Abstract
Acanthocephalans are attractive candidates as model organisms for studying the ecology and co-evolutionary history of parasitic life cycles in the marine ecosystem. Adding to earlier molecular analyses of this taxon, a total of 36 acanthocephalans belonging to the classes Archiacanthocephala (3 species), Eoacanthocephala (3 species), Palaeacanthocephala (29 species), Polyacanthocephala (1 species) and Rotifera as outgroup (3 species) were analyzed by using Bayesian Inference and Maximum Likelihood analyses of nuclear 18S rDNA sequence. This data set included three re-collected and six newly collected taxa, Bolbosoma vasculosum from Lepturacanthus savala, Filisoma rizalinum from Scatophagus argus, Rhadinorhynchus pristis from Gempylus serpens, R. lintoni from Selar crumenophthalmus, Serrasentis sagittifer from Johnius coitor, and Southwellina hispida from Epinephelus coioides, representing 5 new host and 3 new locality records. The resulting trees suggest a paraphyletic arrangement of the Echinorhynchida and Polymorphida inside the Palaeacanthocephala. This questions the placement of the genera Serrasentis and Gorgorhynchoides within the Echinorhynchida and not the Polymorphida, necessitating further insights into the systematic position of these taxa based on morphology.
Introduction
The endoparasitic phylum Acanthocephala Kohlreuther, 1771 consists of about 1,150 species, belonging to 125 genera [1] and 19 families [2]. They are characterized by an evertable proboscis as the attachment organ, sexual dimorphism, males with cement glands and an uterine bell in females. Unique is the syndermatic tegument, placing the acanthocephalans, also confirmed by molecular studies, sister to the Rotifera [3], [5]. Recent classifications distinguish the four classes Archiacanthocephala, Eoacanthocephala, Palaeacanthocephala and Polyacanthocephala [2], [6]–[10], with a majority of 62.7% of the species primarily infecting aquatic hosts [1]. Around 57% species of the Acanthocephala belong to the Palaeacanthocephala [1] with the two orders Echinorhynchida and Polymorphida. They show the highest species diversity and are the most common acanthocephalans of marine teleost fish.
Earliest molecular data of the Acanthocephala were based on a single acanthocephalan taxon used as an outgroup to estimate the phylogenetic position of the Chaetognatha amongst the Metazoa [11]. The first molecular phylogenetic analyses inside the Acanthocephala [12] confirmed the major taxonomic grouping of the traditional classifications. There, Palaeacanthocephala placed close to the Eoacanthocephala, with the Archiacanthocephala being the most basal taxon. The bird parasitic Archiacanthocephala and Eoacanthocephala (parasites of fish, amphibians and reptiles) appeared on different branches on the resulting rDNA tree [13], [14], indicating independent evolution. Furthermore, the phylogenetic analyses suggested very complex evolutionary and taxonomic relationships among the species [12]. With their relatively small number of species, a conserved two-host (arthropod–vertebrate) life cycle, and corroborated phylogenetic relationships to a free-living sister group (the Rotifera), the acanthocephalans are attractive candidates as model organisms for studying the ecology and co-evolutionary history of parasitic life cycles in marine ecosystem. However, with many genera having only a single representative, few researchers collected specimens for molecular studies. With poor representation especially of marine taxa, the phylogenetic relationships within this interesting phylum are far from getting resolved.
Most previous analyses of acanthocephalan phylogenetic relationships have been based exclusively on nuclear small subunit (SSU) ribosomal DNA (rDNA). This highly conserved region is best suited for an analysis of the upper level phylogeny. García-Valera and Nadler [4], [9] analyzed a total of 21 acanthocephalan species, including 3 Archiacanthocephala, 2 Eoacanthocephala, 15 Palaeacanthocephala and 1 Polyacanthocephala. The purpose of the present study was to add new sequence data especially of marine fish parasitic taxa, providing a better resolution inside the Palaeacanthocephala. This is a prerequisite for a better understanding of this taxon, also enabling a better taxonomic placement and morphological identification of the species within this group. Marine acanthocephalans from different sources were collected, morphologically identified, and analyzed for the nearly complete 18S rDNA. Five of these species have not been included in molecular phylogenetic analyses before (Bolbosoma vasculosum, Filisoma rizalinum, Rhadinorhynchus prists, R. lintoni and Serrasentis sagittifer). The available sequence data of 29 Palaeacanthocephala, 3 Eoacanthocephala, 3 Archiacanthocephala, a single Polyacanthocephala, and three from Rotifera as outgroup were analyzed by Bayesian Inference and Maximum Likelihood. Implications for the phylogeny of the marine acanthocephalans are discussed.
Results
Species identification and data set
All collected acanthocephalans (Table 1) were identified to species level by using morphological characters and existing keys [2], [7], [15]–[19], [28]–[30]. Of the resulting host-parasite combinations, Filisoma rizalinum and Rhadinorhynchus lintoni are new host and locality records. We have sequenced nearly the complete 18S rRNA gene, using cloning techniques to obtain strong sequencing signals for the entire gene (Figure 1). Identical sequences that represent different host or geographic isolates of a particular species were only included once in the phylogenetic analyses. They, however, provide molecular information on the host specificity and zoogeography of the studied acanthocephalan species. The SSU rDNA sequences were newly generated for 13 taxa and added to the published data set (GenBank). Analyses of this dataset (excluding sites containing gaps) of 40 taxa in Bayesian Inference had considerable similarity to the Maximum Likelihood tree. The SSU sequence length in the constructed alignment ranged from 1,649 (Plagiorhynchus cylindraceus) to 2,090 (Polyacanthorhynchus caballeroi) bp (Table 2). Nucleotide frequencies were 0.2544 (A), 0.1965 (C), 0.2657 (G) and 0.2834 (T). The proportion of invariable sites equaled 0.1605 and the distribution of gamma shape parameter (Gd) was 0.5669 (Table 3).
Table 1. Newly collected acanthocephalans.
| Species | Host | Source |
| Bolbosoma vasculosum | Lepturacanthus savala | Java, Indonesia |
| Pomphorhynchus laevis | Platichthys flesus | Baltic Sea |
| Pomphorhynchus laevis | Rutilus rutilus | Lippe River, NRW, Germany |
| Echinorhynchus gadi | Gadus morhua | Baltic Sea |
| Echinorhynchus gadi | Macrourus berglax | Irminger Sea, Greenland |
| Echinorhynchus gadi | Platichthys flesus | Baltic Sea |
| Filisoma rizalinum | Scatophagus argus | Java, Indonesia |
| Rhadinorhynchus prists | Gempylus serpens | Java, Indonesia |
| Rhadinorhynchus lintoni | Selar crumenophthalmus | Oahu, Hawaii |
| Serrasentis sagittifer | Johnius coitor | Java, Indonesia |
| Southwellina hispida | Epinephelus coioides | Java, Indonesia |
Some species with identical sequence data have been collected from different hosts.
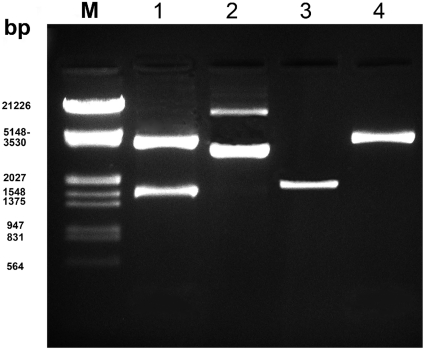
Figure 1. Electrophoretic analysis of restriction mapping.
Lane 1 shows the pCR®2.1-TOPO® vector (3923 bp) above, which is cleaved by EcoRI (Invitrogen, Karlsruhe) and below the amplified 18S rRNA gene fragment (1724 bp) with the correct orientation (lane 2 with the incorrect orientation). The controls show the amplified PCR product (lane 3) and vector without insert (lane 4). M: marker λ-EcoRI +HindIII-Marker-Mix 3 (Fermentas, St. Leon-Rot) 1 µg.
Table 2. Acanthocephala and Rotifera specimen information and GenBank accession numbers.
| Species | Family | Host | 18S-rDNA | Length bp | Aligned |
| Acanthocephaloides propinquus | Arythmacanthidae | Gobius bucchichii | AY830149 | 1727 | 1657 |
| Acanthocephalus dirus | Echinorhynchidae | Asselus aquaticus | AY830151 | 1724 | 1654 |
| Acanthocephalus lucii | Echinorhynchidae | Perca fluviatilis | AY830152 | 1725 | 1655 |
| Arythmorhynchus brevis | Polymorphidae | Nycticorax nycticorax | AF064812 | 1784 | 1694 |
| Bolbosoma vasculosum | Polymorphidae | Lepturacanthus savala | this study | 1739 | 1653 |
| Corynosoma enhydri | Polymorphidae | Enhydra lutris | AF001837 | 1747 | 1651 |
| Corynosoma magdaleni | Polymorphidae | Phoca hispida botnica | EU267803 | 1722 | 1653 |
| Echinorhynchus gadi | Echinorhynchidae | Macrourus berglax | this study | 1745 | 1659 |
| Echinorhynchus truttae | Echinorhynchidae | Thymallus thymallus | AY830156 | 1729 | 1659 |
| Filisoma bucerium | Cavisomidae | Kyphosus elegans | AF064814 | 1744 | 1655 |
| Filisoma rizalinum | Neoechinorhynchidae | Scatophagus argus | this study | 1741 | 1652 |
| Floridosentis mugilis | Neoechinorhynchidae | Mugil cephalus | AF064811 | 1760 | 1668 |
| Gorgorhynchoides bullocki | Rhadinorhynchidae | Eugerres plumieri | AY830154 | 1720 | 1651 |
| Koronacantha mexicana | Illiosentidae | Pomadasys leuciscus | AY830157 | 1688 | 1665 |
| Koronacantha pectinaria | Illiosentidae | Microlepidotus brevipinnis | AF092433 | 1761 | 1673 |
| Leptorhynchoides thecatus | Rhadinorhynchidae | Lepomis cyanallus | AF001840 | 1758 | 1663 |
| Macracanthorhynchus ingens | Oligacanthorhynchidae | Procyon lotor | AF001844 | 1765 | 1669 |
| Moniliformis moniliformis | Moniliformidae | Rattus rattus | Z19562 | 1769 | 1668 |
| Neoechinorhynchus crassus | Neoechinorhynchidae | Catostomus commersoni | AF001842 | 1773 | 1677 |
| Neoechinorhynchus saginata | Neoechinorhynchidae | not applicable | AY830150 | 1745 | 1675 |
| Oligacanthorhynchus tortuosa | Oligacanthorhynchidae | Didelphis virginiana | AF064817 | 1767 | 1671 |
| Plagiorhynchus cylindraceus | Plagiorhynchidae | Armadillidium vulgare | AF001839 | 1745 | 1649 |
| Polyacanthorhynchus caballeroi | Polyacanthorhynchidae | Caiman yacare | AF388660 | 2176 | 2090 |
| Polymorphus altmani | Polymorphidae | Enhydra lutris | AF001838 | 1745 | 1649 |
| Polymorphus minutus | Polymorphidae | Gammarus pulex | EU267806 | 1720 | 1651 |
| Pomphorhynchus laevis | Pomphorhynchidae | Rutilus rutilus | this study | 1742 | 1656 |
| Pomphorhynchus tereticollis | Pomphorhynchidae | Gammarus pulex | AY423347 | 1662 | 1656 |
| Pseudocorynosoma anatrium | Polymorphidae | Bucephala albeola | EU267801 | 1723 | 1654 |
| Pseudocorynosoma constrictum | Polymorphidae | Anas clypeata | EU267800 | 1723 | 1654 |
| Pseudoleptorhynchoides lamothei | Rhadinorhynchidae | Ariopsis guatemalensis | EU090950 | 1748 | 1663 |
| Rhadinorhynchus lintoni | Rhadinorhynchidae | Selar crumenophthalmus | this study | 1740 | 1653 |
| Rhadinorhynchus pristis | Rhadinorhynchidae | Gempylus serpens | this study | 1744 | 1656 |
| Serrasentis sagittifer | Rhadinorhynchidae | Platycephalus arenarius | this study | 1741 | 1654 |
| Southwellina hispida | Polymorphidae | Tigrisoma mexicanum | EU267807 | 1730 | 1661 |
| Southwellina hispida | Polymorphidae | Epinephelus coioides | this study | 1747 | 1661 |
| Transvena annulospinosa | Transvenidae | Anampses neoguinaicus | AY830153 | 1693 | 1656 |
| Rotifera | |||||
| Asplanchna sieboldi | Asplanchnidae | Free-living | AF092434 | 1728 | 1663 |
| Brachionus patulus | Branchionidae | Free-living | AF154568 | 1745 | 1656 |
| Lecane bulla | Lecanidae | Free-living | AF154566 | 1733 | 1668 |
Table 3. Tree statistics for rDNA data set.
| Total characters | Uninformative-characters | Constant characters | Informative characters | CI | Tree length | -ln likelihood | Pinv | Gd | |
| ML | 2191 | 259 | 1224 | 708 | 0.547 | 2.866 | 16191.7480 | 0.1605 | 0.5669 |
Numbers of informative characters, consistency index (CI) and tree length refer to parsimony inference. Proportion of invariable sites (Pinv), shape of gamma distribution (Gd) and –ln Likelihood refer to Maximum Likelihood Inference.
Phylogenetic analyses
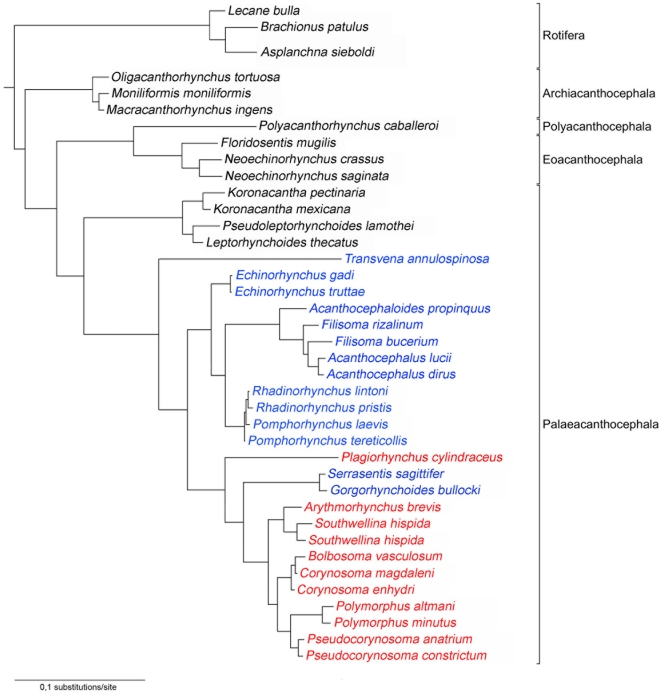
Bayesian Inference analysis yielded a single tree (Figure 2) with the same general topology as the ML result. Using this model the respective clades received high support in the ML bootstrap analysis. The topology of the BI tree depicts paraphyly of the Palaeacanthocephala.
Figure 2. Bayesian consensus phylogram for Acanthocephala relationship based on the SSU rDNA data set.
Rotifera is used as outgroup, acanthocephalans are classified as indicated on the right site of the graphic. This tree illustrates the hypothesis that the order Echinorhynchida (blue) and Polymorphida (red) have a paraphyletic arrangement. The branch length scale is the number of substitutions per site.
Maximum-Likelihood analysis yielded a single best tree with a likelihood score of 16191.7480, a consistency index (CI) of 0.547 and a length of 2,866 steps (Table 2). The -ln likelihood score for the first alternative topology was 16182.22431. Based on the results the Kishino-Hasegawa (KH) [18] and Shimodaira-Hasegawa (SH) [19] tests were implemented in PAUP* using full optimization and 100 bootstrap replicates. Bootstrap values (<50%) are given on equivalent branches of the ML tree. The phylogenetic tree of the phylum Acanthocephala (Figure 2) is subdivided into four classes and the Rotifera as outgroup. The tree begins with the Archiacanthocephala as the earliest divergent clade, followed by the Polyacanthocephala and the Eoacanthocephala as sistertaxa, and the Palaeacanthocephala as the most derived clade. The Palaeacanthocephala show the highest diversity inside the class, presenting the orders Echinorhynchida and Polymorphida in a paraphyletic arrangement. All analyses support the current hypothesis separating four classes [((Eoacanthocephala, Polyacanthocephala) Palaeacanthocephala) (Archiacanthocephala), (Rotifera)], by Maximum-Likelihood trees and Bayesian Inference.
Defining morphological characters of the Archiacanthocephala are proboscis hooks in spirals, a single ligament sac in the females, and 8 cement glands in the males. The second clade consists of the Polyacanthocephala sister to the Eoacanthocephala. The Polyacanthocephala with the single genus Polyacanthorhynchus have 2 distinct ligament sacs in the females, and 2 elongate pyriform to tubular cement glands with giant nuclei in the males. The Eoacanthocephala with the representative Neoechinorhynchus are characterized by 2 ligament sacs in the females and a single cement gland in the males. The Palaeacanthocephala separate into the order Echinorhynchida as the original and the Polymorphida as the more derived taxon. The Echinorhynchida have an aspinosed trunk and a short neck. The cement glands of the males are divided into 2 or more compact or tubular lobes, and the females have eggs with polar prolongations of the middle shell. The final hosts are marine or aquatic fishes. The earliest divergent clade of the Echinorhynchida includes Koronacantha, Pseudoleptorhynchoides and Leptorhynchoides, which belong to the families Illosentidae and Rhadinorhynchidae. Koronacantha has an elongate proboscis with a heavy cuticular coating, cuticular body spines, genital spines are present in both sexes, the males have 8 cement glands, and the heavy, strongly recurved hooks in the shape of an inverted apostrophe with roots that are simple but exaggerated in size with a small hook. Pseudoleptorhynchoides and Leptorhynchoides have both, a cylindrical aspinose trunk, a cylindrical and elongated proboscis, and the males have 8 tubular cement glands. The next echinorhynchid taxon, Transvena annulospinosa, appears separate from the other 2 major clades. Transvena can be distinguished from all other Acanthocephala genera by having a combination of a single ring of small spines on its trunk near or at the junction between the neck and the trunk, and hooks which decrease in length from the apex to the base of the proboscis. The males have 2 pyriform or tubular cement glands. The next echinorhynchid clades lacks the 2 genera Serrasentis and Gorgorhynchoides (members of the echinorhynchids based on traditional classifications) (cp. Figures 3C,D), which appear in the polymorphid clade (Figure 2). Echinorhynchus is separated from the genera Acanthocephaloides, Acanthocephalus, and Filisoma, that form a sister group to Rhadinorhynchus and Pomphorhynchus. All these acanthocephalans are characterized by a slender cylindrical proboscis with many alternating longitudinal rows of homeomorphous hooks, the lack of surface hooks, and 4–6 cement glands in the males. The proboscis of Rhadinorhynchus shows a basal hook annulus, which is rudimentary in Pomphorhynchus and is absent in all other genera (Figures 3A,B,E). They are mainly fish parasites in the aquatic environment, including the common and widely distributed marine genus Rhadinorhynchus. The second clade of the Palaeacanthocephala consists of the Polymorphida, including the two echinorhynchid genera Serrasentis and Gorgorhynchoides. The most basal genus is the polymorphid Plagiorhynchus cylindraceus followed by a clade with the two echinorhynchids Serrasentis sagittifer and Gorgorhynchoides bullocki. The genus Plagiorhynchus can be distinguished from the remaining clade by the cylindrical fusiform aspinose trunk, slender lemnisci, and 6 elongate reniform or tubular cement glands in the males. The order Polymorphida is commonly characterized by the possession of alternating rows of hooks on the fusiform to globular proboscis, a mainly spinose trunk, surface hooks arranged in patterns, predominantly 2–4 cement glands in the males, and the final host specificity (adults in birds and mammals, juveniles in fishes, amphibians, and reptiles). The echinorhynchid genera Serrasentis and Gorgorhynchoides appear sister to the most derived monophyletic clade within the Palaeacanthocephala, within the polymorphids (Figure 2). According to morphology they demonstrate some polymorphid morphological characters, such as the spinose trunk and the rather globular, short calviform proboscis with longitudinal rows of variable numbers of hooks. While in Gorgorhynchoides the presence of trunk spines is limited to the anterior portion, Serrasentis has a trunk with unique ventral transverse rows of spines which are fused to form a comb-like structure (Figures 3C,D). The males have 6 clubbed cement glands (G. bullocki), and 4 elongate pyriform cement glands (S. sagittifer), which leads to the assignment into the Echinorhynchida based on morphology. Both genera occur mainly in fishes, rarely in amphibians, and in reptiles. The most derived genera within the present phylogenetic analyses belong to the Polymorphida, with the genera Arhythmorhynchus and Southwellina sister to Polymorphus, Pseudocorynosoma, Bolbosoma, and Corynosoma. While Arhythmorhynchus is characterized by an extremely long slender, anterior swollen trunk covered with a single field of spines, an usually enlarged cylindrical proboscis with greatly enlarged ventral hooks in the middle, and 2 (or 4) cement glands in the males, the genus Southwellina has a short trunk with spines that are arranged in 2 fields, and 4 tubular cement glands. Both parasitize birds as final hosts. Bolbosoma and Corynosoma are characterized by a small to medium sized body with a clubbed trunk, anteriorly swollen and armed with numerous regularly arranged spines. Bolbosoma is formed in the shape of a bulb, and is armed with spines that form 2 complete rings (see Figure 3F). The proboscis is calviform or conical, followed by a short neck, and the males have 2 tubular long cement glands. The trunk of Corynosoma is flattened on one side and forms a fore and a hind trunk. The spines are arranged within a single field, the proboscis is cylindrical, also followed by a short neck, and males have 6 pyriform or rarely tubular cement glands. Both genera use amphipods as intermediate, fishes as paratenic, and marine mammals as final hosts. Polymorphus and Pseudocorynosoma both show a spindle-shaped body armed with spines that are arranged in a single field, and a cylindrical or ovoid proboscis. Polymorphus has a small anterior spinose trunk, a cylindrical proboscis increasing in size proximally, a distinct neck region, and 4 tubular cement glands in the males. They prefer aquatic or semi aquatic birds, occasionally mammals, as final hosts. Pseudocorynosoma has a spindle-shaped body with a slight constriction, separating the fore and the hind trunk. Numerous spines that cover the most anterior part of the fore trunk are symmetrically distributed on the ventral and dorsal sides. In addition, a single field of spines is surrounding the genital pore. The proboscis has a slightly swollen region, followed by a truncated cone-shaped neck which is longer than wide [20], [22]. The males show 4 or 6 tubular cement glands. Pseudocorynosoma is using waterfowls as definitive hosts and amphipods as intermediate hosts.
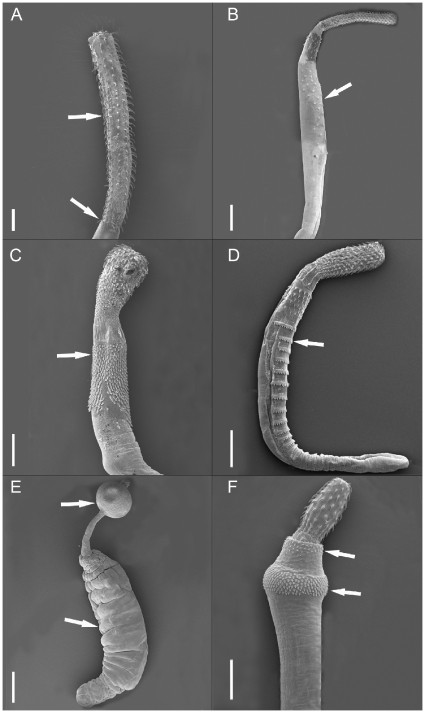
Figure 3. SEM (scanning electron microscope) micrographs of Palaeacanthocephala.
(A) Proboscis of male Rhadinorhynchus pristis from Gempylus serpens (Indonesia, Indian Ocean) armed with regular hooks a and basal hook annulus. (B) Praesoma of female R. lintoni from Selar crumenophthalmus (Hawaii, Pacific) with irregular arrangement of trunk hooks. (C) Praesoma of Gorgorhynchoides golvani from Platycephalus arenarius (Indonesia, Indian Ocean) regular arrangement of surface hooks. (D) Habitus of Serrasentis sagittifer from Platycephalus arenarius (Indonesia, Indian Ocean) with hooks are transformed into strong plates arranged as combs. (E) Habitus of Pomphorhynchus laevis from Platichthys flesus (Baltic Sea) shows any trunk hooks on bulb, neck and trunk. (F) Praesoma of Bolbosoma vasculosum from Lepturacanthus savala (Indonesia, Indian Ocean) formed in the shape of a bulb, and armed with regular hooks which are arranged in two rings. Scale bars: A 400 µm, B, D, F, 200 µm, E 100 µm.
Discussion
The present study is the most detailed phylogenetic analyses of the Acanthocephala so far based on SSU rDNA, especially of the class Palaeacanthocephala. Earlier studies of acanthocephalans combining data sets of both, SSU and LSU (large subunit, already demonstrated similar results to the SSU alone [9]. Our data set adds to the most recent analyses of acanthocephalan relationships by Garey et al. [12] and García-Varela and Nadler [9]. We can support the notion that the acanthocephalans are monophyletic in origin, and separate into four distinct classes [2], [8], [9]. The Archiacanthocephala (Figure 2), parasites of birds and terrestrial vertebrates, are the earliest divergent lineage of acanthocephalans which utilize terrestrial vertebrates as intermediate hosts. More derived follow the Polyacanthocephalans as parasites of fishes and crocodiles, sister to the Eoacanthocephalans (in fish, amphibians and reptiles) from the aquatic environment. This result is consistent with the hypothesis that the Polyacanthocephala represent a different class within the phylum Acanthocephala. The more derived Palaeacanthocephala, including the Echinorhynchida and Polymorphida, are arranged in a paraphyletic assemblage. Both orders demonstrate high morphological diversity, which may explain why traditional identification keys have distinguished among the taxa according to their final hosts [1], [2]. The order Echinorhynchida infects teleost fishes, occasionally amphibians and reptiles whereas the Polymorphida include parasites of reptiles (rarely), birds, and marine mammals. The Echinorhynchida so far separate into 10 families and 339 valid species. The Polymorphida include only three families and a total of 255 valid species (Centrorhynchidae with two genera and 75 species; Plagiorhynchidae with 3 subfamilies and 8 genera and 53 species; Polymorphidae with 9 genera and 127 species). Consequently, these species rich taxa include 83 genera and 594 species of Acanthocephalans, mainly from the aquatic environment (Integrated Taxonomic Information System).
Herlyn et al. [14] for the first time described paraphyly within the Palaeacanthocephala, indicating independent evolution within these widely distributed taxa. Similarly, molecular and morphological studies so far indicated that the family Rhadinorhynchidae is paraphyletic or polyphyletic, and that the genera should be reexamined and reclassified by using morphological, ecological, and molecular characters [9], [21], [22], in agreement with the cladistic studies by García-Varela and Nadler [9] and Herlyn et al. [23]. The present analyses place the two species Serrasentis sagittifer (Rhadinorhynchidae) and Gorgorhynchoides bullocki (Rhadinorhynchidae), both Echinorhynchida, into the Polymorphida. Neither species demonstrates any morphological similarity. Conspicuous are the trunk hooks of Serrasentis that are arranged within rows (comb-like), and the presence of four cement glands in the males. Gorgorhynchoides has trunk hooks on its praesoma and six cement gland in the males (Gorgorhynchoides golvani from Platycephalus arenarius, Indonesia, Indian Ocean, see Figure 3). Most interesting is the position of the polymorphid Plagoirhynchus cyndraecus, which is arranged between the Echinorhynchida and Polymorphida. This species uses birds as final hosts. The cylindrical trunk also has anterior hooks around a small bulb, and the males have also six cement glands. According to traditional classifications, this result questions the relationship of Serrasentis and Gorgorhynchoides to the other echinorhynchids. While only some echinorhynchid acanthocephalans have mainly irregularly arranged surface hooks on the trunk, the herewith recognized character of regularly arranged hooks on the trunk is one of the most common features within the polymorphids.
Recent morphological assessment led to incongruent conclusions, due to difficulties in finding morphological characters that distinguish taxa, and to the partly subjective character states that often lack homologies with the outgroup [21]. According to García-Valera and Nadler [9], many families have been diagnosed based on character combinations rather than shared derived features. For several species, only a single record exists, caused by difficulties in sampling especially from the marine environment and in confirming the life cycles experimentally [1]. Most previous molecular approaches include too few acanthocephalan sequences, owed to difficult and/or biased sampling, to allow more detailed conclusions on the phyletic status of the acanthocephalan subclades [12], [14], [24], [25]. Nevertheless, with their relatively small number of species, a conserved two-host (arthropod–vertebrate) life cycle that involves paratenic hosts in the most derived clade, and the phylogenetic relationship to a free-living sister group, acanthocephalans are attractive candidates as model organisms for studying host-parasite co-evolution. For example, the species distribution within the host illustrates that fish and birds are the most widely used definitive hosts, followed by mammals. It is, however, interesting to note that the oldest group of vertebrates, the fish, is not utilized by significantly more species than the youngest groups, the birds and mammals [1], indicating expansive adaptive radiation in these newly explored host groups.
We are aware that the presented molecular phylogeny of the Acanthocephala is not yet comprehensive, and needs to be tested and validated by future studies. This requires further taxon sampling and ideally the inclusion of additional molecular markers. However, our data also demonstrate the preliminary nature of the acanthocephalan classification in general, especially of the derived echinorhynchids, the most common acanthocephalans in fish. We suggest that the current state of knowledge warrants the identification of further morphological characters for a better understanding of the acanthocephalan diversity, perhaps best driven by more in-depth molecular phylogenetic studies. This will enable the mapping of more morphological characters onto the molecular trees, and redefining the higher level classification of the Acanthocephala.
Acanthocephalans are attractive candidates as model organisms for studying the ecology and co-evolutionary history of parasitic life cycles in the marine ecosystem. However, the lack of phylogenetic studies and taxonomic identification of especially marine Acanthocephala prevents detailed comparison to other endoparasites. We do hope that our study will iniciate future research on the species composition, zoogeography and evolution of the phylum Acanthocephala, allowing comparisons to be made on the ecology of this taxon and other species groups such as the nematodes and cestodes that have diversified under similar conditions.
Materials and Methods
Ethics statement
An approval by a review board institution or ethics committee was not necessary, because all the fish in the current study were obtained in different locations from fishermen selling fresh fish for consumption or were collected during regularly fishery cruises.
Collection of specimens
Acanthocephalan specimens were collected between 2001 and 2008 from their naturally infected vertebrate hosts (Table 1). The isolated parasites were washed in saline solution before fixation in 70% ethanol or absolute ethanol for molecular studies. The metasoma was used for molecular rDNA analyses, while the praesoma was processed for scanning electron microscopy (SEM). In other cases, the praesoma was stained in Mayer's acetic carmine, mounted in Canada balsam and identified using the common keys and original papers [26]–[28]. Molecular vouchers or voucher specimens were deposited in The Natural History Museum Berlin. A list of taxa, their place of origin and deposition numbers is given in Table 1.
Nucleic acid isolation, polymerase chain reaction and sequencing
Genomic DNA was extracted from individual specimens using a commercial extraction kit (Peqlab, Erlangen). The region of nuclear rDNA was amplified using polymerase chain reaction (PCR). Nearly complete SSU rDNA (∼1.800 bp) regions were amplified after Garey et al. [12] (94°C 4-min initial denaturing followed by 30 cycles: 94°C 30 s, 60°C 30 s, 72°C 90 s) using primers corresponding to conserved regions at the extreme ends of the 18S rRNA gene (5′-AGATTAAGCCATGCATGCGTAAG-3′ and 5′-TGATCCTTCTGCAGGTTCACCTAC-3′), cloned into pCR®2.1-TOPO® vector (Invitrogen, Karlsruhe) and used to transform competent Escherichia coli (TOP 10, Invitrogen, Karlsruhe). Positive clones were identified by blue/white selection, and target inserts of white colonies were confirmed by PCR of bacterial DNA extracts. Liquid cultures for minipreps were grown in Luria broth containing 50 µg/µl of ampicillin following plasmid purification on the next day (MBI Fermentas, St. Leon-Rot). Orientation of cloned inserts was controlled by restriction mapping using in 1% agarose gel (Figure 1). Both strands of the 18S rDNA were sequenced completely in both directions after Sanger et al. [29] by Seqlab (Göttingen) using M13 universal primers (forward (−20): 5′ -GTAAAACGACGGCCAG-3′, reverse: 5′ -CAGGAAACAGCTATGAC-3′) of Invitrogen. Site polymorphisms were recorded only when both alternative nucleotide peaks were present in all sequence reactions representing both DNA strands. The sequences have been deposited in GenBank as given in Table 2.
Alignment and phylogenetic analyses of sequence data
Sequences of the 18S rRNA gene of 11 sampled host-parasite combinations (Table 1) were aligned together with those from GenBank (Table 2), and included a total of 3 outgroup (Rotifera, belonging to the two major classes) and 36 ingroup (Acanthocephala) taxa (Table 2), representing the classes Archiacanthocephala (with three of four orders: Moniliformida, Gigantorhynchida and Oligacanthorhynchida), Eoacanthocephala (with one of two orders: Neoechinorhynchida) and Palaeacanthocephala (with two of two orders: Echinorhynchida and Polymorphida). The sequences were initially aligned using Clustal_X [30] and adjusted by eye. Based on these 40 sequences alignment had 2190 characters, 1902 were parsimony-informative. The complete alignment is available from the corresponding author upon request.
Phylogenetic trees were constructed using Bayesian Inference (BI) conducted with MrBayes v3.1.2 [31] and Maximum Likelihood (ML) with PAUP* v4.0b10 [32]. For BI, likelihood settings were set to nst = 6, rates = gamma, the nucleotide substitution model of evolution was the general time reversible (GTR) model [33], with invariable sites (+I) and rate heterogeneity (+G) [34] suggested as the best fitting model by Modeltest version 3.8 [35] based on Akaike Information Criterion (AIC). Four chains (one cold, three heated temp = 0.2) were run for 1,000,000 generations and sampled every 100 generations, whereas the 40,000 generations were discarded as ‘burnin’. For the calculated consensus tree a value of 0.95% and higher was considered having good statistical support.
For ML analyses, the same model parameters were used and heuristic searches were preset by nearest-neighbor-interchange (NNI), branch swapping was performed until the topology remained unchanged. Bootstrapping with 100 replicates was performed and the results were plotted onto the best known likelihood tree. Based on dataset BI analyses phylogenetic tree were reconstructed by TreeGraph [36] (Figure 2).
Acknowledgments
We thank Sonja Kleinertz, Anika Rohde and Stefan Theisen for providing samples. Two anonymous referees helped to significantly improve the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The present study was financially supported by BiK-F (LOEWE) and by the German Research Council (DFG PA 664/4-2, 6-1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kennedy CR. Ecology of the Acanthocephala. 2006. pp. 1–240. Cambridge University Press New York.
- 2.Amin OM. Key to the families and subfamilies of Acanthocephala, with erection of a new class (Polyacanthocephala) and a new order (Polyacanthorhynchida). J Parasitol. 1987;73:1216–1219. [PubMed] [Google Scholar]
- 3.Zrzavý J. Gastrotricha and metazoan phylogeny. Zool Scr. 2002;32:61–81. [Google Scholar]
- 4.García-Varela M, Nadler S. Phylogenetic relationships among Syndermata inferred from nuclear and mitochondrial gene sequences. Mol Phyl Evol. 2006;40:61–72. doi: 10.1016/j.ympev.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Witek A, Herlyn H, Meyer A, Boell L, Bucher G, et al. EST based phylogenomics of Syndermata questions monophyly of Eurotatoria. BMC Evol Biol. 2008;345:1–11. doi: 10.1186/1471-2148-8-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bullock WL. Morphological features as tool and pitfall in acanthocephalan systematics. In: Schmidt GD, editor. Problems in systematic of parasites. University Park Press Baltimore; 1969. pp. 9–45. [Google Scholar]
- 7.Amin OM. Crompton DWT, Nickol BB, editors. Classification. Biology of the Acanthocephala. 1985. pp. 27–72. Cambridge University Press, London.
- 8.García-Varela M, Cummings MP, Perez-Ponce de León G, Gardner SL, Lacletteam JP. Phylogenetic analysis based on 18S ribosomal RNA gene sequences supports the existence of class Polyacanthocephala (Acanthocephala). Mol Phyl Evol. 2002;23:288–292. doi: 10.1016/S1055-7903(02)00020-9. [DOI] [PubMed] [Google Scholar]
- 9.García-Varela M, Nadler S. Phylogenetic relationships of Palaeacanthocephala (Acanthocephala) inferred from SSU and LSU rDNA gene sequences. J Parasitol. 2005;91:1401–1409. doi: 10.1645/GE-523R.1. [DOI] [PubMed] [Google Scholar]
- 10.Taraschewski H. Rhode K, editor. Acanthocephala (thorny or spiny-headed worms). Marine Parasitology. 2005. pp. 116–121. CSIRO Publishing, Collingwood.
- 11.Telford MJ, Holland PW. The phylogenetic affinities of the chaetognaths: A molecular analysis. Mol Biol Evol. 1993;10:660–676. doi: 10.1093/oxfordjournals.molbev.a040030. [DOI] [PubMed] [Google Scholar]
- 12.Garey JR, Near TJ, Nonnemacher MR, Nadler SA. Molecular evidence for Acanthocephala as a subtaxon Rotifera. Mol Phyl Evol. 1996;43:287–292. doi: 10.1007/BF02338837. [DOI] [PubMed] [Google Scholar]
- 13.Near TJ, Garey JR, Nadler SA. Phylogenetic relationships of the acanthocephalan inferred from 18S ribosomal DNA sequences. Mol Phyl Evol. 1998;10:287–298. doi: 10.1006/mpev.1998.0569. [DOI] [PubMed] [Google Scholar]
- 14.Herlyn H, Piskurek O, Schmitz J, Ehlers U, Zischlera H. The syndermatan phylogeny and the evolution of acanthocephalan endoparasitism as inferred from 18S rDNA sequences. Mol Phyl Evol. 2003;26:155–164. doi: 10.1016/s1055-7903(02)00309-3. [DOI] [PubMed] [Google Scholar]
- 15.Wayland MT, Gibson DI, Sommerville C. Echinorhynchus salmonis Müller, 1784 (Acanthocephala: Echinorhynchidae) from the Bothnian Bay, Baltic Sea: morphological variability and radial asymmetry of proboscis hooks. Syst Parasitol. 2004;58:149–158. doi: 10.1023/b:sypa.0000029419.07989.1a. [DOI] [PubMed] [Google Scholar]
- 16.Petrochenko VI. Acanthocephala of domestic and wild animals. 1971. pp. 1–478. Academy of Sciences of the USSR. Moscow.
- 17.Amin OM, Nahhas FM. Acanthocephala of marine fishes off Fiji Islands, with descriptions of Filisoma longcementglandatus n.sp., Neorhadinorhynchus macrospinosus n.sp. (Cavisomidae), and gravid females of Rhadinorhynchus johnstoni (Rhadinorhynchidae); and keys to species of the genera Filisoma and Neorhadinorhynchus. J Parasitol. 1994;80:768–774. [PubMed] [Google Scholar]
- 18.Kishino H, Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. Mol Phyl Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 19.Shimodaira H, Hasegawa M. Multiple comparison of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- 20.Aznar FJ, Perez-Ponce de León G, Raga JA. Status of Corynosoma (Acanthocephala: Polymorphidae) based on anatomical, ecological, and phylogenetic evidence, with the erection of Pseudocorynosoma n. gen. J Parasitol. 2006;92:548–64. doi: 10.1645/GE-715R.1. [DOI] [PubMed] [Google Scholar]
- 21.Monks S. Phylogeny of the Acanthocephala based on morphological characters. Syst Parasitol. 2001;48:81–115. doi: 10.1023/a:1006400207434. [DOI] [PubMed] [Google Scholar]
- 22.García-Varela M, González-Oliver A. The systematic position of Leptorhynchoides (Kostylew, 1924) and Pseudoleptorhynchoides (Salgado-Maldonado, 1976), inferred from nuclear and mitochondrial DNA gene sequences. J Parasitol. 2008;94:959–962. doi: 10.1645/GE-1420.1. [DOI] [PubMed] [Google Scholar]
- 23.Herlyn H, Martini N, Ehlers U. Organisation of the praesoma of Paratenuisentis ambiguus (Van Cleave, 1921) (Acanthocephala: Eoacanthocephala), with special reference to the lateral sense organs and musculature. Syst Parasitol. 2001;50:105–116. doi: 10.1023/a:1011925516086. [DOI] [PubMed] [Google Scholar]
- 24.Giribet G, Distel DL, Polz M, Sterrer W, Wheeler W. Triploblastic relationships with emphasis on the acoelomates and the position of Gnathostomulida, Cycliophora, Plathelminthes, and Chaetognatha: a combined approach of 18S rDNA sequences and morphology. Syst Biol. 2000;49:539–562. doi: 10.1080/10635159950127385. [DOI] [PubMed] [Google Scholar]
- 25.Welch BD. Bayesian and maximum likelihood analyses of rotifer-acanthocephalan relationships. Hydrobiol. 2005;546:47–54. [Google Scholar]
- 26.Yamaguti S. Systema Helminthum, Volume V: Acanthocephala. 1963. pp. 1–423. Interscience Publishers John Wiley and Sons, New York.
- 27.Golvan YJ. Systematique des acanthocephales (Acanthocephala, Rudolphi 1801). 1969. pp. 1–373. L'ordre des Palaeacanthocephala Meyer 1931. La superfamille des Echinorhynchoidea (Cobbold 1876) Golvan et Houin, 1963. Série A, Zoologie 57. Mémoires du Muséum National d'Histoire Naturelle, Paris.
- 28.Bhattacharya SB. Handbook of Indian Acanthocephala. 2007. pp. 1–226. Zoological Survey of India, Kolkata.
- 29.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. P Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acid Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 32.Swofford DL. PAUP* 4.0: Phylogenetic Analysis Using Parsimony (*and other methods). 2002. Sinauer Associates, Sunderland.
- 33.Rodríguez F, Oliver JF, Marin A, Medina JR. The general stochastic model of nucleotide substitution. J Theor Biol. 1990;142:485–501. doi: 10.1016/s0022-5193(05)80104-3. [DOI] [PubMed] [Google Scholar]
- 34.Yang Z. Estimating the patterns of nucleotides substitution. J Mol Evol. 1994;39:105–111. doi: 10.1007/BF00178256. [DOI] [PubMed] [Google Scholar]
- 35.Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 36.Stöver BC, Müller KF. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BioMed Central Bioinformatics. 2010;11, 7 doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]