Abstract
In this paper, a new method of one-pot biosynthesizing of gold nanoparticles (GNPs), using chloroplasts as reductants and stabilizers is reported. The as-prepared GNPs were characterized by ultraviolet visible spectroscopy, transmission electron microscopy, X-ray powder diffraction, and Fourier transform infrared spectroscopy (FTIR). The cytotoxicity of the GNPs was evaluated using the 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) method against gastric mucous cell line GES-1 and gastric cancer cell line MGC-803. Rhodamine 6G as a Raman probe was used for investigating surface-enhanced Raman spectroscopy (SERS) enhancement of GNPs. The transmission electron microscopy results indicated that the GNPs were spherical in structure and almost 20 nm in diameter. Ultraviolet visible spectroscopy exhibited an absorption peak at 545 nm. The GNPs exhibited high crystallinity, with the (111) plane as the predominant orientation, clarified by X-ray powder diffraction. In addition, a potential mechanism was proposed to interpret the formation process of GNPs, mainly based on the analysis of FTIR results. The FTIR spectrum confirmed that the GNPs were carried with N–H groups. Toxicological assays of as-prepared GNPs revealed that the green GNPs were nontoxic. SERS analysis revealed that the GNPs without any treatment could substantially enhance the Raman signals of rhodamine 6G. The Raman enhancement factor was calculated to be nearly 1010 orders of magnitude. In conclusion, the GNPs with good biocompatibility and excellent SERS effect were successfully synthesized using chloroplasts. These biogenetic GNPs have great potential for ultrasensitive detection of biomarkers in vitro and in vivo based on SERS.
Keywords: green synthesis, room temperature, gold nanospheres, MTT, SERS
Introduction
Recently, a hot spot of interest in the green nanotechnology area has been work on the development of a simple, ecofriendly, and safe approach for synthesizing metallic nanoparticles (MNPs). Biomaterials have been considered an attractive resource for green synthesis due to their diversity, regenerative ability, and safety.1–4 Among various MNPs, gold nanoparticles (GNPs) have been extensively studied based on their unique optoelectronic and electrochemical properties. The unique properties of GNPs have been potentially of interest for biological applications including sensors,5,6 catalysts,7 optical probes,8–10 tumor photothermal therapy,11 biomedical engineering,12 and surface-enhanced Raman scattering (SERS).13–15 Today, biosynthesis of GNPs using plants or plant extracts,16,17 fungus,18–20 and bacteria,21,22 has been actively exploited. However, all the above-mentioned reports involved excessively complicated biological components in the reaction process, expensive biological resources, and laborious culture conditions in the laboratories. Subsequently, the separation and purification of MNPs were also relatively difficult and time consuming. Green synthesis of GNPs using organelles may be an exciting alternative to methods currently available. When Shama et al23 bioreduced Au3+ to Au0 nanoparticles in plants, they discovered an interesting phenomena that many GNPs surrounded organelles. Additionally, Beattie and Haverkamp24 demonstrated that the site of the most abundant reduction of Au3+ metal salts to Au0 nanoparticles in plants were chloroplasts. This prompted the authors of the present paper to investigate the possibility of biosynthesizing GNPs using chloroplasts as biological templates. Chloroplasts are plentiful in the world of plants as well as the easiest to separate and collect and no tedious culture work is required in the laboratory. Furthermore, the structure of chloroplasts has been studied in detail. Chloroplasts are one of the most important organelles that synthesize carbohydrates via photosynthesis. After synthesizing in chloroplasts, the resulting organic compounds are transported to other parts of the plant, providing nutrients and energy for the plant’s growth. Therefore, chloroplasts are often known as the nutrient manufacturers and energy conversion compartments of plants.25 Chloroplasts have a unique structure known as “grana,” which are stacks of thylakoid discs. Aside from their unique structure, chloroplasts contain particular pigments (such as chlorophyll and carotenoids) and electron transfer chains for photosynthesis, both of which are located on the grana. Chlorophyll a is one kind of charged compound, and is the location of redox reaction and electron transportation and chlorophyll b is an aldehyde compound, while carotenoids are tetraterpenoid organic pigments. The authors hypothesize that the effective utilization of various phytochemicals that contain functional groups – such as tetraterpenoid, aldehyde, and carboxy – within the protein frameworks and in combination with the presence of saccharides coming from the photosynthesis of chloroplasts, will provide synergistic chemical reduction power for the reduction of gold salts into their corresponding nanoparticles.4,2,24 Since chloroplasts are very similar in higher plants, the chloroplasts from Trifolium, a plant which is widespread throughout the authors’ campus, were collected to synthesize GNPs. As far as the authors are aware, this paper is the first report of a simple, one-spot, cost-effective, and ecofriendly strategy for the biosynthesis of GNPs based on chloroplasts at room temperature. The in vitro toxicity of prepared GNPs was also evaluated by 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays based on normal human gastric mucosa cell lines and gastric cancer cell lines co-cultured with different concentrations of GNPs. Meanwhile, the enhanced SERS effect of the prepared GNPs was investigated using rhodamine 6G (R6G) as a Raman probe. The unique route of synthesized GNPs based on the template of chloroplasts has excellent potential applications in the near future, such as for green synthesis of nanoparticles and the ultrasensitive detection of biomarkers.
Materials and methods
Reagents
Chloroauric acid (HAuCl4), sodium chloride (NaCl), sodium hydroxide (NaOH), and R6G were purchased from Sigma-Aldrich (St Louis, MO). All reagents were commercial grade and were used without further purification. Water used in the experiment was ultrapure grade. Trifolium was collected from the campus of the Shanghai Jiao Tong University, Shanghai, People’s Republic of China. The following instruments were used: homogenizer (PRO200; Ginotech, Shanghai, People’s Republic of China); universal microplate spectrophotometer (μQuant, MQX200; BioTek Instruments Inc, Winooksi, VT); ultraviolet (UV) spectrophotometer (UV2550; Shimadzu, Tokyo, Japan); Raman microscope (Senterra R200-L; Bruker Optics, Ettlingen, Germany); transmission electron microscope (JEM-2100EX; JEOL, Tokyo, Japan); X-ray diffractometer (D/max-RC; Rigaku Co, Tokyo, Japan); Fourier transform spectrometer (EQUINOX 55; Bruker Optics, Bruck, Germany); INCA energy spectrometer (Oxford Instruments, Abingdon, UK).
Separation of chloroplasts
The chloroplasts used for the reduction of Au3+ ions to Au0 were isolated by combining 3 g of thoroughly washed Trifolium leaves with distilled water and isotonic solution (0.35 mol/L NaCl). Then the leaves were chopped finely and homogenized for 3–5 min at 5000 rpm in 10 mL of isotonic solution. The homogenate was filtered through six layers of gauze. The filtrate was then centrifuged at 1000 rpm/min for 2 minutes for collecting supernatant. The first supernatant was centrifuged at 3000 rpm for 5 minutes to obtain the second supernatant. Finally, the second supernatant was again centrifuged at 3000 rpm/min for 20 minutes for harvesting chloroplasts. The isolated chloroplasts were redispersed with 1 mL isotonic solution and stored at 4°C for further study. The supernatant of the chloroplast suspension was subjected to UV visible (UV-Vis) absorbance analysis. If no absorption peak occurred at the range of 250–290 nm, this demonstrated that intact chloroplasts were isolated. The separate experiments were carried out at 4°C.
Synthesis of GNPs
The chloroplast stock suspension was diluted 100× with distilled water before further experimentation. The ratio of the volume between the diluted chloroplast solution and the HAuCl4 solution was 2:1 for preparation of the GNPs. The pH value of the HAuCl4 solution was adjusted to around 7.5 by adding 1 M NaOH solution. Typically, 5 mL of diluted chloroplast solution was introduced into 2.5 mL of 10 mM HAuCl4 water solution. The final volume of the reaction solution was adjusted to 15 mL using deionized water under vigorous stirring for 24–36 hours in a 25°C water bath. The color changes of the reaction solution were observed from light green color to faint pink and finally to a purple color, which indicated the formation of GNPs.
Characterization of as-prepared GNPs
UV-Vis absorbance spectroscopy
UV-Vis absorption spectroscopy was used to monitor the plasmon absorption of the produced GNPs. UV-Vis spectroscopic measurements were operated at a resolution of 1 nm at a range of 200–800 nm with a scanning speed of 1856 nm/min.
Transmission electron microscopy measurements and X-ray diffraction analysis
Samples of chloroplast-mediated GNPs were prepared by dropping the reaction mixtures over carbon-coated copper grids and allowing the solution to evaporate. Then, transmission electron microscopy (TEM) measurements were performed on the JEM-2100EX transmission electron microscope. The X-ray diffraction pattern (XRD) of asprepared GNPs was confirmed by using the Dmax-RC X-ray diffractometer with Cu-Kα1 by radiation with γ = 1.540 Å. The scanning was done in the region of the 2θ angle from 30° to 80° with a step of 0.02° and a 2-second time constant for each step. The chemical composition of the as-prepared products was determined by energy-dispersive analysis of X-ray spectroscopy using the INCA energy spectrometer.
Fourier transform infrared spectroscopy analysis
Fourier transform infrared spectroscopy analysis (FTIR) measurements were made after complete reduction of AuCl4− ions by the chloroplast solution, and then the reaction solution was centrifuged at 14,000 rpm for 20 minutes to isolate the GNPs from free proteins or other compounds present in the solution. The collected GNPs were redispersed in water prior to FTIR analysis, and centrifuged again at 14,000 rpm for 20 minutes to isolate the GNPs from traces of free proteins or any other biomolecules present in the solution. Then the GNPs were freeze-dried. The frozen GNP powder (0.3 mg) was ground thoroughly with 1 mg KBr for FTIR analysis on an EQUINOX 55 spectrum instrument in the diffuse transmittance mode operating at a resolution of 4 cm−1 over 4000–400 cm−1. Meanwhile, chloroplasts were freeze-dried for FTIR analysis under the same conditions as GNPs.
Cytotoxicity of as-prepared GNPs
A series of various concentrations of as-prepared GNP solutions were prepared with cell media (25 μg/mL, 50 μg/mL, 100 μg/mL, 125 μg/mL, and 150 μg/mL). An MTT assay was performed on the human normal gastric mucous cell line GES-1 and gastric cancer cell line MGC-803. Briefly, GES-1 and MGC-803 cells were seeded into 96-well plates at a density of 2 × 106 cells/mL and 100 μL/well, respectively. Cells were cultured in an RPMI-1640 medium supplemented with 10% newborn calf serum, 100 U/mL penicillin, and 0.1 mg/mL streptomycin. The 96-well plates were placed into a conventional incubator with 5% CO2 at 37°C overnight for cell adherence. After that, cells were treated with different concentrations of GNP solution with 100 μL/well and continuously cultivated for 36 hours. Then MTT assays were performed at 570 nm using the universal microplate spectrophotometer. The wells that were untreated with GNPs were treated as 100% viable.
SERS
For SERS measurements, 500 μL of R6G water solution of 20 μM was added into a centrifugal tube with 1 mg of GNPs. The centrifugal tube was allowed to stay for 4 hours under ultrasound. Then the R6G-absorbed GNPs were washed twice with distilled water and dispersed in 50 μL water. An aliquot suspension of 10 μL was dropped on to a silicon wafer. The dropped solution spread evenly into a circle. After evaporation of the water, the sample was subjected to Raman measurement. As the control, 10 mM R6G solution was used alone and this was also subjected to Raman analysis. All experiments were carried out at room temperature in the same conditions.
Results and discussion
Characterization of as-prepared GNPs
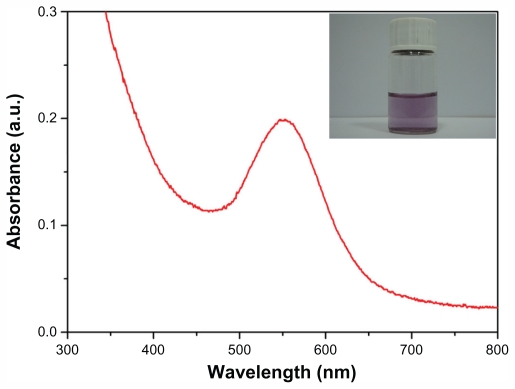
In the course of preparing GNPs, the color of the green reaction mixture gradually changed to purple, which suggested the formation of GNPs. The reduction of Au3+ during reaction with the chloroplast solution could be easily followed by UV-Vis spectroscopy. UV-Vis spectra of as-prepared GNPs displayed a single but strong surface plasmon resonance (SPR) band at about 545 nm (Figure 1), which demonstrated that the shapes of prepared nanoparticles were isotropic.26 Otherwise, the anisotropic particles of GNPs would have two or three SPR bands, depending on their shape. The SPR band of GNPs was a consequence of their small size, but bands can be influenced by numerous factors. In particular, solvent and surface functionalization were important contributors to the exact frequency and intensity of the SPR band.27 Therefore, the symmetrical SPR band suggested that prepared GNPs were approximately a spherical shape, which could be further corroborated by the TEM image (Figure 2A). The biogenetic GNPs were spherical with a diameter of almost 20 nm. According to Mie theory, an SPR band at 545 nm usually indicates that GNPs will be >60 nm. However, TEM analysis of the particles in this study showed that the size of GNPs was almost 20 nm. The difference in results between UV-vis and TEM images is because the average radii of GNPs formed within plants depends on the proportion of plants to the initial gold content.28 Actually, it should be noted that the diameters of biologically prepared GNPs in the solution were far bigger than the diameters checked by TEM. Biomolecules on the surface of GNPs cannot be observed under TEM, but those capping biomolecules can strongly affect the spectra of UV-Vis absorption of GNP solution, which presents a red shift in the SPR band of GNPs.4,12,29
Figure 1.
UV-visible absorption spectra of as-prepared gold nanoparticles. Inset photograph shows the reaction solution.
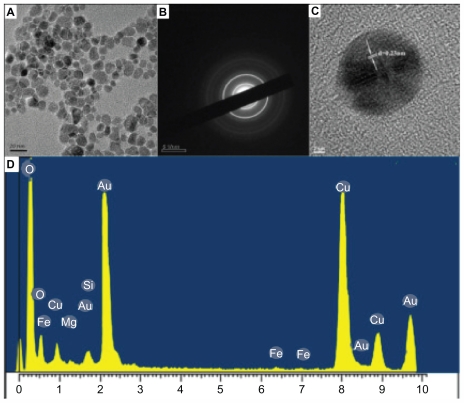
Figure 2.
(A) Representative transmission electron microscopy (TEM) image of as-prepared gold nanoparticles; (B) selected areas electron diffraction pattern corresponding to A; (C) high-resolution TEM image; (D) energy dispersive X-ray spectroscopy measurement profiles.
The crystallinity and crystal structure of GNPs were certificated by high-resolution TEM (HR-TEM) and XRD analysis. The typical selected areas electron diffraction (SAED) pattern with bright circular rings (Figure 2B) suggested that the as-prepared GNPs were highly crystalline. As shown in Figure 2C, the typical HR-TEM image of GNPs had clear lattice fringes with a spacing of 0.23 nm illustrating that the growth of GNPs occurred preferentially on the (111) plane. The interplanar distance of the (111) plane of as-prepared GNPs was in agreement with the (111) d-spacing of bulk Au (0.2355 nm).27 The spot-profile energy-dispersive analysis of X-ray spectroscopy of the Au particles manifested strong signals from Au atoms along with weak signals from Mg and Fe (Figure 2D). These weak signals of Mg and Fe were attributed to chlorophyll and iron sulfur proteins in chloroplasts.
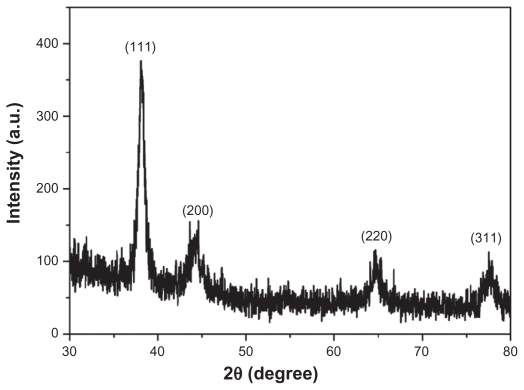
The XRD spectra (Figure 3) also confirmed the highly crystalline nature of as-prepared GNPs. Bragg reflections at 2θ were recorded from 30°C–80°C, which could be indexed on the basis of the face-centered cubic Au structure. No spurious diffraction peaks because of crystallographic impurities were found in XRD spectra, which strongly indicated that the as-prepared GNPs were of high purity. The diffraction peaks, which appeared at 38.1°, 44.5°, 64.8°, and 78.8°, were ascribed to the (111), (200), (220), and (311) planes, respectively. An overwhelmingly strong diffraction peak was the (111) facet, while the three other facets were much weaker. It is noteworthy that the ratio of intensity between the (200) and (111) diffraction peaks was much lower than the standard value (0.07 versus 0.52). Also, the ratio between the intensity of the (220) and (111) diffraction peaks was lower than the standard value (0.05 versus 0.33). These observations suggested that the (111) plane was the predominant orientation of as-prepared GNPs.30 The selected areas electron diffraction patterns of the GNPs (Figure 2B), corresponding to the face-centered cubic crystalline structure, further corroborated the results of XRD measurement.
Figure 3.
X-ray powder diffraction spectrum of as-prepared gold nanoparticles.
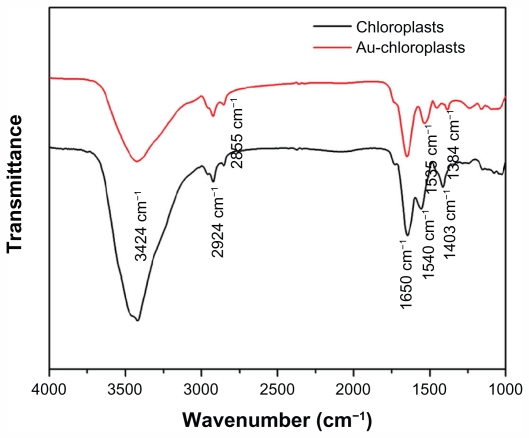
FTIR analysis was carried out to identify the possible biomolecules involved in the reduction of Au3+ and also the capping agents responsible for the stability of the biogenetic GNP solution (Figure 4). The intense broad band at 3424 cm−1, both in chloroplasts and as-prepared GNPs, was due to stretching and bending vibrations of O–H groups in the water.31 The two small peaks at 2924 cm−1 and 2855 cm−1 were assigned to methylene symmetric and antisymmetric vibrations of the hydrocarbons in proteins or enzymes.17,32 There were two prominent and sharp resonances both in chloroplasts (1650 cm−1 and 1540 cm−1) and as-prepared GNPs (1650 cm−1 and 1535 cm−1). The absorption peak at 1650 cm−1 was N–H stretching of amide I, which was characteristic of the stretch mode of the carbonyl group coupled to the amide linkage.33 This suggested that proteins bind to GNPs through free amine groups. The presence of the other prominent absorbance at 1535 cm−1 in the FTIR spectrum of GNPs was considered in the N–H stretching vibration of amide II linkages of polypeptides or proteins.34 The existence of amide I and amide II linkages showed that glutathiones capped on the surface of GNPs.19,35 In addition, it is also well-known that proteins can bind to GNPs through cysteine residues as well as amine groups in lysine residues.32 The peak at 1403 cm−1 of chloroplasts could be ascribed to the COO− symmetric stretching.18 The stretch at 1384 cm−1 corresponded to C=C or C–N stretching vibrations of aromatic amines.36 According to the FTIR pattern, there were no significant shifts in the 2924 cm−1 and 2855 cm−1 band between chloroplasts and GNPs, which implied that proteins acted as capping agents in the course of the formation of GNPs. The N–H stretching vibration of amide II linkages (1540 cm−1) shifts to lower wavenumbers (1535 cm−1) followed by the disappearance of the 1403 cm−1 resonance, which demonstrated that proteins might be responsible for reducing of Au3+ to Au0 nanoparticles. Thereby, it was believed that the capping agents also played a role as reducing agents to realize one-step reduction of Au3+ to Au0 nanoparticles.
Figure 4.
Fourier transform infrared spectroscopy spectra of chloroplasts and as-prepared gold nanoparticles scanning at a range of 4000–1000 cm−1.
Cytotoxicity of as-prepared GNPs
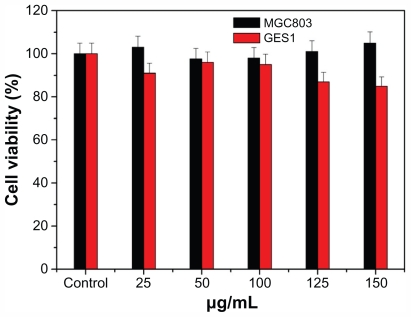
In vitro cytotoxicity of as-prepared GNPs was determined by MTT method on GES-1 cells and MGC-803 cells. Untreated cells and cells treated with different test doses of GNPs (25 μg/mL, 50 μg/mL, 100 μg/mL, 125 μg/mL, and 150 μg/mL) were used to estimate the biocompatibility of GNPs. After 36 hours of treatment with GNPs, both GES-1 and MGC-803 cells retained better viability at >85%, even up to the 150 μg/mL test dose (Figure 5), which suggested good biocompatibility of the as-prepared GNPs. The GNPs with good biocompatibility could bring new opportunities for ultrasensitive detection, molecular imaging, and therapy.
Figure 5.
MTT assays of normal human gastric cells GES-1 and human gastric cancer cells MGC-803 with different concentrations of gold nanoparticles.
SERS
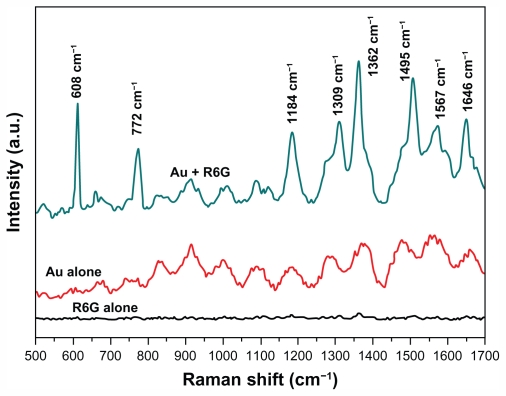
SERS effects occurred usually because of the very strong electromagnetic field and “hot spots” of the metal colloid cluster. The ability to induce SERS activity was dependent on two key factors: (1) resonant surface plasmon excitation of a metal substrate and (2) close proximity of analytic molecules to the metal substrate surface. If the surface of the metal substrate was not able to absorb analytes, then SERS effects would be difficult to induce. Obviously, the surface chemistry of metal substrate was considered as one critical factor for SERS activity because the analyte molecule must be within 0–4 nm of the substrate surface or the electromagnetic field to have an effect. Effectively, the molecule must bind to the metal surface to achieve SERS enhancement, a nontrivial problem for many molecules. For example, R6G, which is a common probe used for testing SERS activity on Ag but is generally difficult to detect on Au substrates because R6G does not strongly interact with Au surfaces, and Au substrates have been found to be less sensitive to R6G.37 However, it is worth mentioning that unique surface properties of as-prepared GNPs in this report were capable of extreme absorption of R6G and exhibited strong SERS effects without any further treatment. SERS activity is successfully demonstrated for R6G in Figure 6. Upon addition of the probe molecule R6G, there was a substantial Raman shift in bands of 1646 cm−1, 1567 cm−1, 1495 cm−1, 1362 cm−1, 1309 cm−1, and 1184 cm−1 when contrasted with GNPs and R6G alone using the Raman shift. These Raman signals arose from the totally symmetric modes of in-plane C–C stretching vibrations of R6G.35 The Raman band at 772 cm−1 was assigned to the C–H out-of-plane bending vibration of R6G. And the shift at 608 cm−1 was assigned to the in-plane bending vibration of the R6G CCC ring.38,39 These vibrational frequencies were similar to solution resonance Raman and bulk surface-enhanced Raman spectra of R6G (within 3–4 cm−1) but prominently increasing in intensity. The enhancement factor was calculated as ~1010 orders of magnitude. According to Schwartzberg’s report,14 sulfur groups on the surface of Au substrates allowed the binding and detecting of many molecules by SERS, including R6G and biological molecules. So it was inferred that the uniquely strong binding character for R6G was due to the high binding affinity of many molecules to sulfur species on the surface of GNPs. The sulfur species were supposedly from capping proteins, including cysteine residues, iron sulfur proteins, and glutathione in chloroplasts. This conclusion was typically consistent with the results of FTIR analysis, which further demonstrated that proteins, especially glutathiones, capped and stabilized the GNPs. The authors believe that the unique surface character allowed easy binding and detection of a variety of biomolecules. It could be as powerful as a SERS substrate used for trace biomarkers ultrasensitive detection.
Figure 6.
Raman spectra. 10 mM rhodamine 6G (R6G); Au; Au+R6G. Excitation wavelength of 633 nm and a power of 2 mW at the sample were used with a typical accumulation time of 10 seconds.
Conclusion
In summary, a one-pot, economic, and ecofriendly method of biosynthesizing GNPs was successfully established using chloroplasts as biological templates. The synthesized GNPs were a spherical shape, 20 nm in diameter. The chloroplasts acted in dual roles as reducing agents and stabilizers in the course of the formation of GNPs. This green chemistry approach is amenable to large scale commercial production. One major advantage of GNPs is the strongly enhanced SERS ability for R6G without any treatment. Furthermore, the prepared GNPs were confirmed to have good biocompatibility. The authors believe that the superior SERS ability of GNPs has great potential in many areas, including cancer diagnosis, biosensors, and ultrasensitive detection. Further study is necessary to determine the mechanism of SERS activity and bioreduction of the GNPs.
Acknowledgments
This work is supported by the National Key Basic Research Program (973 Project) (2010CB933901), National 863 Hi-tech Project (2007AA022004), Important National Science and Technology Specific Projects (2009ZX10004-311), National Natural Scientific Fund (No. 20771075 and No. 20803040), Special Project for Nanotechnology from Shanghai (No. 1052nm04100), New Century Excellent Talent of the Ministry of Education of China (NCET-08-0350), and Shanghai Science and Technology Fund (10XD1406100). Supported by Project 2011021031-3 (supported by Natural Science Foundation of Shanxi) and Shanghai Jiao Tong University Innovation Fund for Postgraduates (Z-340-011).
Footnotes
Disclosure
The authors claim no conflicts of interest in this work.
References
- 1.Kong Y, Chen J, Gao F, et al. A multifunctional ribonuclease-A-conjugated CdTe quantum dot cluster nanosystem for synchronous cancer imaging and therapy. Small. 2010;6(21):2367–2373. doi: 10.1002/smll.201001050. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Yang D, Kong Y, Wang X, Pandoli O, Gao G. Synergetic antibacterial effects of silver nanoparticles@aloe vera prepared via a green method. Nano Biomed Eng. 2010;2(4):252–257. [Google Scholar]
- 3.Yang DP, Chen S, Huang P, et al. Bacteria-template synthesized silver microspheres with hollow and porous structures as excellent SERS substrate. Green Chem. 2010;12:2038–2042. [Google Scholar]
- 4.Shukla R, Nune SK, Chanda N, et al. Soybeans as a phytochemical reservoir for the production and stabilization of biocompatible gold nanoparticles. Small. 2008;4(9):1425–1436. doi: 10.1002/smll.200800525. [DOI] [PubMed] [Google Scholar]
- 5.Yang DP, Cui DX. Advances and prospects of gold nanorods. Chem Asian J. 2008;3(12):2010–2022. doi: 10.1002/asia.200800195. [DOI] [PubMed] [Google Scholar]
- 6.Kim Y, Johnson RC, Hupp JT. Gold nanoparticle-based sensing of “spectroscopically silent” heavy metal ions. Nano Lett. 2001;1(4):165–167. [Google Scholar]
- 7.Huang J, Wang W, Lin L, et al. A general strategy for the biosynthesis of gold nanoparticles by traditional Chinese medicines and their potential application as catalysts. Chem Asian J. 2009;4(7):1050–1054. doi: 10.1002/asia.200900064. [DOI] [PubMed] [Google Scholar]
- 8.Kim CK, Kalluru RR, Singh JP, et al. Gold-nanoparticle-based miniaturized laser-induced fluorescence probe for specific DNA hybridization detection: studies on size-dependent optical properties. Nanotechnology. 2006;17:3085–3093. [Google Scholar]
- 9.Ray PC, Darbha GK, Ray A, Hardy W, Walker J. A gold-nanoparticle-based fluorescence resonance energy transfer probe for multiplexed hybridization detection: accurate identification of bio-agents DNA. Nanotechnology. 2007;18:1–6. [Google Scholar]
- 10.Ao L, Gao F, Pan B, He R, Cui D. Fluoroimmunoassay for antigen based on fluorescence quenching signal of gold nanoparticles. Anal Chem. 2006;78(4):1104–1106. doi: 10.1021/ac051323m. [DOI] [PubMed] [Google Scholar]
- 11.Li Z, Huang P, Zhang X, et al. RGD-conjugated dendrimer-modified gold nanorods for in vivo tumor targeting and photothermal therapy. Mol Pharmaceutics. 2009;7(1):94–104. doi: 10.1021/mp9001415. [DOI] [PubMed] [Google Scholar]
- 12.Cu D, Zhang H, Wang K, et al. Gold nanoparticles enhance efficiency of in vitro gene transcription-translation system. Nano Biomed Eng. 2011;3(2):120–125. [Google Scholar]
- 13.Nikoobakht B, El-Sayed MA. Surface-enhanced Raman scattering studies on aggregated gold nanorods. J Phys Chem A. 2003;107(18):3372–3378. [Google Scholar]
- 14.Schwartzberg AM, Grant CD, Wolcott A, et al. Unique gold nanoparticle aggregates as a highly active surface-enhanced Raman scattering substrate. Journal Phys Chem B. 2004;108(50):19191–19197. [Google Scholar]
- 15.Boisselier E, Astruc D. Gold nanoparticles in nanomedicine: preparations, imaging, diagnostics, therapies and toxicity. Chem Soc Rev. 2009;38(6):1759–1782. doi: 10.1039/b806051g. [DOI] [PubMed] [Google Scholar]
- 16.Kumar V, Yadav SK. Plant-mediated synthesis of silver and gold nanoparticles and their applications. J Chem Technol Biot. 2009;84(2):151–157. [Google Scholar]
- 17.Rajasekharreddy P, Usha Rani P, Sreedhar B. Qualitative assessment of silver and gold nanoparticle synthesis in various plants: a photobiological approach. J Nanopart Res. 2010;12(5):1711–1721. [Google Scholar]
- 18.Xie J, Lee JY, Wang DIC, Ting YP. High-yield synthesis of complex gold nanostructures in a fungal system. J Phys Chem C. 2007;111(45):16858–16865. [Google Scholar]
- 19.Shankar SS, Ahmad A, Pasricha R, Sastry M. Bioreduction of chloroaurate ions by geranium leaves and its endophytic fungus yields gold nanoparticles of different shapes. J Mater Chem. 2003;13(7):1822–1826. [Google Scholar]
- 20.Mukherjee P, Senapati S, Mandal D, et al. Extracellular synthesis of gold nanoparticles by the fungus Fusarium oxysporum. Chem Bio Chem. 2002;3(5):461–463. doi: 10.1002/1439-7633(20020503)3:5<461::AID-CBIC461>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 21.He S, Zhang Y, Guo Z, Gu N. Biological synthesis of gold nanowires using extract of Rhodopseudomonas capsulata. Biotechnol Prog. 2008;24(2):476–480. doi: 10.1021/bp0703174. [DOI] [PubMed] [Google Scholar]
- 22.Feng Y, Yu Y, Wang Y, Lin X. Biosorption and bioreduction of trivalent aurum by photosynthetic bacteria Rhodobacter capsulatus. Curr Microbiol. 2007;55(5):402–408. doi: 10.1007/s00284-007-9007-6. [DOI] [PubMed] [Google Scholar]
- 23.Sharma NC, Sahi SV, Nath S, Parsons JG, Gardea-Torresdey JL, Pal T. Synthesis of plant-mediated gold nanoparticles and catalytic role of biomatrix-embedded nanomaterials. Environ Sci Technol. 2007;41(14):5137–5142. doi: 10.1021/es062929a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beattie IR, Haverkamp RG. Silver and gold nanoparticles in plants: sites for the reduction to metal. Metallomics. 2011;3(6):628–632. doi: 10.1039/c1mt00044f. [DOI] [PubMed] [Google Scholar]
- 25.Jensen R, Bassham J. Photosynthesis by isolated chloroplasts. Proc Nat Acad Sci U S A. 1966;56(4):1095–1101. doi: 10.1073/pnas.56.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao J, Bender CM, Murphy CJ. Dependence of the gold nanorod aspect ratio on the nature of the directing surfactant in aqueous solution. Langmuir. 2003;19(21):9065–9070. [Google Scholar]
- 27.Philip D. Synthesis and spectroscopic characterization of gold nanoparticles. Spectrochim Acta A. 2008;71(1):80–85. doi: 10.1016/j.saa.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez E, Parsons JG, Peralta-Videa JR, et al. Potential of Chilopsis linearis for gold phytomining: using XAS to determine gold reduction and nanoparticle formation within plant tissues. Int J Phytoremediation. 2007;9(2):133–147. doi: 10.1080/15226510701232807. [DOI] [PubMed] [Google Scholar]
- 29.Zhen G, Tang Z, Xi D, et al. Preparation of PyDDP-modified gold nanoparticles and mechanism for red shift in the surface plasma resonance absorption. Acta Phys Chim Sin. 2004;20(6):647–650. [Google Scholar]
- 30.Kannan P, Abraham John S. Synthesis of mercaptothiadiazole-functionalized gold nanoparticles and their self-assembly on Au substrates. Nanotechnology. 2008;19:1–10. doi: 10.1088/0957-4484/19/8/085602. [DOI] [PubMed] [Google Scholar]
- 31.Gao G, Wu H, Chen M, Zhang L, Yu B, Xiang L. Synthesis of size- and shape-controlled cuo assemblies. J Electrochem Soc. 2011;158(3):K69–73. [Google Scholar]
- 32.Gole A, Dash C, Ramakrishnan V, et al. Pepsin-gold colloid conjugates: preparation, characterization, and enzymatic activity. Langmuir. 2001;17(5):1674–1679. [Google Scholar]
- 33.Yang J, Eom K, Lim EK, et al. In situ detection of live cancer cells by using bioprobes based on Au nanoparticles. Langmuir. 2008;24(21):12112–12115. doi: 10.1021/la802184m. [DOI] [PubMed] [Google Scholar]
- 34.Dong A, Huang P, Caughey WS. Redox-dependent changes in beta.-extended chain and turn structures of cytochrome c in water solution determined by second derivative amide I infrared spectra. Biochemistry. 1992;31(1):182–189. doi: 10.1021/bi00116a027. [DOI] [PubMed] [Google Scholar]
- 35.Foyer CH, Halliwell B. The presence of glutathione and glutathione reductase in chloroplasts: a proposed role in ascorbic acid metabolism. Planta. 1976;133(1):21–25. doi: 10.1007/BF00386001. [DOI] [PubMed] [Google Scholar]
- 36.Faramarzi MA, Forootanfar H. Biosynthesis and characterization of gold nanoparticles produced by laccase from Paraconiothyrium variabile. Colloids Surf B Biointerfaces. 2011;87(1):23–27. doi: 10.1016/j.colsurfb.2011.04.022. [DOI] [PubMed] [Google Scholar]
- 37.Gupta R, Weimer W. High enhancement factor gold films for surface enhanced Raman spectroscopy. Chem Phys Lett. 2003;374((3–4)):302–306. [Google Scholar]
- 38.Nie S, Emory SR. Probing single molecules and single nanoparticles by surface-enhanced Raman scattering. Science. 1997;275:1102–1106. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- 39.Hildebrandt P, Stockburger M. Surface-enhanced resonance Raman spectroscopy of rhodamine 6G adsorbed on colloidal silver. J Phys Chem. 1984;88(24):5935–5944. [Google Scholar]