Abstract
Chronic rhinosinusitis (CRS) is a common inflammatory disease of the sinonasal cavity mediated, in part, by polymicrobial communities of bacteria. Recent molecular studies have confirmed the importance of Streptococcus pneumoniae and nontypeable Haemophilus influenzae (NTHi) in CRS. Here, we hypothesize that interaction between S. pneumoniae and NTHi mixed-species communities cause a change in bacterial virulence gene expression. We examined CRS as a model human disease to validate these polymicrobial interactions. Clinical strains of S. pneumoniae and NTHi were grown in mono- and co-culture in a standard biofilm assay. Reverse transcriptase real-time PCR (RTqPCR) was used to measure gene expression of key virulence factors. To validate these results, we investigated the presence of the bacterial RNA transcripts in excised human tissue from patients with CRS. Consequences of physical or chemical interactions between microbes were also investigated. Transcription of NTHi type IV pili was only expressed in co-culture in vitro, and expression could be detected ex vivo in diseased tissue. S. pneumoniae pyruvate oxidase was up-regulated in co-culture, while pneumolysin and pneumococcal adherence factor A were down-regulated. These results were confirmed in excised human CRS tissue. Gene expression was differentially regulated by physical contact and secreted factors. Overall, these data suggest that interactions between H. influenzae and S. pneumoniae involve physical and chemical mechanisms that influence virulence gene expression of mixed-species biofilm communities present in chronically diseased human tissue. These results extend previous studies of population-level virulence and provide novel insight into the importance of S. pneumoniae and NTHi in CRS.
Introduction
The current paradigm of one organism causing disease has been valuable in treating acute infections, however, it has become clear that many chronic bacterial infections are comprised of mixed-species microbial communities. In a healthy state, microbial species exist interdependently among each other and with the host. However, this balance is delicate, and a shift in relative abundances within these complex communities can lead to chronic infection [1]–[6]. Jenkinson and Lamont first described the idea of ‘community virulence’ in the context of oral microbial communities [5]. This paper introduced the idea that interactions among specific microbial communities produce increased or attenuated virulence profiles. In accord with this observation, Ehrlich and colleagues established ‘bacterial plurality’ as a new rubric for understanding chronic infections. Bacterial plurality asserts that bacteria within a population display multiple phenotypes and have multiple genotypes (supragenome), which arise by horizontal gene transfer [1]–[3], [7]. Often, communities of bacteria acting as multicellular organisms express virulence factors that planktonic bacteria do not [3]. Population-level virulence is important in chronic infections including cystic fibrosis, dental caries, otitis media and chronic rhinosinusitis [6], [8]–[11]. Here, we examine chronic rhinosinusitis (CRS) as a model human disease to describe polymicrobial interactions.
Chronic rhinosinusitis is a common disease of the sinonasal tract characterized by symptoms of inflammation lasting for >12 weeks. Several studies have demonstrated the polymicrobial nature of CRS using 454 sequencing and 16S fluorescence in situ hybridization (FISH) [12]–[15]. Recent molecular studies of CRS have confirmed the importance of the respiratory pathogens Streptococcus pneumoniae and Haemophilus influenzae [12], [16]–[18]. S. pneumoniae is a member of the normal flora of the nasopharynx in up to 60% of all healthy children, and carriage rates can approach 100% in some developing countries [4], [19]. Nevertheless, S. pneumoniae is implicated in otitis media, pneumonia, and meningitis. Non-typeable H. influenzae (NTHi) are nonencapsulated strains found naturally in the human respiratory tract [20]. NTHi is implicated in several diseases including otitis media (chronic and acute) and chronic rhinosinusitis (for a comprehensive review of NTHi see reference [21]). Since S. pneumoniae and H. influenzae are implicated in otorhinolaryngologic disease, an increased understanding of their interspecies interactions is important.
S. pneumoniae has several virulence factors that regulate nasopharyngeal colonization and host tissue damage. Pyruvate oxidase (spxB) metabolism results in H2O2 production, which aids in colonization of the host and confers a competitive advantage with respect to NTHi when establishing microbial communities [22], [23]. Pneumolysin (ply) is a hemolytic pore-forming toxin that is important in respiratory infection and host cell damage [22], [24]. Pneumococcal adherence factor A (pavA) binds host fibronectin and is important for bacterial adherence in the upper airway and damaged host tissue [25], [26]. Although not explored in this study, the pneumococcal capsule is an important virulence determinant and can now be differentiated into over 90 serotypes, which can indicate invasiveness and virulence of S. pneumoniae [27], [28]. NTHi type IV pili (pilA) are important for nasopharyngeal colonization in vivo [29], [30]. Type IV pili are involved in biofilm formation and allow NTHi to establish itself within the host resulting in prolonged infection. NTHi biofilms have been implicated in chronic otitis media [31], and recently, chronic rhinosinusitis [16].
In this study, we demonstrate regulation of gene expression between S. pneumoniae and NTHi during polymicrobial biofilm growth and confirmed in vivo gene expression from excised human CRS tissue. Furthermore, we defined the effects of physical contact and secreted factors on these community interactions. These data support the current hypotheses regarding population-level virulence in chronic polymicrobial disease since virulence factors important for colonization and bacterial persistence were up-regulated in mixed species communities. Since NTHi and S. pneumoniae can be cultured from the sinuses using standard clinical techniques, it is relatively easy to identify patients who are co-infected with both organisms. In the future, designing drugs or vaccines that interfere with colonization of the sinuses by S. pneumoniae and NTHi will impact the severity of CRS disease by reducing biofilm abundance.
Methods
Bacterial growth conditions
Clinical strains of Streptococcus pneumoniae, and nontypeable Haemophilus influenzae were obtained from the respiratory tract of hospitalized patients. S. pneumoniae were grown on tryptic soy (TS) agar and in TS broth. H. influenzae were grown on chocolate agar (BD) and brain heart infusion broth supplemented with 2% bovine hemoglobin (Sigma Aldrich) and Isovitalix enrichment agent (BD). All cultures were grown at 37°C under ambient oxygen conditions.
Growth curve of S. pneumoniae and H. influenzae
Mono- or co-cultures were grown for each species of bacteria in appropriate media overnight. Cultures were diluted 1∶100 into fresh media. Optical density (600nm) was recorded on an Eppendorf spectrophotometer. Readings were taken every hour for a total of 10 hours. Bacteria were diluted, plated on chocolate agar and TS agar, and CFU calculated at 0 minutes, 255min, 485min, and 600min after inoculation with overnight cultures of each species.
Biofilm growth
Biofilms were grown in 24-well polystyrene plates (Falcon) as previously described [32]. For co-cultures, an equal volume of each diluted log-phase culture was added to each well to achieve a total volume of 500 µl. The plates were covered and incubated at 37°C without agitation for 24 hours. Biofilm cells were harvested by sonication for 30 seconds after inverting the plates to remove unattached cells. Biofilm cells were collected in 1.7 mL eppendorf tubes, centrifuged to pellet, and stored in RNAlater for RNA extractions, below.
RNA extractions for reverse transcriptase
RNA was extracted using a RiboPure-Bacteria kit (Applied Biosystems) per manufacturer protocol. Following extraction, RNA was treated with DNase I (Applied Biosystems) to remove any DNA contaminants. RNA concentration and purity was determined by measuring absorbance at both 260 nm and 280 nm on a NanoDrop ND-2000 spectrophotometer (Thermo Scientific). For cDNA synthesis, RNA was normalized to 1 ng/µl. First strand cDNA was synthesized using a High Capacity cDNA reverse transcription kit (Applied Biosystems) per manufacturer protocol. A negative control containing the reaction components without reverse transcriptase was included to ensure no DNA contamination was present.
Relative gene expression
Gene expression was determined using a SYBRgreen (Applied Biosystems) real time PCR assay. Gene specific primers were designed in the open reading frame of each gene of interest, Table 1. The 20 µl reactions consisted of 1X SYBRgreen mastermix, 1 µM each primer, and 2 µl of cDNA. Real time cycler conditions were 1X of 50°C for 2 min, 1X of 95°C for 2 min, 40X of 95°C for 15 sec, and 60°C for 1 min, 1X of 72°C for 5m, followed by a melt curve of 71X at 5 degree intervals for 5 seconds each. No reverse transcriptase (NRT), and a no template control (NTC) were included to verify the reaction mixtures were contaminant-free. A positive control of pure DNA from each species was included to ensure the reaction conditions were appropriate and gene amplification occurred during each run. We used the ΔΔCt method to determine relative gene expression of the gene of interest in co-culture v. mono-culture biofilms normalized to expression of the 16S rRNA gene for each organism. This allowed for visualization of up or down regulation of a particular gene when two species were in co-culture. Results are reported as ΔΔCt and graphed appropriately. Fold change was calculated by using the exponential 2ΔΔCt. Significance was determined using Student's t-test. A p≤0.05 was considered statistically significant.
Table 1. List of primer sequences and target gene function.
| Organism | Primer ID | Primer Sequence 5′-3′ | Gene Target | Gene Function | Reference/ Accession Number |
| Haemophilus influenzae | HFLU-F | TCCTAAGAAGAGCTCAGAGAT | 16s | Ribosomal RNA | [59] |
| Haemophilus influenzae | HFLU-R | TGATCCAACCGCAGGTTCC | 16s | ||
| Haemophilus influenzae | pilA-F | CTATATACACATAATTCCACATCAGCCTTA | pilA | Colonization, biofilm formation | [30], [60] |
| Haemophilus influenzae | pilA-R | CCACCATCGCAATTCCTTCTT | pilA | ||
| Streptococcus pneumoniae | STPN-F | CTGCGTTGTATTAGCTAGTTGGTG | 16s | Ribosomal RNA | [61] |
| Streptococcus pneumoniae | STPN-R | TCCGTCCATTGCCGAAGATTC | 16s | ||
| Streptococcus pneumoniae | spxB-F | ATTCGGCGGCTCAATCGGGG | spxB | Production of H2O2 | NC_008533.1 |
| Streptococcus pneumoniae | spxB-R | CAGCACGGCAGGCTTCGTCA | spxB | ||
| Streptococcus pneumoniae | Ply2-F | CTACCCGATGAGTTTGTTGTT | ply | Cytolysin | [61] |
| Streptococcus pneumoniae | Ply2-R | TCCAGGATAGAGGCGACT | ply | ||
| Streptococcus pneumoniae | pavA-F | GGTCGCATCCAGAAAATC | pavA | Adherence and inflammation | [61] |
| Streptococcus pneumoniae | pavA-R | AGAAAGGAGCAGGCGATG | pavA |
Genes for NTHi and S. pneumoniae are listed along with primers sequences (5′-3′), gene target, function, and GenBank accession number, if applicable.
Scanning electron microscopy
NTHi was grown in mono and co-culture for 24h in 4-chamber glass slides. Following incubation, bacteria were fixed with 4% paraformaldehyde for 4h at room temperature. Slides were dried in ethanol, desiccated overnight, and sputtered with gold for SEM. Mono-culture NTHi and co-culture NTHi and S. pneumoniae biofilms were observed at 10,000X magnification.
Ethics Statement
Tissue samples from human sinuses were obtained from the Hospital of the University of Pennsylvania. A total of 10 patients undergoing functional endoscopic sinus surgery (FESS) were solicited. The Hospital of the University of Pennsylvania Institutional Review Board specifically approved this study under the approval number #800614. Written informed consent was provided by study participants.
RNA isolation from excised human sinus tissue
Following biopsy, tissue was stored in RNAlater and shipped to NAU for analysis. A total of 7 CRS and 3 non-CRS patients were used for these studies. Total bacterial RNA was extracted from the excised human sinus tissue using the RiboPure – Bacteria with the following modifications to manufacturer protocol. Approximately 25 mg of tissue was homogenized using 0.1 mm zirconia beads and one 5mm steel bead on a Qiagen TissueLyser in RNAwiz (Applied Biosystems) at 4oC. The homogenate was transferred to a new tube and 0.2 volumes of chloroform added. The RNA mixture was incubated at room temperature for 10 min. Following incubation, the mixture was centrifuged and the top layer containing partially purified RNA removed. Ethanol was added and the mixture placed directly on a spin column provided with the kit. RNA was washed 3X, and eluted. Purified RNA was treated with DNase I and quantified as described above.
Ex vivo gene expression
The presence of bacterial RNA was determined by universal 16S primers in a SYBRgreen assay at a 1 µM concentration. The primers were 1406f, 5′-??? TGYACACACCGCCCGT-3′ (universal, 16S rRNA gene), and 23Sr, 5′-??? GGGTTBCCCCATTCRG-3′ (bacterial-specific, 23S rRNA gene) [33]. Relative bacterial gene expression was determined as described above.
Physical contact co-culture assay
To determine the role of physical contact in the production of community virulence factors, we utilized 0.22 µM tissue culture inserts as described previously [34]. To ensure biofilm confluence on the tissue culture insert, bacteria were stained with Syto-9 and visualized using fluorescence microscopy (data not shown). Biofilms were incubated for 24 hours and RNA isolated as described above. cDNA was used to assay for gene expression and ΔΔCt are reported below.
Treatment of biofilm bacteria with supernatant fluid
To investigate whether polymicrobial-specific gene regulation was mediated by secreted factors, co-culture biofilms of H. influenzae and S. pneumoniae were grown in 24-well polystyrene plates (Falcon) as previously described. After 24 hours, supernatant fluid (SNF) was extracted into sterile 50 mL conical tubes (Fisher Scientific) and centrifuged at 4,000x rpm for 20 minutes to remove cellular components. The SNF was then decanted into a new, sterile 50 mL conical tube and filter sterilized (Millipore, 0.22 µm pore diameter). Mono-culture biofilms were inverted to remove the spent nutrient-rich media and the sterilized, conditioned SNF was then introduced and the biofilms incubated at 37°C for an additional 24 hours. Cells were harvested by sonication for 30 seconds, and RNA was extracted as described above. Complementary DNA was synthesized and used in rtPCR assays for genetic analysis. Controls of nutrient deprived media were employed to rule out nutrient deprivation as a main factor in gene expression changes.
Statistical methods
For rtPCR analysis, mean Ct values were calculated across trials. Three independent trials were conducted unless otherwise stated. Standard deviation was reported and Student's t-test was used to determine significance compared to control. Significance was determined by comparing raw Ct values to the number of PCR cycles used in each assay. Co-cultures were normalized to mono-culture expression of each gene by the following equation, ΔΔCt = ((µCt(mono-culture) – µCt(co-culture)) –(µCt(16s monoculture)- µCt(16s coculture)), where µ is the mean. Fold change was calculated by using the exponential 2 ΔΔCt. Ct differences were considered significant when p≤0.05.
Results
Planktonic growth is not inhibited in co-culture conditions
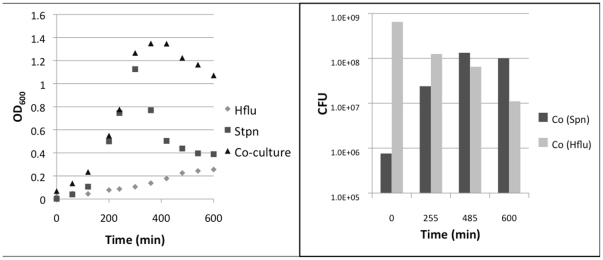
To confirm that gene regulation observed in co-culture biofilms was not a product of cellular death or inhibition of growth, we measured the optical density of mono- and co-culture bacteria over 10 hours of planktonic growth. These studies demonstrated that growth inhibition did not occur in co-culture conditions (Figure 1). Conversely, the curve suggested a synergistic interaction in the planktonic state, since co-culture bacteria grew more vigorously. Colony forming units were calculated periodically throughout the growth curve beginning at time 0, directly after inoculation with S. pneumoniae and NTHi. Both NTHi and S. pneumoniae survived to 10h, well into the stationary phase of growth, indicating that co-existence can occur in vitro.
Figure 1. Growth curve and colony forming units of planktonic mono and co-cultures of NTHi and S. pneumoniae.
Planktonic NTHi and S. pneumoniae were grown in sBHI for 10h. Optical density (600nm) and CFU were recorded. Growth inhibition was not detected in co-culture by this method.
H. influenzae type IV pili is expressed only in co-culture conditions
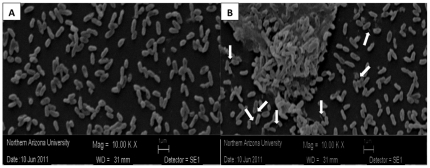
To investigate gene regulation when H. influenzae was cultured with S. pneumoniae, the transcript for NTHi type IV pili (pilA) and was examined and compared to expression of NTHi 16S rRNA as a reference gene. Type IV pili are important for nasopharyngeal colonization and in vivo growth of H. influenzae [30], [35]. Since pilA is important for early biofilm formation, mono- and co-culture biofilms were grown for 4h and 24h and relative gene expression was calculated. Following each co-culture experiment, bacteria were plated to ensure growth of both organisms after 4 and 24 hours (data not shown). We found that pilA was only expressed in co-culture conditions (Table 2) at both 4h and 24h (p = 0.001, and 0.013, respectively). For this study we examined S. pneumoniae and NTHi mediated induction of pilA, however, other studies in our lab suggest that this not a species-specific effect (data not shown). Scanning electron images of NTHi mono-culture biofilm (Figure 2, panel A) and co-culture with S. pneumoniae (Figure 2, panel B) were also observed at 10,000X to identify pili. There is an increase in pili-like structures from the rod-shaped cells (NTHi) in the co-culture setting, whereas no pili-like structures could be detected in mono-culture after 24h of growth.
Table 2. Real time PCR analysis of expression of NTHi type IV pili.
| 2ΔΔCt (Fold change from reference gene) | ||
| 4h | 24h | |
| pilA | 9.33E+07* | 9.17E+07* |
| (+ 1.94) | (+ 2.53) | |
NTHi were grown in mono- and co-culture biofilms and expression of 16s rRNA reference gene and pilA are recorded and the ΔΔCt calculated. Fold change, or 2ΔΔCt, from the reference gene are displayed in panel B. Standard deviation is presented, and changes are considered significant when p≤0.05, determined by Students t-test. Significance is indicated by *.
Figure 2. Scanning electron microscopy of NTHi mono-culture and co-culture with S. pneumoniae show increase in pili-like structures in co-culture.
NTHi was grown for 24h in mono- and co-culture with S. pneumoniae and SEM was used to identify pili-like structures. While no pili could be identified at 10,000X magnification in mono-culture, at least 7 structures were identified in co-culture with S. pneumoniae.
S. pneumoniae virulence genes are differentially regulated in co-culture with H. influenzae
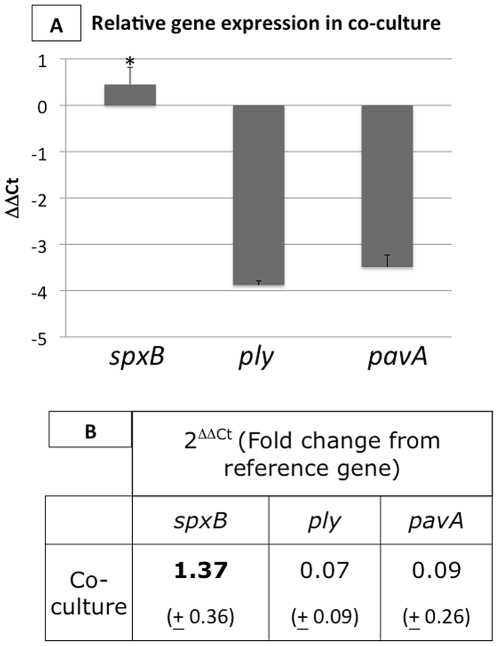
To further define the products of interactions between S. pneumoniae and H. influenzae, we examined the transcription of several S. pneumoniae virulence factors in mono- and co-culture. Genes of interest included pneumolysin (ply), pneumococcal adherence factor A (pavA), and pyruvate oxidase (spxB). These virulence factors are important in nasopharyngeal colonization in vivo, and have been implicated in respiratory tract infection [23], [24], [36]. For a reference of normal gene expression, streptococcal 16S rRNA gene was also included. Notably, spxB was up-regulated when in co-culture with H. influenzae for 24h (Fold change, 1.37, p = 0.001), while the other virulence factors, ply and pavA, were down-regulated (p = 001, and p = 0.012, respectively). The ΔΔCt values of co-culture compared to mono-culture are demonstrated in Figure 3 panel A, and fold change is described in Figure 3 panel B.
Figure 3. Real time PCR for expression of S. pneumoniae housekeeping and virulence genes show differential gene regulation in co-culture with NTHi.
S. pneumoniae were grown in mono- and co-culture biofilms and expression of spxB, ply, and pavA are recorded and normalized to expression of the 16s rRNA gene using the ΔΔCt method. Up and down-regulation are displayed as ΔΔCt in panel A. Fold change from the reference gene are displayed in panel B. Changes are considered significant when p≤0.05. Significance is indicated by *. Here, we show significant up-regulation of spxB in co-culture with NTHi, and down-regulation of ply, and pavA.
Virulence gene regulation occurs in complex communities in vivo
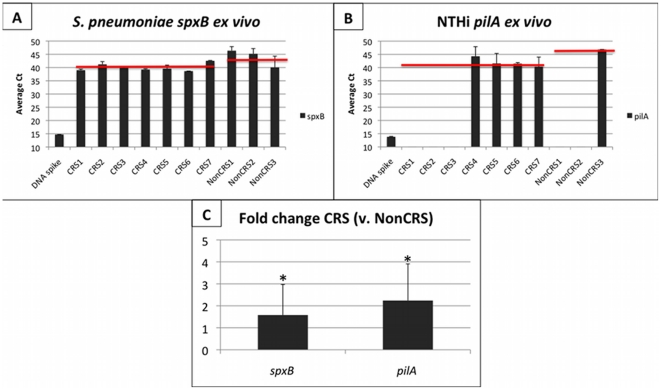
To investigate whether these genes were expressed in diseased human sinus tissue from patients with chronic rhinosinusitis (CRS), we isolated bacterial RNA from excised human sinus tissue. Although the communities present in the sinuses are notably complex [12], we hypothesized that the interactions observed in vitro are likely to occur in vivo in a chronic disease setting. Both pilA and spxB were expressed in CRS associated microbial communities, (Figure 4). Reference 16S genes from each organism were included to calculate ΔΔCt and fold change (Figure 4C). S. pneumoniae spxB was present in 7/7 CRS patients and was up-regulated in CRS tissue (average Ct = 40.02) compared to non-CRS control (average Ct = 43.87). When normalized to the expression of S. pneumoniae 16S rRNA, spxB was expressed 1.58x greater in CRS vs. non-CRS patient tissue samples. Expression of spxB in important for nasopharyngeal colonization in vivo, so expression in samples without NTHi is not surprising, especially in the presence of polymicrobial communities in CRS tissue. NTHi pilA was expressed in 4/7 CRS patients, and in 1/3 non-CRS patients. Expression of pilA was significantly increased in CRS (average Ct = 41.93) compared to non-CRS tissue (average Ct = 46.85, p = 0.003). When normalized to the 16S reference gene expression, pilA was expressed 2.24x greater in CRS vs. non-CRS tissue.
Figure 4. Real time PCR for S. pneumoniae and NTHi virulence gene transcripts from CRS and non-CRS excised tissue.
Transcripts for virulence factors of S. pneumoniae and NTHi were observed in human tissue from 7 CRS and 3 non-CRS patients (x-axis). A positive control of the appropriate DNA spike was included. Panels A and B show the average Ct for spxB (A) and pilA (B) genes, and the average among all positive samples in CRS and non-CRS (red bar). Panel C displays the fold change (2ΔΔCt) of spxB and pilA relative to the appropriate 16s rRNA gene expression in CRS vs. non-CRS tissue. This figure shows the presence of S. pneumoniae spxB in 7/7 CRS and 3/3 non-CRS; however, expression was significantly greater in CRS patients (P = 0.002). NTHi pilA was demonstrated in 4/7 CRS samples and 1/3 non-CRS samples. Expression of pilA was significantly greater in CRS than non-CRS patients. Standard deviation is presented and significance (P<0.05) is indicated by *, determined by students t-test.
Physical contact regulates S. pneumoniae and NTHi gene expression
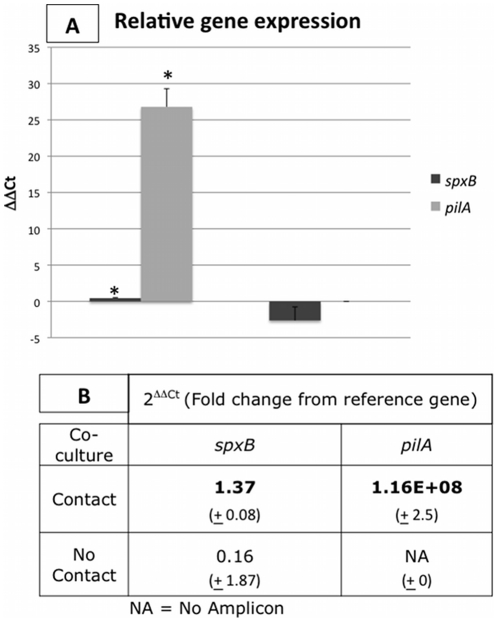
To investigate whether physical contact played a role in virulence gene regulation, we grew co-cultures of both organisms separated by a trans-well tissue culture insert [34]. Genes that were up-regulated in co-culture were included in these studies. When physical contact was allowed, spxB was expressed 1.37x greater in co-culture with NTHi (Figure 5). Interestingly, S. pneumoniae spxB was slightly down-regulated in the absence of physical contact, although this result was not statistically significant. NTHi expression of pilA was induced only when physical contact with S. pneumoniae was allowed. No gene expression of pilA was observed when NTHi was grown in mono-culture or in the trans-well assay with S. pneumoniae.
Figure 5. Physical contact regulates the expression of NTHi pilA, not S. pneumoniae spxB.
Virulence factors pilA and spxB were assayed for when S. pneumoniae and NTHi were in mono-culture, co-culture with physical contact (dark gray bars), and in co-culture without physical contact (light gray bars). The ΔΔCt is presented in Panel A. Fold change from 16s gene expression is displayed in Panel B. Expression of pilA was induced only when physical contact was allowed. S. pneumoniae spxB was only significantly up-regulated when physical contact was allowed (p = 0.002). Standard deviation is presented and significance (P<0.05) is indicated by *, determined by students t-test.
Soluble factors in supernatant fluid from co-culture biofilms influence mono-culture gene expression
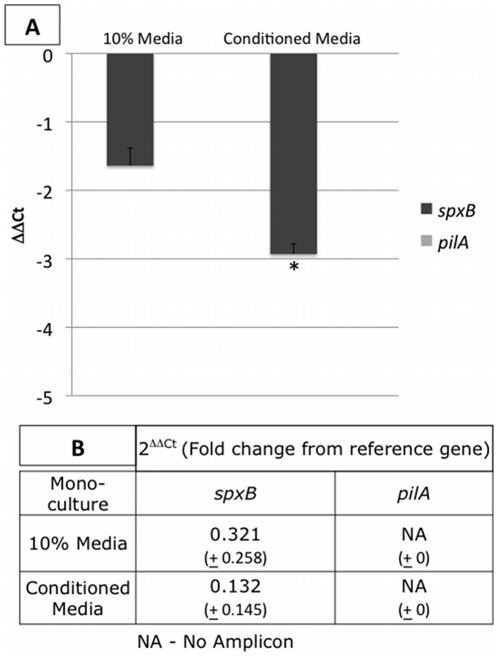
To assess whether soluble factors produced during co-culture growth influenced mono-culture gene expression, we employed a conditioned media assay (Figure 6, experimental details provided above). To ensure that the effects on gene expression were not solely due to nutrient starvation, we included a 10% media control. We found that expression of pilA was not induced by soluble factors, suggesting that secreted factors do not regulate type IV expression in NTHi in this assay system. S. pneumoniae expression of spxB suppressed in the presence of conditioned media, which may indicate a negative interaction on cell growth or metabolic activity when only soluble factors from co-cultures are present. While it is clear that nutrient starvation decreased expression of spxB, media conditioned with co-culture soluble factors was significantly different (p = 0.0002), suggesting that some down-regulation can be attributed to secreted factors in the co-culture setting.
Figure 6. Chemical factors secreted in co-culture conditions regulate the expression of key virulence genes.
Conditioned media from S. pneumoniae/NTHi 24h co-cultures was used to treat 24h single species biofilms (Conditioned Media column). Cells were grown in 100% media, and treated with 100%, or 10% media to distinguish the effects of nutrient depletion. The ΔΔCt is presented in Panel A. Fold change from 16s gene expression is displayed in Panel B. Conditioned media did not induce pilA expression. S. pneumoniae spxB and the 16s rRNA genes were down-regulated in the presence of CM compared to the media controls. Student's t-test determined significant down-regulation of spxB only when conditioned media was used.
Discussion
In this study, we investigated the regulation of virulence determinants in S. pneumoniae and H. influenzae clinical isolates as a function of community interactions. While competition between strains of S. pneumoniae and NTHi has been observed [37], [38], our analyses did not demonstrate any measurable competition that would compromise these data. Rather, growth curve and CFU analysis of dual NTHi and S. pneumoniae cultures suggested an additive effect. These discrepancies could be explained by the use of vastly different strains of NTHi and S. pneumoniae between these studies. For example, Weiser and colleagues demonstrated an inverse relationship between S. pneumoniae and NTHi colonization in vitro, largely due to S. pneumoniae production of pyruvate oxidase, using the typed laboratory strain P294 (S. pneumoniae serotype 4) [37]. However, the isolates of S. pneumoniae and NTHi used in the studies presented here were clinical strains directly from the human nasopharynx. It should be noted that due to the clinical nature of these strains, S. pneumoniae was not fully characterized and serotype information was not available. We recognize the enormous intraspecies heterogeneity of S. pneumoniae, and appreciate this limitation of these studies. Over 90 pneumococcal serotypes varying in capsular structure have been identified, and associations between serotypes and colonization of the upper respiratory tract have been described [39]. For example, in a large-scale epidemiological study of acute otitis media in Germany, serotypes 19F and 3 were the most commonly isolated [40], while serotypes 1, 5, 6, 14, 19F, and 23F were most commonly isolated from invasive pneumococcal disease [41]. While serotype information would help to clarify disparities in the literature, we feel that the strains used in these studies represent potential in vivo interactions.
The frank S. pneumoniae virulence factors streptococcal pneumolysin and pneumococcal adherence factor A were down-regulated in co-culture with H. influenzae. Streptococcal pneumolysin is a cholesterol dependent cytolysin and is an important virulence determinant of pneumonia [42]. Pneumococcal adherence factor A is required for optimal adherence to host cells and is responsible for increased virulence in a mouse model [25], [26]. These results were expected because these factors are often associated with acute infections in humans. Chronic rhinosinusitis, and other biofilm-mediated diseases, are not characterized by frank epithelial damage from bacterial virulence factors, but by prolonged inflammation caused by persistence of pathogenic microbial communities.
Expression of streptococcal pyruvate oxidase (spxB), an enzyme that is important in the production of hydrogen peroxide, was up-regulated in the co-culture setting. Hydrogen peroxide production has been extensively studied in S. pneumoniae as a means for nasal colonization [23], [43]–[45]. Pyruvate oxidase also confers a competitive advantage for nasopharyngeal colonization in vivo partly because S. pneumoniae is resistant to self-produced H2O2 killing while other species are susceptible during stages of early colonization [23], [44]. Interestingly, Allegrucci and colleagues demonstrated that maximal production of spxB occurs in mature single-species biofilms (6 days) [46]. Our studies show an increase of spxB transcription in 24h co-culture biofilms, suggesting that streptococcal virulence is intensified with respect to colonization and competition in co-culture. Other streptococcal virulence factors play a role in nasopharyngeal colonization including the capsule, the adhesion molecule phosphorylcholine (choP), choline-binding proteins, and exoglycosidases [26]. We are currently investigating the role of these genes in streptococcal community virulence in vitro and ex vivo.
H. influenzae virulence factor type IV pili (pilA) was expressed only in mixed-species biofilms. Type IV pili are important in biofilm formation, epithelial colonization and colonization of the upper respiratory tract. Prior to the study by Bakaletz and colleagues (2005), NTHi were not considered to produce functional type IV pili [47]. This was, in part, because single species cultures of NTHi in vitro do not express important attachment proteins when grown on common growth media. However, Bakaletz et al. visualized type IV pili under defined growth conditions. Recently, Jurcisek and colleagues (2007) demonstrated that type IV pili have an important role in biofilm formation and epithelial attachment in vivo [29]. Here, we show pilA expression is inducible in co-culture in vitro and is expressed in chronically diseased sinus tissue. These studies support recent findings by Swords and colleagues that NTHi interacts with pneumococci to promote biofilm formation in a chronic infection [48]. While further study is necessary to define the implications of NTHi pilA and S. pneumoniae spxB in disease, these data provide an important first step to understanding the mechanisms of polymicrobial persistence in disease. Likely, when in the same niche, S. pneumoniae and NTHi aid each other to adhere to the host epithelia and in the eventual establishment of a polymicrobial community that is recalcitrant to antibiotic treatment.
We confirmed the presence of S. pneumoniae and NTHi virulence transcripts in excised sinus tissue from 7 CRS and 3 non-CRS patients. Non-CRS patients were selected based on the absence of inflammation in the sinonasal cavity but are not classically ‘healthy’ patients. Nonetheless, transcription of S. pneumoniae spxB was increased in CRS compared to non-CRS tissue, regardless of the presence of NTHi. While our in vitro assays show that interaction with NTHi increased expression of streptococcal pyruvate oxidase, it is likely that the complex microbial communities in the human sinus also interact to increase gene expression of S. pneumoniae spxB (data not shown). Transcription of H. influenzae pilA is indicative of physical interactions among multispecies communities of microbes since it was only transcribed in the presence of another species in vitro. Importantly, we did not rule out host-pathogen interactions as a contributing factor to increased virulence gene expression in sinus tissue specimens. Recent studies have suggested that metabolic stress, components of the host immune system, and microenvironments within the host can influence bacterial virulence and invasiveness [49]-[52]. CRS is broadly characterized by a common endpoint of sinonasal inflammation. This response is notably complex, and results in an influx of inflammatory cells, effector molecules and small cationic peptides that likely influence bacterial gene expression [53]–[55]. In addition to the presence of polymicrobial communities in the diseased sinus, host-pathogen interactions may account for the increase of S. pneumoniae spxB expression even in the absence of NTHi. In future studies, we aim to elucidate the mechanisms that contribute to pathogenesis of S. pneumoniae and NTHi in CRS. Additionally, further investigation of the disease state of these patients is underway to see if NTHi with type IV pili and S. pneumoniae spxB are correlated with more severe or recalcitrant CRS disease. Importantly, these results confirmed our in vitro analysis and demonstrated their clinical relevance in diseased tissue.
Bacteria have several mechanisms to mediate interactions in a community setting. Secreted factors, which may include quorum sensing molecules, secondary metabolites, carbohydrates, or proteins, can influence gene regulation in microbial communities. To test for this interaction, while not ruling out additional factors, we employed a conditioned media assay, described above. To rule out changes in gene expression due to nutrient depletion, we included a depleted (10%) media control. These data clearly show that NTHi pilA was not inducible by soluble components alone. Interestingly, S. pneumoniae spxB was down-regulated in the presence of conditioned media. This shows that soluble factors alone have a depressive effect on this virulence gene. This may be important when S. pneumoniae co-infects with another species, but does not share a niche. In this scenario, pyruvate oxidase metabolism would not be necessary as a competitive factor. S. pneumoniae and NTHi have been shown to co-infect, but form separate mono-species biofilms, in a tightly controlled chinchilla model of otitis media [48]. However, it is unlikely that this would occur in a complex chronic infection such as CRS, where there are several co-infecting species in addition to the host's normal flora. Importantly, while we have not completely ruled out the potential importance of secreted factors, we show that these factors alone cannot induce expression.
A second mechanism we examined was the effect of direct physical contact. NTHi pilA was expressed when physical contact occurred, suggesting physical interaction was important for expression of type IV pili. When physical contact was blocked, pilA expression was not induced. This may be similar to the interaction with host cells in in vivo infection models [29]. Since type IV pili are required for twitching motility, colonization, and biofilm formation, understanding these mechanisms may aid in treatment of many otolaryngologic and respiratory diseases. For example, Bakaletz and colleagues have demonstrated that immunization against NTHi type IV pili confers significant protection against NTHi infection in a chinchilla model of otitis media [56]–[58]. Suppression of spxB in S. pneumoniae when exposed to conditioned media reveal that secreted factors from H. influenzae has a negative effect on virulence in S. pneumoniae. Additionally, physical contact alone did not regulate spxB expression. However, when physical contact and secreted factors were coupled in a standard co-culture assay, maximum expression was observed. These data suggest that interactions between S. pneumoniae and NTHi are more complex than a single mechanism and warrant additional studies.
Overall, these data suggest that interactions between H. influenzae and S. pneumoniae involve physical (adherence) and chemical (secreted) mechanisms that influence virulence gene expression of mixed-species biofilm communities present in chronically diseased human tissue. The findings generated from these studies contribute to the understanding of the complex and clinically significant interactions between S. pneumoniae and NTHi in upper respiratory and otolaryngologic disease. While further investigation of the specific mechanisms that regulate community virulence are needed to better characterize the function of microbial communities in the context of polymicrobial disease, these studies may influence the development of new diagnostics and therapeutics. This will lead to more directed treatment of CRS and other chronic upper respiratory diseases worsened by the presence of S. pneumoniae and NTHi.
Acknowledgments
We thank the surgical team at University of Pennsylvania for patient enrollment and expertise in FESS tissue collection. We also thank Therry The (NAU) for handling the SEM specimens, and the members of the Leid Lab for thoughtful discussions and help preparing samples.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by Science Foundation Arizona GRF 0204-08 (http://www.sfaz.org) and the Flight Attendants Medical Research Institute, Clinical Innovator Award #103019. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ehrlich GD, Hiller NL, Hu FZ. What makes pathogens pathogenic. Genome Biol. 2008;9:225. doi: 10.1186/gb-2008-9-6-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehrlich GD, Ahmed A, Earl J, Hiller NL, Costerton JW, et al. The distributed genome hypothesis as a rubric for understanding evolution in situ during chronic bacterial biofilm infectious processes. FEMS Immunol Med Microbiol. 2010;59:269–279. doi: 10.1111/j.1574-695X.2010.00704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu FZ, Ehrlich GD. Population-level virulence factors amongst pathogenic bacteria: relation to infection outcome. Future Microbiol. 2008;3:31–42. doi: 10.2217/17460913.3.1.31. [DOI] [PubMed] [Google Scholar]
- 4.Henriques-Normark B, Normark S. Commensal pathogens, with a focus on Streptococcus pneumoniae, and interactions with the human host. Exp Cell Res. 2010;316:1408–1414. doi: 10.1016/j.yexcr.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13:589–595. doi: 10.1016/j.tim.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. Interspecies interactions within oral microbial communities. Microbiol Mol Biol Rev. 2007;71:653–670. doi: 10.1128/MMBR.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehrlich GD, Hu FZ, Shen K, Stoodley P, Post JC. Bacterial plurality as a general mechanism driving persistence in chronic infections. Clin Orthop Relat Res. 2005. pp. 20–24. [DOI] [PMC free article] [PubMed]
- 8.Brogden KA, Guthmiller JM, Taylor CE. Human polymicrobial infections. Lancet. 2005;365:253–255. doi: 10.1016/S0140-6736(05)17745-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheadle WG, Spain DA. The continuing challenge of intra-abdominal infection. Am J Surg. 2003;186:15S–22S; discussion 31S-34S. doi: 10.1016/j.amjsurg.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Rogers GB, Hart CA, Mason JR, Hughes M, Walshaw MJ, et al. Bacterial diversity in cases of lung infection in cystic fibrosis patients: 16S ribosomal DNA (rDNA) length heterogeneity PCR and 16S rDNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol. 2003;41:3548–3558. doi: 10.1128/JCM.41.8.3548-3558.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellis ME, Watson B, Mandal BK, Dunbar EM, Craske J, et al. Micro-organisms in gastroenteritis. Arch Dis Child. 1984;59:848–855. doi: 10.1136/adc.59.9.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stephenson MF, Mfuna L, Dowd SE, Wolcott RD, Barbeau J, et al. Molecular characterization of the polymicrobial flora in chronic rhinosinusitis. J Otolaryngol Head Neck Surg. 2010;39:182–187. [PubMed] [Google Scholar]
- 13.Mantovani K, Bisanha AA, Demarco RC, Tamashiro E, Martinez R, et al. Maxillary sinuses microbiology from patients with chronic rhinosinusitis. Braz J Otorhinolaryngol. 2010;76:548–551. doi: 10.1590/S1808-86942010000500002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niederfuhr A, Kirsche H, Riechelmann H, Wellinghausen N. The bacteriology of chronic rhinosinusitis with and without nasal polyps. Arch Otolaryngol Head Neck Surg. 2009;135:131–136. doi: 10.1001/archoto.2008.531. [DOI] [PubMed] [Google Scholar]
- 15.Chan J, Hadley J. The microbiology of chronic rhinosinusitis: results of a community surveillance study. Ear Nose Throat J. 2001;80:143–145. [PubMed] [Google Scholar]
- 16.Sanderson AR, Leid JG, Hunsaker D. Bacterial biofilms on the sinus mucosa of human subjects with chronic rhinosinusitis. Laryngoscope. 2006;116:1121–1126. doi: 10.1097/01.mlg.0000221954.05467.54. [DOI] [PubMed] [Google Scholar]
- 17.Healy DY, Leid JG, Sanderson AR, Hunsaker DH. Biofilms with fungi in chronic rhinosinusitis. Otolaryngol Head Neck Surg. 2008;138:641–647. doi: 10.1016/j.otohns.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Foreman A, Psaltis AJ, Tan LW, Wormald PJ. Characterization of bacterial and fungal biofilms in chronic rhinosinusitis. Am J Rhinol Allergy. 2009;23:556–561. doi: 10.2500/ajra.2009.23.3413. [DOI] [PubMed] [Google Scholar]
- 19.Cheung YB, Zaman SM, Nsekpong ED, Van Beneden CA, Adegbola RA, et al. Nasopharyngeal carriage of Streptococcus pneumoniae in Gambian children who participated in a 9-valent pneumococcal conjugate vaccine trial and in their younger siblings. Pediatr Infect Dis J. 2009;28:990–995. doi: 10.1097/INF.0b013e3181a78185. [DOI] [PubMed] [Google Scholar]
- 20.St Geme JW, 3rd The pathogenesis of nontypable Haemophilus influenzae otitis media. Vaccine. 2000;19(Suppl 1):S41–50. doi: 10.1016/s0264-410x(00)00277-2. [DOI] [PubMed] [Google Scholar]
- 21.Hardy GG, Tudor SM, St Geme JW, 3rd The pathogenesis of disease due to nontypeable Haemophilus influenzae. Methods Mol Med. 2003;71:1–28. doi: 10.1385/1-59259-321-6:01. [DOI] [PubMed] [Google Scholar]
- 22.Orihuela CJ, Gao G, Francis KP, Yu J, Tuomanen EI. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J Infect Dis. 2004;190:1661–1669. doi: 10.1086/424596. [DOI] [PubMed] [Google Scholar]
- 23.Regev-Yochay G, Trzcinski K, Thompson CM, Lipsitch M, Malley R. SpxB is a suicide gene of Streptococcus pneumoniae and confers a selective advantage in an in vivo competitive colonization model. J Bacteriol. 2007;189:6532–6539. doi: 10.1128/JB.00813-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadioglu A, Taylor S, Iannelli F, Pozzi G, Mitchell TJ, et al. Upper and lower respiratory tract infection by Streptococcus pneumoniae is affected by pneumolysin deficiency and differences in capsule type. Infect Immun. 2002;70:2886–2890. doi: 10.1128/IAI.70.6.2886-2890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmes AR, McNab R, Millsap KW, Rohde M, Hammerschmidt S, et al. The pavA gene of Streptococcus pneumoniae encodes a fibronectin-binding protein that is essential for virulence. Mol Microbiol. 2001;41:1395–1408. doi: 10.1046/j.1365-2958.2001.02610.x. [DOI] [PubMed] [Google Scholar]
- 26.Kadioglu A, Weiser JN, Paton JC, Andrew PW. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol. 2008;6:288–301. doi: 10.1038/nrmicro1871. [DOI] [PubMed] [Google Scholar]
- 27.Crook DW. Capsular type and the pneumococcal human host-parasite relationship. Clin Infect Dis. 2006;42:460–462. doi: 10.1086/499248. [DOI] [PubMed] [Google Scholar]
- 28.Sjostrom K, Spindler C, Ortqvist A, Kalin M, Sandgren A, et al. Clonal and capsular types decide whether pneumococci will act as a primary or opportunistic pathogen. Clin Infect Dis. 2006;42:451–459. doi: 10.1086/499242. [DOI] [PubMed] [Google Scholar]
- 29.Jurcisek JA, Bakaletz LO. Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J Bacteriol. 2007;189:3868–3875. doi: 10.1128/JB.01935-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jurcisek JA, Bookwalter JE, Baker BD, Fernandez S, Novotny LA, et al. The PilA protein of non-typeable Haemophilus influenzae plays a role in biofilm formation, adherence to epithelial cells and colonization of the mammalian upper respiratory tract. Mol Microbiol. 2007;65:1288–1299. doi: 10.1111/j.1365-2958.2007.05864.x. [DOI] [PubMed] [Google Scholar]
- 31.Ehrlich GD, Veeh R, Wang X, Costerton JW, Hayes JD, et al. Mucosal biofilm formation on middle-ear mucosa in the chinchilla model of otitis media. JAMA. 2002;287:1710–1715. doi: 10.1001/jama.287.13.1710. [DOI] [PubMed] [Google Scholar]
- 32.Leid JG, Willson CJ, Shirtliff ME, Hassett DJ, Parsek MR, et al. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J Immunol. 2005;175:7512–7518. doi: 10.4049/jimmunol.175.11.7512. [DOI] [PubMed] [Google Scholar]
- 33.Fisher MM, Triplett EW. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl Environ Microbiol. 1999;65:4630–4636. doi: 10.1128/aem.65.10.4630-4636.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leid JG, Kerr M, Selgado C, Johnson C, Moreno G, et al. Flagellar-mediated biofilm defense mechanisms of Pseudomonas aeruginosa against host derived lactoferrin. Infect Immun. 2009. [DOI] [PMC free article] [PubMed]
- 35.Morton DJ, Smith A, VanWagoner TM, Seale TW, Whitby PW, et al. Lipoprotein e (P4) of Haemophilus influenzae: role in heme utilization and pathogenesis. Microbes Infect. 2007;9:932–939. doi: 10.1016/j.micinf.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pracht D, Elm C, Gerber J, Bergmann S, Rohde M, et al. PavA of Streptococcus pneumoniae modulates adherence, invasion, and meningeal inflammation. Infect Immun. 2005;73:2680–2689. doi: 10.1128/IAI.73.5.2680-2689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pericone CD, Overweg K, Hermans PW, Weiser JN. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect Immun. 2000;68:3990–3997. doi: 10.1128/iai.68.7.3990-3997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettigrew MM, Gent JF, Revai K, Patel JA, Chonmaitree T. Microbial interactions during upper respiratory tract infections. Emerg Infect Dis. 2008;14:1584–1591. doi: 10.3201/eid1410.080119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prevention CfDCa. Pneumococcal disease Epidemiology and prevention of vaccine-preventable diseases; Prevention CfDCa, editor 2007.
- 40.van der Linden M, Reinert RR. Serotype distribution in pneumococcal acute otitis media with ruptured tympanic membrane or sepsis in Germany. Eur J Clin Microbiol Infect Dis. 2010;29:749–754. doi: 10.1007/s10096-010-0945-8. [DOI] [PubMed] [Google Scholar]
- 41.Johnson HL, Deloria-Knoll M, Levine OS, Stoszek SK, Freimanis Hance L, et al. Systematic evaluation of serotypes causing invasive pneumococcal disease among children under five: the pneumococcal global serotype project. PLoS Med. 2010;7 doi: 10.1371/journal.pmed.1000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirst RA, Gosai B, Rutman A, Guerin CJ, Nicotera P, et al. Streptococcus pneumoniae deficient in pneumolysin or autolysin has reduced virulence in meningitis. J Infect Dis. 2008;197:744–751. doi: 10.1086/527322. [DOI] [PubMed] [Google Scholar]
- 43.Spellerberg B, Cundell DR, Sandros J, Pearce BJ, Idanpaan-Heikkila I, et al. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol Microbiol. 1996;19:803–813. doi: 10.1046/j.1365-2958.1996.425954.x. [DOI] [PubMed] [Google Scholar]
- 44.Regev-Yochay G, Trzcinski K, Thompson CM, Malley R, Lipsitch M. Interference between Streptococcus pneumoniae and Staphylococcus aureus: In vitro hydrogen peroxide-mediated killing by Streptococcus pneumoniae. J Bacteriol. 2006;188:4996–5001. doi: 10.1128/JB.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Battig P, Muhlemann K. Influence of the spxB gene on competence in Streptococcus pneumoniae. J Bacteriol. 2008;190:1184–1189. doi: 10.1128/JB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allegrucci M, Hu FZ, Shen K, Hayes J, Ehrlich GD, et al. Phenotypic characterization of Streptococcus pneumoniae biofilm development. J Bacteriol. 2006;188:2325–2335. doi: 10.1128/JB.188.7.2325-2335.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bakaletz LO, Baker BD, Jurcisek JA, Harrison A, Novotny LA, et al. Demonstration of Type IV pilus expression and a twitching phenotype by Haemophilus influenzae. Infect Immun. 2005;73:1635–1643. doi: 10.1128/IAI.73.3.1635-1643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weimer KE, Armbruster CE, Juneau RA, Hong W, Pang B, et al. Coinfection with Haemophilus influenzae promotes pneumococcal biofilm formation during experimental otitis media and impedes the progression of pneumococcal disease. J Infect Dis. 2010;202:1068–1075. doi: 10.1086/656046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alverdy J, Holbrook C, Rocha F, Seiden L, Wu RL, et al. Gut-derived sepsis occurs when the right pathogen with the right virulence genes meets the right host: evidence for in vivo virulence expression in Pseudomonas aeruginosa. Ann Surg. 2000;232:480–489. doi: 10.1097/00000658-200010000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bishop JL, Finlay BB. Friend or foe? Antimicrobial peptides trigger pathogen virulence. Trends Mol Med. 2006;12:3–6. doi: 10.1016/j.molmed.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 51.Luo G, Niesel DW, Shaban RA, Grimm EA, Klimpel GR. Tumor necrosis factor alpha binding to bacteria: evidence for a high-affinity receptor and alteration of bacterial virulence properties. Infect Immun. 1993;61:830–835. doi: 10.1128/iai.61.3.830-835.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu LR, Zaborina O, Zaborin A, Chang EB, Musch M, et al. Surgical injury and metabolic stress enhance the virulence of the human opportunistic pathogen Pseudomonas aeruginosa. Surg Infect (Larchmt) 2005;6:185–195. doi: 10.1089/sur.2005.6.185. [DOI] [PubMed] [Google Scholar]
- 53.Schleimer RP, Kato A, Peters A, Conley D, Kim J, et al. Epithelium, inflammation, and immunity in the upper airways of humans: studies in chronic rhinosinusitis. Proc Am Thorac Soc. 2009;6:288–294. doi: 10.1513/pats.200808-088RM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tieu DD, Kern RC, Schleimer RP. Alterations in epithelial barrier function and host defense responses in chronic rhinosinusitis. J Allergy Clin Immunol. 2009;124:37–42. doi: 10.1016/j.jaci.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Psaltis AJ, Wormald PJ, Ha KR, Tan LW. Reduced levels of lactoferrin in biofilm-associated chronic rhinosinusitis. Laryngoscope. 2008;118:895–901. doi: 10.1097/MLG.0b013e31816381d4. [DOI] [PubMed] [Google Scholar]
- 56.Novotny LA, Adams LD, Kang DR, Wiet GJ, Cai X, et al. Epitope mapping immunodominant regions of the PilA protein of nontypeable Haemophilus influenzae (NTHI) to facilitate the design of two novel chimeric vaccine candidates. Vaccine. 2009;28:279–289. doi: 10.1016/j.vaccine.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kennedy BJ, Novotny LA, Jurcisek JA, Lobet Y, Bakaletz LO. Passive transfer of antiserum specific for immunogens derived from a nontypeable Haemophilus influenzae adhesin and lipoprotein D prevents otitis media after heterologous challenge. Infect Immun. 2000;68:2756–2765. doi: 10.1128/iai.68.5.2756-2765.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Novotny LA, Clements JD, Bakaletz LO. Transcutaneous immunization as preventative and therapeutic regimens to protect against experimental otitis media due to nontypeable Haemophilus influenzae. Mucosal Immunol. 2011;4:456–467. doi: 10.1038/mi.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abdeldaim GM, Stralin K, Kirsebom LA, Olcen P, Blomberg J, et al. Detection of Haemophilus influenzae in respiratory secretions from pneumonia patients by quantitative real-time polymerase chain reaction. Diagn Microbiol Infect Dis. 2009;64:366–373. doi: 10.1016/j.diagmicrobio.2009.03.030. [DOI] [PubMed] [Google Scholar]
- 60.Mason KM, Munson RS, Jr, Bakaletz LO. Nontypeable Haemophilus influenzae gene expression induced in vivo in a chinchilla model of otitis media. Infect Immun. 2003;71:3454–3462. doi: 10.1128/IAI.71.6.3454-3462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gupta R, Shah P, Swiatlo E. Differential gene expression in Streptococcus pneumoniae in response to various iron sources. Microb Pathog. 2009;47:101–109. doi: 10.1016/j.micpath.2009.05.003. [DOI] [PubMed] [Google Scholar]