5–5 min. Analysis was done using an in house program (Array Analyze, available at www.activemotif.com/catalog/667.html or from the authors upon request). False positives were identified by manual inspection and by calculation of the average binding intensity to all spots carrying a particular modification. False negatives were identified by manual inspection. For quantitative description of the quality of an antibody, we use the specificity factor, which is defined as the ratio of the average binding intensity to all peptides carrying a particular modification and the average binding intensity to all peptides not having that modification. It gives an overall measure of the binding specificity of the antibody taking into account false positives and negatives. The antibodies used in this study were arbitrarily chosen to represent different positions on the histone tails and types of modifications. Different batches of the same antibody may yield different results.
Conclusions
Antibody specificity analysis on histone peptide arrays is a reliable, quantitative, inexpensive and fast method. It should become a regular quality control procedure conducted by antibody suppliers and users, in particular if expensive downstream analytic procedures like deep sequencing are applied. The method provides a very detailed binding pattern also including effects of multiple modifications. Purified peptides may be used to confirm key results and determine quantitative binding data. It is recommended to determine the antibody specificity before conducting a complicated or expensive experiment because specificities may vary with batches, in particular for the polyclonal antibodies. Detailed knowledge about the cross-reactivity and the secondary PTMs leading to false negatives will allow researchers to implement necessary control experiments in biological studies. Hence, detailed specificity information of an antibody will allow more reliable interpretation of biological experiments using these antibodies.
Acknowledgements
Thanks are due to Sandra Becker for technical support. This work has been supported in part by the NIH DK08267 grant.
Supplementary Material
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg AD, Allis CD, Bernstein E. Epigenetics: landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 4.Trojer P, Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature? Mol Cell. 2007;28:1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 5.Zilberman D, Henikoff S. Genome-wide analysis of DNA methylation patterns. Development. 2007;134:3959–3965. doi: 10.1242/dev.001131. [DOI] [PubMed] [Google Scholar]
- 6.Beck S, Rakyan VK. The methylome: approaches for global DNA methylation profiling. Trends Genet. 2008;24:231–237. doi: 10.1016/j.tig.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Mendenhall EM, Bernstein BE. Chromatin state maps: new technologies, new insights. Curr Opin Genet Dev. 2008;18:109–115. doi: 10.1016/j.gde.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lennartsson A, Ekwall K. Histone modification patterns and epigenetic codes. Biochim Biophys Acta. 2009;1790:863–868. doi: 10.1016/j.bbagen.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Goens G, Rusu D, Bultot L, Goval JJ, Magdalena J. Characterization and quality control of antibodies used in ChIP assays. Methods Mol Biol. 2009;567:27–43. doi: 10.1007/978-1-60327-414-2_2. [DOI] [PubMed] [Google Scholar]
- 10.Frank R. Spot-synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron. 1992;48:9217. [Google Scholar]
- 11.Wenschuh H, Volkmer-Engert R, Schmidt M, Schulz M, Schneider-Mergener J, Reineke U. Coherent membrane supports for parallel microsynthesis and screening of bioactive peptides. Biopolymers. 2000;55:188–206. doi: 10.1002/1097-0282(2000)55:3<188::AID-BIP20>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 12.Frank R. The SPOT-synthesis technique. Synthetic peptide arrays on membrane supports—principles and applications. J Immunol Methods. 2002;267:13–26. doi: 10.1016/s0022-1759(02)00137-0. [DOI] [PubMed] [Google Scholar]
- 13.Hilpert K, Winkler DF, Hancock RE. Peptide arrays on cellulose support: SPOT synthesis, a time and cost efficient method for synthesis of large numbers of peptides in a parallel and addressable fashion. Nat Protoc. 2007;2:1333–1349. doi: 10.1038/nprot.2007.160. [DOI] [PubMed] [Google Scholar]
- 14.Volkmer R. Synthesis and application of peptide arrays: quo vadis SPOT technology. Chembiochem. 2009;10:1431–1442. doi: 10.1002/cbic.200900078. [DOI] [PubMed] [Google Scholar]
- 15.Reineke U, Sabat R. Antibody epitope mapping using SPOT peptide arrays. Methods Mol Biol. 2009;524:145–167. doi: 10.1007/978-1-59745-450-6_11. [DOI] [PubMed] [Google Scholar]
- 16.Wu C, Li SS. CelluSpots: a reproducible means of making peptide arrays for the determination of SH2 domain binding specificity. Methods Mol Biol. 2009;570:197–202. doi: 10.1007/978-1-60327-394-7_8. [DOI] [PubMed] [Google Scholar]
- 17.Tegge WJ, Frank R. Analysis of protein kinase substrate specificity by the use of peptide libraries on cellulose paper (SPOT-method) Methods Mol Biol. 1998;87:99–106. doi: 10.1385/0-89603-392-9:99. [DOI] [PubMed] [Google Scholar]
- 18.Hilpert K, Hansen G, Wessner H, Schneider-Mergener J, Hohne W. Characterizing and optimizing protease/peptide inhibitor interactions, a new application for spot synthesis. J Biochem. 2000;128:1051–1057. doi: 10.1093/oxfordjournals.jbchem.a022833. [DOI] [PubMed] [Google Scholar]
- 19.Panse S, Dong L, Burian A, Carus R, Schutkowski M, Reimer U, et al. Profiling of generic anti-phosphopeptide antibodies and kinases with peptide microarrays using radioactive and fluorescence-based assays. Mol Divers. 2004;8:291–299. doi: 10.1023/b:modi.0000036240.39384.eb. [DOI] [PubMed] [Google Scholar]
- 20.Rathert P, Zhang X, Freund C, Cheng X, Jeltsch A. Analysis of the substrate specificity of the Dim-5 histone lysine methyltransferase using peptide arrays. Chem Biol. 2008;15:5–11. doi: 10.1016/j.chembiol.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rathert P, Dhayalan A, Murakami M, Zhang X, Tamas R, Jurkowska R, et al. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol. 2008;4:344–346. doi: 10.1038/nchembio.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nady N, Min J, Kareta MS, Chedin F, Arrowsmith CH. A SPOT on the chromatin landscape? Histone peptide arrays as a tool for epigenetic research. Trends Biochem Sci. 2008;33:305–313. doi: 10.1016/j.tibs.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 23.Bua DJ, Kuo AJ, Cheung P, Liu CL, Migliori V, Espejo A, et al. Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks. PloS One. 2009;4:e6789. doi: 10.1371/journal.pone.0006789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Briant DJ, Murphy JM, Leung GC, Sicheri F. Rapid identification of linear protein domain binding motifs using peptide SPOT arrays. Methods Mol Biol. 2009;570:175–185. doi: 10.1007/978-1-60327-394-7_6. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Jurkowska R, Soeroes S, Rajavelu A, Dhayalan A, Bock I, et al. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 2010;38:4246–4253. doi: 10.1093/nar/gkq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winkler DF, Hilpert K, Brandt O, Hancock RE. Synthesis of peptide arrays using SPOT-technology and the Celluspots-method. Methods Mol Biol. 2009;570:157–174. doi: 10.1007/978-1-60327-394-7_5. [DOI] [PubMed] [Google Scholar]
- 27.Reineke U, Kramer A, Schneider-Mergener J. Antigen sequence- and library-based mapping of linear and discontinuous protein-protein-interaction sites by spot synthesis. Curr Top Microbiol Immunol. 1999;243:23–36. doi: 10.1007/978-3-642-60142-2_2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.