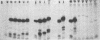Abstract
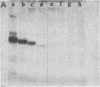
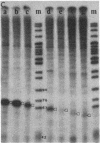
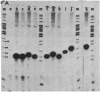
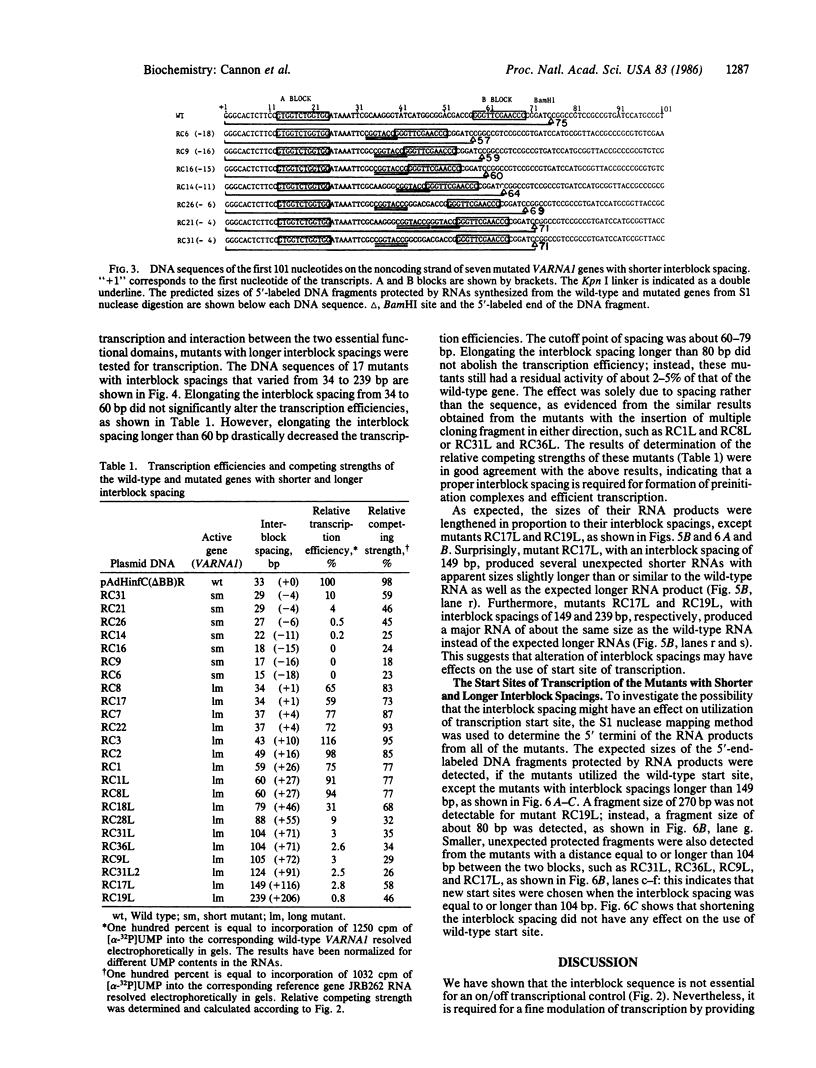
The internal transcriptional control region (ITCR) of VARNA1 gene consists of a 33-base-pair (bp) interblock sequence and two 12-bp sequence blocks that are highly conserved in most of the genes transcribed by RNA polymerase III. To define the functions of and study the interactions between the two blocks, we have constructed mutants with altered interblock sequence or spacing for transcription. The results of transcription efficiencies and competing strengths indicated that the interblock sequence was dispensable and the A and B blocks were essential for transcription control. One of the major functions of the interblock sequence was to maintain an optimal spacing for an intimate interaction between the two essential blocks. Shortening or elongating the interblock spacing in the mutants beyond this range drastically decreased the transcription efficiencies and competing strengths of these mutated genes. To further study how the interaction between the two blocks leads to initiation, the start sites and sizes of RNA products of the mutants were determined. When the interblock spacing was less than 105 bp, the wild-type start site was dictated by the A block after an interaction with the B block through proteins. However, when the interblock spacing was longer than 105 bp, several new start sites located closer to the B block were preferentially used. This suggests that new start sites may be dictated by the B block when its interaction with the A block is weakened by longer spacing. The mechanisms of interaction between the bipartite domain in this gene leading to initiation are different from those in tRNAs and Alu-family RNA genes.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Ciliberto G., Raugei G., Costanzo F., Dente L., Cortese R. Common and interchangeable elements in the promoters of genes transcribed by RNA polymerase iii. Cell. 1983 Mar;32(3):725–733. doi: 10.1016/0092-8674(83)90058-2. [DOI] [PubMed] [Google Scholar]
- DeFranco D., Sharp S., Söll D. Identification of regulatory sequences contained in the 5'-flanking region of Drosophila lysine tRNA2 genes. J Biol Chem. 1981 Dec 10;256(23):12424–12429. [PubMed] [Google Scholar]
- Dingermann T., Sharp S., Schaack J., Söll D. Stable transcription complex formation of eukaryotic tRNA genes is dependent on a limited separation of the two intragenic control regions. J Biol Chem. 1983 Sep 10;258(17):10395–10402. [PubMed] [Google Scholar]
- Fowlkes D. M., Shenk T. Transcriptional control regions of the adenovirus VAI RNA gene. Cell. 1980 Nov;22(2 Pt 2):405–413. doi: 10.1016/0092-8674(80)90351-7. [DOI] [PubMed] [Google Scholar]
- Fuhrman S. A., Engelke D. R., Geiduschek E. P. HeLa cell RNA polymerase III transcription factors. Functional characterization of a fraction identified by its activity in a second template rescue assay. J Biol Chem. 1984 Feb 10;259(3):1934–1943. [PubMed] [Google Scholar]
- Guilfoyle R., Weinmann R. Control region for adenovirus VA RNA transcription. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3378–3382. doi: 10.1073/pnas.78.6.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar A. B., Martin P. L., Roeder R. G. Transcription of class III genes: formation of preinitiation complexes. Science. 1983 Nov 18;222(4625):740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Perez-Stable C., Ayres T. M., Shen C. K. Distinctive sequence organization and functional programming of an Alu repeat promoter. Proc Natl Acad Sci U S A. 1984 Sep;81(17):5291–5295. doi: 10.1073/pnas.81.17.5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaack J., Sharp S., Dingermann T., Söll D. Transcription of eukaryotic tRNA genes in vitro. II. Formation of stable complexes. J Biol Chem. 1983 Feb 25;258(4):2447–2453. [PubMed] [Google Scholar]
- Shenk T. Transcriptional control regions: nucleotide sequence requirements for initiation by RNA polymerase II and III. Curr Top Microbiol Immunol. 1981;93:25–46. doi: 10.1007/978-3-642-68123-3_3. [DOI] [PubMed] [Google Scholar]
- Söderlund H., Pettersson U., Vennström B., Philipson L., Mathews M. B. A new species of virus-coded low molecular weight RNA from cells infected with adenovirus type 2. Cell. 1976 Apr;7(4):585–593. doi: 10.1016/0092-8674(76)90209-9. [DOI] [PubMed] [Google Scholar]
- Thimmappaya B., Jones N., Shenk T. A mutation which alters initiation of transcription by RNA polymerase III on the Ad5 chromosome. Cell. 1979 Dec;18(4):947–954. doi: 10.1016/0092-8674(79)90207-1. [DOI] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann R., Jaehning J. A., Raskas H. J., Roeder R. G. Viral RNA synthesis and levels of DNA-dependent RNA polymerases during replication of adenovirus 2. J Virol. 1975 Jan;17(1):114–126. doi: 10.1128/jvi.17.1.114-126.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann R., Raskas H. J., Roeder R. G. Role of DNA-dependent RNA polymerases II and III in transcription of the adenovirus genome late in productive infection. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3426–3439. doi: 10.1073/pnas.71.9.3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. J. Adenovirus DNA-directed transcription of 5.5S RNA in vitro. Proc Natl Acad Sci U S A. 1978 May;75(5):2175–2179. doi: 10.1073/pnas.75.5.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. J., Cannon R. E. An economical large scale procedure to purify E. coli amplifiable plasmids for DNA sequencing, in vitro transcription and in vitro mutagenesis. Experientia. 1985 Nov 15;41(11):1488–1490. doi: 10.1007/BF01950053. [DOI] [PubMed] [Google Scholar]
- Wu G. Faithful transcription of adenovirus 5.5 S RNA gene by RNA polymerase III in a human KB cell-free extract. J Biol Chem. 1980 Jan 10;255(1):251–258. [PubMed] [Google Scholar]