Abstract
Recent studies as well as theoretical models of error processing assign fundamental importance to the brain's dopaminergic system. Research about how the electrophysiological correlates of error processing—the error-related negativity (ERN) and the error positivity (Pe)—are influenced by variations of common dopaminergic genes, however, is still relatively scarce. In the present study, we therefore investigated whether polymorphisms in the DAT1 gene and in the DRD4 gene, respectively, lead to interindividual differences in these error processing correlates. One hundred sixty participants completed a version of the Eriksen Flanker Task while a 26-channel EEG was recorded. The task was slightly modified in order to increase error rates. During data analysis, participants were split into two groups depending on their DAT1 and their DRD4 genotypes, respectively. ERN and Pe amplitudes after correct responses and after errors as well as difference amplitudes between errors and correct responses were analyzed. We found a differential effect of DAT1 genotype on the Pe difference amplitude but not on the ERN difference amplitude, while the reverse was true for DRD4 genotype. These findings are in line with predictions from theoretical models of dopaminergic transmission in the brain. They furthermore tie results from clinical investigations of disorders impacting on the dopamine system to genetic variations known to be at-risk genotypes.
Introduction
Electrophysiological research extensively investigates the generation of two correlates of error processing: One component, the error-related negativity (ERN), is a defined negative event-related potential (ERP) with an amplitude of up to 10 µV that appears in the response-locked EEG 50 to 100 ms after an erroneous response [1], [2]. The ERN has been theorized to reflect the activity of an error processing system that detects incorrect motor commands via a central processing pathway [3] without using sensory or proprioceptive information [2]. A second component, the error positivity (Pe), is a slow positive EEG potential that peaks 200 to 400 ms after an error is committed [1], [4]. The Pe has been suggested to represent the conscious detection of an erroneous response [3], [5], an adaptation of response strategy, or the subjective or emotional evaluation of the error [4].
An influential theoretical model by Holroyd and Coles [6] links the ERN to processes of reinforcement learning: This model holds that when an error occurs, the basal ganglia send a dopamine(DA)-mediated negative reinforcement learning signal to the anterior cingulate cortex (ACC), thereby generating the ERN.
Given the hypothesized role of DA transmission in the generation of the ERN, several studies investigated error processing in patients with Huntington's Disease (HD) and Parkinson's Disease (PD), both conditions with altered dopaminergic transmission. Beste and colleagues [7] reported reduced ERN amplitudes in HD patients with decreased amplitudes correlating with genetic disease load, thereby supporting the importance of intact striatal functioning for error processing. Results for PD patients are conflicting [8], [9], [10], which might in part be explained by an undesired side effect of dopaminergic PD medication on error processing [11].
Error processing has also been investigated in patients with psychiatric disorders, with most research focusing on attention deficit hyperactivity disorder (ADHD), a disorder which comprises altered striatal dopamine levels [12]. Many studies report reduced Pe amplitudes in ADHD patients, a reduction of ERN amplitudes, however, does not seem consistent across studies [13], [14], [15], [16].
In healthy volunteers, pharmaceutical alteration of dopaminergic transmission in the basal ganglia by a single dose of d-amphetamine resulted in enlarged ERN amplitudes, which was attributed to d-amphetamine's agonistic effect on dopaminergic transmission [17]. In turn, reducing dopaminergic transmission in the basal ganglia by using the antipsychotic D2/D3 receptor antagonist haloperidol led to a significant reduction of ERN amplitude in healthy participants [11], [18], [19].
So far, only few studies of common genotype variations that influence dopaminergic transmission have been reported and these are partly contradictory (for a review see [20]). The catechol-O-methyltransferase (COMT) polymorphism, for example, is known to influence frontal dopamine levels [21] and was found to affect Pe but not ERN amplitude in one study [22], whereas another study [23] did not yield significant findings. The second study, however, found an effect of a single nucleotide polymorphism (SNP) that supposedly leads to a reduction of dopamine D4 receptors (DRD4) on ERN but not Pe amplitude [23]. In addition, functional magnetic resonance imaging studies investigated genetic polymorphisms influencing dopamine D2 receptor (DRD2) expression: These receptors are prominently expressed in the basal ganglia and genetic variations seem to influence the ability to learn from negative feedback [24], [25].
Since previous research seems to support the hypothesized crucial role of dopaminergic signalling in error processing, we decided to investigate two new polymorphisms of important genes that influence dopaminergic transmission in different areas of the brain:
The gene encoding the dopamine transporter (DAT1, SLC6A3) was found to possess a genetic variation – a 40 nucleotide variable number of tandem repeats (VNTR) polymorphism in the 3′ untranslated region (UTR) – with two common alleles labelled 9-repeat (9R) and 10-repeat (10R), which supposedly influence gene expression and thereby DA reuptake from extracellular space [26], [27], [28], [29], [30]. The direction of this influence, however, is not yet clear, with studies yielding differing results with regard to which VNTR results in greater DAT expression. The DAT is primarily expressed in the striatum with only scarce expression in the prefrontal and medial frontal cortex [31], [32]. Two in vivo SPECT studies of healthy adults showed that the presence of at least one 9R allele led to increased DAT availability in the participants' striatum [33], [34], which might indicate increased striatal dopamine availability in 10/10R carriers.
The dopamine D4 receptor gene (DRD4) also possesses a polymorphism with a 48 bp variable number of tandem repeats (VNTR) in exon III, which can vary from 2 to 11 repeats [35], [36]. An investigation of human post-mortem brain tissue recently showed a trend for reduced DRD4 mRNA expression in samples carrying at least one seven-repeat allele [37]. DRD4 is mainly expressed in the prefrontal cortex with low levels of expression in the basal ganglia [36], [38].
Based on the model by Holroyd and Coles [6] as well as previous research outlined above, it is reasonable to expect some influence of these genes on error processing components: The DAT1 10/10R genotype was linked to smaller Pe amplitudes in children with ADHD, but showed no effect on the ERN [39]; a study of the DRD4 SNP -521C/T found increased ERN amplitudes for homozygous carriers of the T allele, which supposedly leads to less transcriptional efficiency [23].
In our experiment, we examined ERN and Pe amplitudes during a simple reaction time task in an unselected sample of healthy participants and compared amplitudes according to participants' DAT1 and DRD4 genotypes. Based on previous research, we expected the DAT1 10R genotype to be linked to smaller Pe and the DRD4 7R genotype to be linked to increased ERN amplitudes.
Methods
Ethics Statement
Ethical approval was obtained through the Ethical Review Board of the medical faculty of the University of Würzburg (vote 131/04); all procedures involved were in accordance with the 2008 Declaration of Helsinki. Participants gave written informed consent after full explanation of the procedures.
Participants
One hundred sixty subjects (70 male, 90 female) participated in this study. Mean (M) age was 27.02 years, ranging from 20 to 50 years of age (standard deviation SD = 7.28). All participants were right-handed, without any medication, and never treated for neurologic or psychiatric problems. Results from a subsample (n = 56) have been published elsewhere [16], [40].
Participants were stratified into a homozygous 10/10R group (n = 98) and a group carrying at least one 9R allele (9/9R or 9/10R; n = 62) according to their DAT1 VNTR genotype. Additionally, participants were stratified based to their DRD4 VNTR genotype into a 7R group (at least one 7R allele; n = 46) and a no 7R group (n = 114). DAT1 and DRD4 subgroups did not differ significantly with respect to gender, age, symptoms of depression, reaction time for correct answers, number of errors, and number of trials without artefacts per condition (as assessed with t-tests, all p>.05). The severity of ADHD symptoms as assessed with the ADHD Self-Report Scale (ASRS) differed between DAT1 subgroups, with increased values for the 9R group compared to the 10/10R group (t (158) = 2.66, p = .009; see table 1 for details).
Table 1. Mean and standard deviation of demographic data, psychiatric symptoms, reaction time in milliseconds, and artefact-free trials.
| DAT1 | DRD4 | |||
| 9R | 10/10R | no 7R | 7R | |
| n (female) | 62 (31) | 98 (59) | 114 (66) | 46 (24) |
| Age | 27.34 (7.56) | 26.82 (7.14) | 27.11 (7.64) | 26.80 (6.40) |
| Symptoms of depression | 3.29 (3.41) | 4.36 (3.93) | 3.93 (3.92) | 3.98 (3.36) |
| ADHD symptoms | 24.93 ( 8.22 )* | 21.62 ( 7.17 )* | 22.75 (7.54) | 23.23 (8.30) |
| Reaction time | 448.57 (62.81) | 460.50 (52.38) | 455.21 (60.63) | 457.54 (46.35) |
| Number of errors | 59.48 (38.12) | 52.53 (30.78) | 55.15 (34.66) | 55.41 (32.18) |
| Artefact-free trials correct | 261.21 (66.25) | 260.54 (41.50) | 260.89 (53.78) | 260.57 (48.98) |
| Artefact-free trials incorrect | 55.42 (36.26) | 49.94 (30.79) | 51.41 (33.80) | 53.67 (31.26) |
Standard deviations are shown in parentheses; significant differences are marked with *.
Psychological Assessment
Participants completed an 18 item screening questionnaire [41] based on the diagnostic criteria of ADHD as stated in the DSM-IV-TR [42]. It measures the frequency of ADHD symptoms ranging from 0 (‘never’) to 4 (‘very often’). Additionally, the Beck Depression Inventory (BDI, [43]) was used to assess depressive symptoms.
Stimuli and Procedure
We used a modified version of the Eriksen Flanker Task [44] as published before [16], [40]. Since the focus of this study was on neural correlates of error processing, the task was adapted to obtain high error rates. One of four possible combinations of arrows (<<><<, >><>>, ><><>, <><><) was shown in the middle of a 15′ monitor for 125ms. Stimuli were presented in random order, with the probability of occurrence being .25 for each combination. Subjects completed 160 trials twice, with a short break in between, where they had to indicate the direction of the middle arrow as quickly and as accurately as possible with their left (<) and right (>) index finger, respectively. Feedback was given 750 ms after the response by showing a plus sign (correct response), a minus sign (incorrect/no response), or an exclamation mark (correct response outside the reaction time limit) in the centre of the screen for 500 ms. The interstimulus interval varied between 500 ms and 1000 ms. The reaction time limit was determined individually in a practice session – which consisted of 40 trials with a 500 ms time limit – and the mean reaction time over all correct trials was adapted as reaction time limit for the subsequent experimental task. This time limit served to increase error rates. However, all responses (including “late” responses) were subsequently analyzed.
EEG Acquisition and Analyses
Event-related potentials were recorded from 26 Ag/AgCl scalp electrodes. Besides the recording sites specified in the 10–20 system, electrodes were placed on Oz, Fpz, under the right eye and on its outer canthus, and on the left and right mastoids. Impedance was kept below 5 kΩ for all electrodes. The ground electrode was placed between Fz and Fpz. Sampling rate was 1000 Hz, with the amplifier band pass filter set to 0.1–70 Hz and the notch filter set to 50 Hz. All data were recorded in relation to a midline reference placed between Cz and Fz and re-referenced offline to an average reference.
Eye movement artefacts were corrected [45] and response-locked EEG epochs from −200 ms to 500 ms were defined separately for correct and incorrect responses. Artefact-free epochs (no voltage in any channel exceeding ±100 µV or showing drops or rises of more than 100 µV/ms) were averaged: For incorrect responses, an average of 52.06 (SD = 33.01) epochs and for correct responses, an average of 260.80 (SD = 52.29) epochs were averaged.
Baseline correction based on the mean amplitude from −200 ms to 0 ms was implemented, and the time windows to detect ERN and Pe peaks were subsequently determined based on the grand average time curve over all subjects. The ERN was then automatically identified as the negative peak value between −35 ms and 108 ms over the electrode position Cz; the Pe was analogically detected as the positive peak value between 110 ms and 450 ms.
ERP amplitudes were analysed separately for the ERN and the Pe by using an analysis of variance (ANOVA) with the factors condition (correct, incorrect) and genotype (DAT1 (9R group vs. 10/10R group) and DRD4 (no 7R vs. 7R)). Two-sided t-tests were used for post-hoc analyses. For all analyses, p-values below .05 were considered significant.
Genotyping
Genomic DNA was extracted from whole blood samples by salt precipitation according to standard protocols. Genotyping of the DAT1 VNTR was performed by polymerase chain reaction (PCR) and subsequent gel electrophoresis as published previously [46]. Genotyping for the DRD4 VNTR was accomplished using standard PCR procedures modified from a previously published protocol [47]. Further details on protocols are available upon request.
Results
There was a main effect for response time following an erroneous as compared to a correct response (F [1,156] = 90.25, p<.001; post-correct responses: M = 452.21 ms, SD = 56.41; post-error responses: M = 476.90 ms, SD = 68.01), showing significant post-error slowing across all genotype groups. There was no main effect of DAT1 genotype (F [1,156] = .82, p = .366) or DRD4 genotype (F [1,156] = .05, p = .831) and no interaction effects.
Participants gave an average of 267.87 correct responses (SD = 51.13). There was no main effect of DAT1 genotype (F [1,156] = .16, p = .690) or DRD4 genotype (F [1,156] = 1.01, p = .318) as well as no interaction effects.
For the ERN, we found a main effect of condition (F [1,156] = 144.96, p<.001) with incorrect responses leading to more negative values than correct ones and an interaction effect of condition and DRD4 genotype (F [1,156] = 5.37, p = .022), which is discussed below. We did not find an interaction effect of condition and DAT1 genotype (F [1,156] = .40, p = .526), or a three-way interaction effect of condition, DAT1 genotype, and DRD4 genotype (F [1,156] = .66, p = .419). In addition, no main effects of DAT1 genotype (F [1,156] = .73, p = .394) or DRD4 genotype (F [1,156] = .17, p = .685) were found.
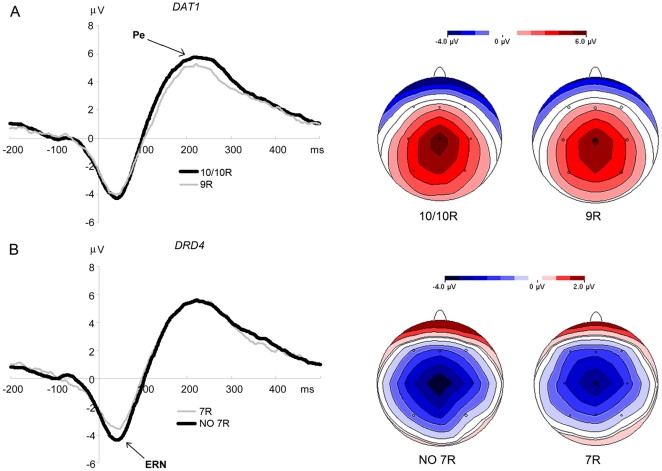
The significant interaction effect of condition and DRD4 genotype can be described by significantly smaller difference amplitudes between the correct and incorrect response conditions in 7R carriers compared to no 7R carriers (t (158) = −2.18, p = .031; see table 2 for M, SD, and post-hoc t-tests; figure 1).
Table 2. Mean and standard deviation of the amplitudes for the different conditions and genotypes.
| DAT1 | DRD4 | |||||
| 9R | 10/10R | t 1-value (p) | no 7R | 7R | t 1-value (p) | |
| ERN | ||||||
| – correct | 1.01 (2.22) | .70 (2.56) | .77 (.442) | .95 (2.34) | .49 (2.66) | 1.08 (.281) |
| – incorrect | −2.78 (3.06) | −3.18 (3.43) | .76 (.451) | −3.24 (3.29) | −2.48 (3.27) | −1.32 (.187) |
| – difference | −3.78 (3.30) | −3.88 (3.23) | .189 (.850) | −4.19 (3.01) | −2.98 (3.65) | −2.18 (.031)* |
| Pe | ||||||
| – correct | 5.16 (3.55) | 4.12 (3.68) | 1.77 (.079)+ | 4.53 (3.41) | 4.52 (4.25) | .02 (.984) |
| – incorrect | 8.96 (3.24) | 8.99 (3.64) | −.05 (.957) | 8.92 (3.45) | 9.12 (3.59) | −.32 (.753) |
| – difference | 3.80 (3.37) | 4.87 (3.36) | −1.97 (.051) + | 4.40 (3.24) | 4.60 (3.78) | −.34 (.731) |
Standard deviations are shown in parentheses; significant differences are marked with *, trends are marked with +; 1 df = 158, two-sided tests.
Figure 1. Grand average difference curves and peak topographies for DAT1 (A) and DRD4 (B) subgroups.
Peak topographies for the ERN are at 40 ms, for the Pe at 218 ms.
For the Pe, we also found a main effect of condition (F [1,156] = 197.70, p<.001) with incorrect responses leading to more positive values than correct ones and an interaction effect of condition and DAT1 genotype (F [1,156] = 4.78, p = .030), which is discussed below. We did not find an interaction effect of condition and DRD4 genotype (F [1,156] = .01, p = .972) or a three-way interaction effect of condition, DAT1 genotype, and DRD4 genotype (F [1,156] = .99, p = .321). Furthermore, no main effects of DAT1 genotype (F [1,156] = .34, p = .562) or DRD4 genotype (F [1,156] <.001, p = .987) were found.
The significant interaction effect can be described by marginally smaller difference amplitudes between the correct and incorrect response conditions in 9R carriers (t (158) = −1.97, p = .051; see table 2). A tendency for increased amplitudes of the Pe to correct reactions in 9R carriers (t (158) = 1.77, p = .079; see table 2 and figure 1) suggests that the interaction effect was mainly but not solely caused by differences in the processing of correct responses.
Discussion
We found the ERN amplitude to be related to participants' DRD4 genotype but not to their DAT1 genotype, whereas the reverse was true for the Pe amplitude. It therefore seems that the DRD4 genotype influences the ERN – defined as the difference wave between incorrect and correct responses – but not the Pe. In turn, DAT1 genotype seems to influence the Pe – also defined as the difference wave – but not the ERN.
According to the model by Holroyd and Coles [6] mentioned above, the amplitude of the ERN should be influenced by phasic changes in dopaminergic transmission in the basal ganglia. Although our results seem to contradict this prediction – DRD4 is mostly expressed in the cortex with very little expression in striatal areas [36], [38] – Rubinstein and colleagues [48] discovered that mice lacking dopamine D4 receptors showed elevated dopamine synthesis and conversion in the striatum. This was possibly caused by glutamatergic transmission from cortical regions and still has to be investigated in humans. It would, however, offer a plausible explanation for our findings.
Based on Bilder and colleagues' [49] model of tonic and phasic dopamine transmission, lower cortical expression of the DRD4 in 7R carriers could result in compensatorily elevated tonic dopamine levels in the striatum. This elevation in tonic striatal dopamine would make 7R carriers less sensitive to changes in phasic dopamine transmission, which could explain their reduced ERN amplitude. A similar argument was proposed by Krämer and colleagues [23], who found an increased ERN amplitude to be related to the DRD4 SNP -521C/T. However, while the impact of this SNP on gene expression is still under debate [50], [51] we can be fairly confident about the transcriptional consequences of the VNTR we investigated.
The results can also be supported from a clinical perspective: ERN amplitude has been connected to self-reported impulsivity with highly impulsive individuals showing lower amplitudes [52]. Impulsivity, on the other hand, was recently linked to the presence of at least one 7R allele [53]. It would therefore seem logical to find lower ERN amplitudes in 7R carriers.
Regarding DAT1, our findings seem to be at odds with a study in children that found smaller Pe amplitudes in 10/10R carriers [39]. The 10R allele has been associated with childhood ADHD [54] which in turn has been associated with decreased Pe amplitudes [13], [14], [55]. Recent studies, however, point to a differential impact of this VNTR in childhood as compared to adulthood [56], [57]: In adults, carrying the 9R allele was associated with persistent ADHD and worse cognitive functioning, respectively.
In addition, a recent SPECT study reported a 9R haplotype which showed significantly higher DAT expression than all other investigated haplotypes [34]. This led the authors to suggest that reported higher striatal dopamine transporter availability in 9R carriers could mainly be caused by this specific subgroup. It might therefore be interesting and more informative to investigate Pe amplitudes in participants carrying this haplotype and compare them to all other haplotypes.
The findings of van de Giessen and colleagues [34] underscore the importance of basic research into the effects of VNTRs and SNPs as well as haplotypes on gene expression. While the interpretation of our results relies on the findings of these studies, reports on the effects of a certain genetic variation on gene expression are still often contradictory.
Limitations of the present study are linked to the sample size: Although representing a rather large sample in neurophysiological genetic research, power considerations arise from a general genetic research perspective. The sample size seemed sufficient assuming an intermediate effect size, and effect sizes of Cohen's d = .38 were found for the ERN amplitude depending on participants' DRD4 genotype and of d = .32 for the Pe amplitude depending on participants' DAT1 genotype. A post-hoc power calculation (two-tailed) revealed a probability of detecting a true significant difference of 58 and 50 percent, respectively. For the non-significant findings, effect sizes were d≤.06, indicating that these results represent non-meaningful and practically irrelevant effects.
Furthermore, it would have been interesting to investigate gene-gene interactions. However, this procedure would have resulted in four – partly very small – subgroups, which would likely obscure the effects. Even though this could therefore not be implemented, both our DAT1 and DRD4 groups are very comparable with regard to demographic data, psychiatric symptoms, reaction time, and artefact-free trials. The only group difference was found for the DAT1 9R and 10/10R groups with regard to their symptoms of ADHD. This is in line with the current literature [56], [57], and further helps to tie clinical findings of reduced Pe amplitudes in ADHD to an identified genetic risk factor.
Although we did not find any results on the behavioral level, our study is able to link previously observed error processing deficits in impulsive and ADHD participants to genetic variants that are known to contribute to these disorders.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by the German Research Foundation (DFG; HE 4531/1-1 and RTG 1253/1). This publication was funded by the German Research Foundation and the University of Wuerzburg in the funding programme Open Access Publishing. http://www.dfg.de/index.jsp. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Falkenstein M, Hohnsbein J, Hoormann J, Blanke L. Effects of crossmodal divided attention on late ERP components: II. Error processing in choice reaction tasks. Electroencephalogr Clin Neurophysiol. 1991;78:447–455. doi: 10.1016/0013-4694(91)90062-9. [DOI] [PubMed] [Google Scholar]
- 2.Gehring WJ, Goss B, Coles MGH, Meyer DE, Donchin E. A neural system for error detection and compensation. Psychol Sci. 1993;4:385–390. [Google Scholar]
- 3.Nieuwenhuis S, Ridderinkhof K, Blom J, Band G, Kok A. Error-related brain potentials are differentially related to awareness of response errors: evidence from an antisaccade task. Psychophysiology. 2001;38:752–760. [PubMed] [Google Scholar]
- 4.Falkenstein M, Hoormann J, Christ S, Hohnsbein J. ERP components on reaction errors and their functional significance: a tutorial. Biol Psychol. 2000;51:87–107. doi: 10.1016/s0301-0511(99)00031-9. [DOI] [PubMed] [Google Scholar]
- 5.Endrass T, Reuter B, Kathmann N. ERP correlates of conscious error recognition: aware and unaware errors in an antisaccade task. Eur J Neurosci. 2007;26:1714–1720. doi: 10.1111/j.1460-9568.2007.05785.x. [DOI] [PubMed] [Google Scholar]
- 6.Holroyd CB, Coles MGH. The neural basis of human error processing: reinforcement learning, dopamine, and the error-related negativity. Psychol Rev. 2002;109:679–709. doi: 10.1037/0033-295X.109.4.679. [DOI] [PubMed] [Google Scholar]
- 7.Beste C, Saft C, Andrich J, Gold R, Falkenstein M. Error processing in Huntington's disease. PLoS One. 2006;1 doi: 10.1371/journal.pone.0000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holroyd CB, Praamstra P, Plat E, Coles MGH. Spared error-related potentials in mild to moderate Parkinson's disease. Neuropsychologia. 2002;40:2116–2124. doi: 10.1016/s0028-3932(02)00052-0. [DOI] [PubMed] [Google Scholar]
- 9.Falkenstein M, Hielscher H, Dziobek I, Schwarzenau P, Hoormann J, et al. Action monitoring, error detection, and the basal ganglia: an ERP study. Neuroreport. 2001;12:157. doi: 10.1097/00001756-200101220-00039. [DOI] [PubMed] [Google Scholar]
- 10.Willemssen R, Müller T, Schwarz M, Hohnsbein J, Falkenstein M. Error processing in patients with Parkinson's disease: the influence of medication state. J Neural Transm. 2008;115:461–468. doi: 10.1007/s00702-007-0842-1. [DOI] [PubMed] [Google Scholar]
- 11.Jocham G, Ullsperger M. Neuropharmacology of performance monitoring. Neurosci Biobehav Rev. 2009;33:48–60. doi: 10.1016/j.neubiorev.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Krause KH, Dresel SH, Krause J, Kung HF, Tatsch K. Increased striatal dopamine transporter in adult patients with attention deficit hyperactivity disorder: effects of methylphenidate as measured by single photon emission computed tomography. Neurosci Lett. 2000;285:107–110. doi: 10.1016/s0304-3940(00)01040-5. [DOI] [PubMed] [Google Scholar]
- 13.Jonkman L, van Melis J, Kemner C, Markus C. Methylphenidate improves deficient error evaluation in children with ADHD: an event-related brain potential study. Biol Psychol. 2007;76:217–229. doi: 10.1016/j.biopsycho.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Wiersema J, Van der Meere J, Roeyers H. ERP correlates of impaired error monitoring in children with ADHD. J Neural Transm. 2005;112:1417–1430. doi: 10.1007/s00702-005-0276-6. [DOI] [PubMed] [Google Scholar]
- 15.Liotti M, Pliszka S, Perez R, Kothmann D, Woldorff M. Abnormal brain activity related to performance monitoring and error detection in children with ADHD. Cortex. 2005;41:377–388. doi: 10.1016/s0010-9452(08)70274-0. [DOI] [PubMed] [Google Scholar]
- 16.Herrmann MJ, Mader K, Schreppel T, Jacob CP, Heine M, et al. Neural correlates of performance monitoring in adult patients with attention deficit hyperactivity disorder (ADHD). World J Biol Psychiatry. 2010;11:457–464. doi: 10.1080/15622970902977552. [DOI] [PubMed] [Google Scholar]
- 17.de Bruijn ERA, Hulstijn W, Verkes RJ, Ruigt GSF, Sabbe BGC. Drug-induced stimulation and suppression of action monitoring in healthy volunteers. Psychopharmacology. 2004;177:151–160. doi: 10.1007/s00213-004-1915-6. [DOI] [PubMed] [Google Scholar]
- 18.de Bruijn ERA, Sabbe BGC, Hulstijn W, Ruigt GSF, Verkes RJ. Effects of antipsychotic and antidepressant drugs on action monitoring in healthy volunteers. Brain Research. 2006;1105:122–129. doi: 10.1016/j.brainres.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 19.Zirnheld PJ, Carroll CA, Kieffaber PD, O'Donnell BF, Shekhar A, et al. Haloperidol impairs learning and error-related negativity in humans. J Cogn Neurosci. 2004;16:1098–1112. doi: 10.1162/0898929041502779. [DOI] [PubMed] [Google Scholar]
- 20.Ullsperger M. Genetic association studies of performance monitoring and learning from feedback: the role of dopamine and serotonin. Neurosci Biobehav Rev. 2010;34:649–659. doi: 10.1016/j.neubiorev.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Goldberg TE, Egan MF, Gscheidle T, Coppola R, Weickert T, et al. Executive subprocesses in working memory: relationship to catechol-O-methyltransferase Val158Met genotype and schizophrenia. Arch Gen Psychiatry. 2003;60:889. doi: 10.1001/archpsyc.60.9.889. [DOI] [PubMed] [Google Scholar]
- 22.Frank MJ, D'Lauro C, Curran T. Cross-task individual differences in error processing: neural, electrophysiological, and genetic components. Cogn Affect Behav Neurosci. 2007;7:297–308. doi: 10.3758/cabn.7.4.297. [DOI] [PubMed] [Google Scholar]
- 23.Krämer UM, Cunillera T, Camara E, Marco-Pallares J, Cucurell D, et al. The impact of catechol-O-methyltransferase and dopamine D4 receptor genotypes on neurophysiological markers of performance monitoring. J Neurosci. 2007;27:14190–14198. doi: 10.1523/JNEUROSCI.4229-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein TA, Neumann J, Reuter M, Hennig J, Von Cramon DY, et al. Genetically determined differences in learning from errors. Science. 2007;318:1642. doi: 10.1126/science.1145044. [DOI] [PubMed] [Google Scholar]
- 25.Jocham G, Klein TA, Neumann J, Von Cramon DY, Reuter M, et al. Dopamine DRD2 polymorphism alters reversal learning and associated neural activity. J Neurosci. 2009;29:3695. doi: 10.1523/JNEUROSCI.5195-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuke S, Suo S, Takahashi N, Koike H, Sasagawa N, et al. The VNTR polymorphism of the human dopamine transporter (DAT1) gene affects gene expression. Pharmacogenomics J. 2001;1:152–156. doi: 10.1038/sj.tpj.6500026. [DOI] [PubMed] [Google Scholar]
- 27.Michelhaugh SK, Fiskerstrand C, Lovejoy E, Bannon MJ, Quinn JP. The dopamine transporter gene (SLC6A3) variable number of tandem repeats domain enhances transcription in dopamine neurons. J Neurochem. 2001;79:1033–1038. doi: 10.1046/j.1471-4159.2001.00647.x. [DOI] [PubMed] [Google Scholar]
- 28.Mill J, Asherson P, Browes C, D'Souza U, Craig I. Expression of the dopamine transporter gene is regulated by the 3 ' UTR VNTR: evidence from brain and lymphocytes using quantitative RT-PCR. Am J Med Genet. 2002;114:975–979. doi: 10.1002/ajmg.b.10948. [DOI] [PubMed] [Google Scholar]
- 29.Miller GM, Madras BK. Polymorphisms in the 3 '-untranslated region of human and monkey dopamine transporter genes affect reporter gene expression. Mol Psychiatry. 2002;7:44–55. doi: 10.1038/sj.mp.4000921. [DOI] [PubMed] [Google Scholar]
- 30.VanNess SH, Owens MJ, Kilts CD. The variable number of tandem repeats element in DAT1 regulates in vitro dopamine transporter density. BMC Genet. 2005;6 doi: 10.1186/1471-2156-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis DA, Melchitzky DS, Sesack SR, Whitehead RE, Auh S, et al. Dopamine transporter immunoreactivity in monkey cerebral cortex: regional, laminar, and ultrastructural localization. J Comp Neurol. 2001;432:119–136. doi: 10.1002/cne.1092. [DOI] [PubMed] [Google Scholar]
- 32.Sesack SR, Hawrylak VA, Matus C, Guido MA, Levey AI. Dopamine axon varicosities in the prelimbic division of the rat prefrontal cortex exhibit sparse immunoreactivity for the dopamine transporter. J Neurosci. 1998;18:2697–2708. doi: 10.1523/JNEUROSCI.18-07-02697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Dyck CH, Malison RT, Jacobsen LK, Seibyl JP, Staley JK, et al. Increased dopamine transporter availability associated with the 9-repeat allele of the SLC6A3 gene. J Nucl Med. 2005;46:745–751. [PubMed] [Google Scholar]
- 34.van de Giessen EM, de Win MML, Tanck MWT, van den Brink W, Baas F, et al. Striatal dopamine transporter availability associated with polymorphisms in the dopamine transporter gene SLC6A3. J Nucl Med. 2009;50:45–52. doi: 10.2967/jnumed.108.053652. [DOI] [PubMed] [Google Scholar]
- 35.Schoots O, Van Tol HHM. The human dopamine D4 receptor repeat sequences modulate expression. Pharmacogenomics J. 2003;3:343–348. doi: 10.1038/sj.tpj.6500208. [DOI] [PubMed] [Google Scholar]
- 36.Oak JN, Oldenhof J, Van Tol HHM. The dopamine D-4 receptor: one decade of research. Eur J Pharmacol. 2000;405:303–327. doi: 10.1016/s0014-2999(00)00562-8. [DOI] [PubMed] [Google Scholar]
- 37.Simpson J, Vetuz G, Wilson M, Brookes KJ, Kent L. The DRD4 receptor exon 3 VNTR and 5 ' SNP variants and mRNA expression in human post-mortem brain tissue. Am J Med Genet B. 2010;153B:1228–1233. doi: 10.1002/ajmg.b.31084. [DOI] [PubMed] [Google Scholar]
- 38.Meador-Woodruff JH, Damask SP, Wang JC, Haroutunian V, Davis KL, et al. Dopamine receptor mRNA expression in human striatum and neocortex. Neuropsychopharmacology. 1996;15:17–29. doi: 10.1016/0893-133X(95)00150-C. [DOI] [PubMed] [Google Scholar]
- 39.Althaus M, Groen Y, Wijers AA, Minderaa RB, Kema IP, et al. Variants of the SLC6A3 (DAT1) polymorphism affect performance monitoring-related cortical evoked potentials that are associated with ADHD. Biol Psychol. 2010;85:19–32. doi: 10.1016/j.biopsycho.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 40.Herrmann MJ, Saathoff C, Schreppel TJ, Ehlis AC, Scheuerpflug P, et al. The effect of ADHD symptoms on performance monitoring in a non-clinical population. Psychiatry Res. 2009;169:144–148. doi: 10.1016/j.psychres.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 41.Kessler RC, Adler L, Ames M, Delmer O, Faraone S, et al. The World Health Organization Adult ADHD Self-Report Scale (ASRS): a short screening scale for use in the general population. Psychol Med. 2005;35:245–256. doi: 10.1017/s0033291704002892. [DOI] [PubMed] [Google Scholar]
- 42.American Psychiatric Association (APA) Washington DC: American Psychiatric Association; 2000. Diagnostic and statistical manual of mental disorders, text revision: DSM-IV-TR. [Google Scholar]
- 43.Hautzinger M, Bailer M, Worall H, Keller F. Bern: Huber; 1994. Beck-Depressions-Inventar (BDI). [Google Scholar]
- 44.Eriksen BA, Eriksen CW. Effects of noise letters upon identification of a target letter in a nonsearch task. Percept Psychophys. 1974;16:143–149. [Google Scholar]
- 45.Gratton G, Coles MGH, Donchin E. A new method for off-line removal of ocular artefacts. Electroencephalogr Clin Neurophysiol. 1983;55:468–484. doi: 10.1016/0013-4694(83)90135-9. [DOI] [PubMed] [Google Scholar]
- 46.Hünnerkopf R, Strobel A, Gutknecht L, Brocke B, Lesch KP. Interaction between BDNF Val66Met and dopamine transporter gene variation influences anxiety-related traits. Neuropsychopharmacology. 2007;32:2552–2560. doi: 10.1038/sj.npp.1301383. [DOI] [PubMed] [Google Scholar]
- 47.Ebstein RP, Novick O, Umansky R, Priel B, Osher Y, et al. Dopamine D4 receptor (D4DR) exon III polymorphism associated with the human personality trait of Novelty Seeking. Nat Genet. 1996;12:78–80. doi: 10.1038/ng0196-78. [DOI] [PubMed] [Google Scholar]
- 48.Rubinstein M, Phillips TJ, Bunzow JR, Falzone TL, Dziewczapolski G, et al. Mice lacking dopamine D4 receptors are supersensitive to ethanol, cocaine, and methamphetamine. Cell. 1997;90:991–1001. doi: 10.1016/s0092-8674(00)80365-7. [DOI] [PubMed] [Google Scholar]
- 49.Bilder RM, Volavka J, Lachman HM, Grace AA. The catechol-O-methyltransferase polymorphism: relations to the tonic-phasic dopamine hypothesis and neuropsychiatric phenotypes. Neuropsychopharmacology. 2004;29:1943–1961. doi: 10.1038/sj.npp.1300542. [DOI] [PubMed] [Google Scholar]
- 50.Kereszturi E, Kiraly O, Barta C, Molnar N, Sasvari-Szekely M, et al. No direct effect of the-521 C/T polymorphism in the human dopamine D4 receptor gene promoter on transcriptional activity. BMC Mol Biol. 2006;7 doi: 10.1186/1471-2199-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okuyama Y, Ishiguro H, Nankai M, Shibuya H, Watanabe A, et al. Identification of a polymorphism in the promoter region of DRD4 associated with the human novelty seeking personality trait. Mol Psychiatry. 2000;5:64–69. doi: 10.1038/sj.mp.4000563. [DOI] [PubMed] [Google Scholar]
- 52.Potts G, George M, Martin L, Barratt E. Reduced punishment sensitivity in neural systems of behavior monitoring in impulsive individuals. Neurosci Lett. 2006;397:130–134. doi: 10.1016/j.neulet.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 53.Garcia JR, MacKillop J, Aller EL, Merriwether AM, Wilson DS, et al. Associations between dopamine D4 receptor gene variation with both infidelity and sexual promiscuity. PLoS One. 2010;5 doi: 10.1371/journal.pone.0014162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thapar A, O'Donovan M, Owen MJ. The genetics of attention deficit hyperactivity disorder. Hum Mol Genet. 2005;14:R275–R282. doi: 10.1093/hmg/ddi263. [DOI] [PubMed] [Google Scholar]
- 55.Overtoom CCE, Kenemans JL, Verbaten MN, Kemmer C, van der Molen MW, et al. Inhibition in children with attention-deficit/hyperactivity disorder: a psychophysiological study of the stop task. Biol Psychiatry. 2002;51:668–676. doi: 10.1016/s0006-3223(01)01290-2. [DOI] [PubMed] [Google Scholar]
- 56.Franke B, Vasquez AA, Johansson S, Hoogman M, Romanos J, et al. Multicenter analysis of the SLC6A3/DAT1 VNTR haplotype in persistent ADHD suggests differential involvement of the gene in childhood and persistent ADHD. Neuropsychopharmacology. 2010;35:656–664. doi: 10.1038/npp.2009.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dresler T, Ehlis A-C, Heinzel S, Renner TJ, Reif A, et al. Dopamine transporter (SLC6A3) genotype impacts neurophysiological correlates of cognitive response control in an adult sample of patients with ADHD. Neuropsychopharmacology. 2010;35:2193–2202. doi: 10.1038/npp.2010.91. [DOI] [PMC free article] [PubMed] [Google Scholar]