Abstract
We have assembled transcriptionally active chromatin on 5S DNA plasmids by using a Xenopus oocyte supernatant and the 5S-specific transcription factor IIIA (TFIIIA). In this system, the 5S RNA gene is accurately transcribed at a rapid rate of 50 transcripts per gene per hr. By following the time course of RNA synthesis during chromatin assembly, the dose response to TFIIIA addition, and the effect of novobiocin on the assembled nucleoprotein, we show that there is a strict correlation between transcriptional activity and the generation of torsionally strained DNA supercoils in "dynamic chromatin." Transcription cannot be the cause of the dynamic structure, because the assembly of this chromatin is unaffected by alpha-amanitin levels that completely block RNA polymerase III. Surprisingly, the dynamic chromatin remains transcriptionally active after relaxation with DNA topoisomerase I, which implies that the essential parameter for chromatin transcription is gyration per se, and not its effect on DNA topology.
Full text
PDF

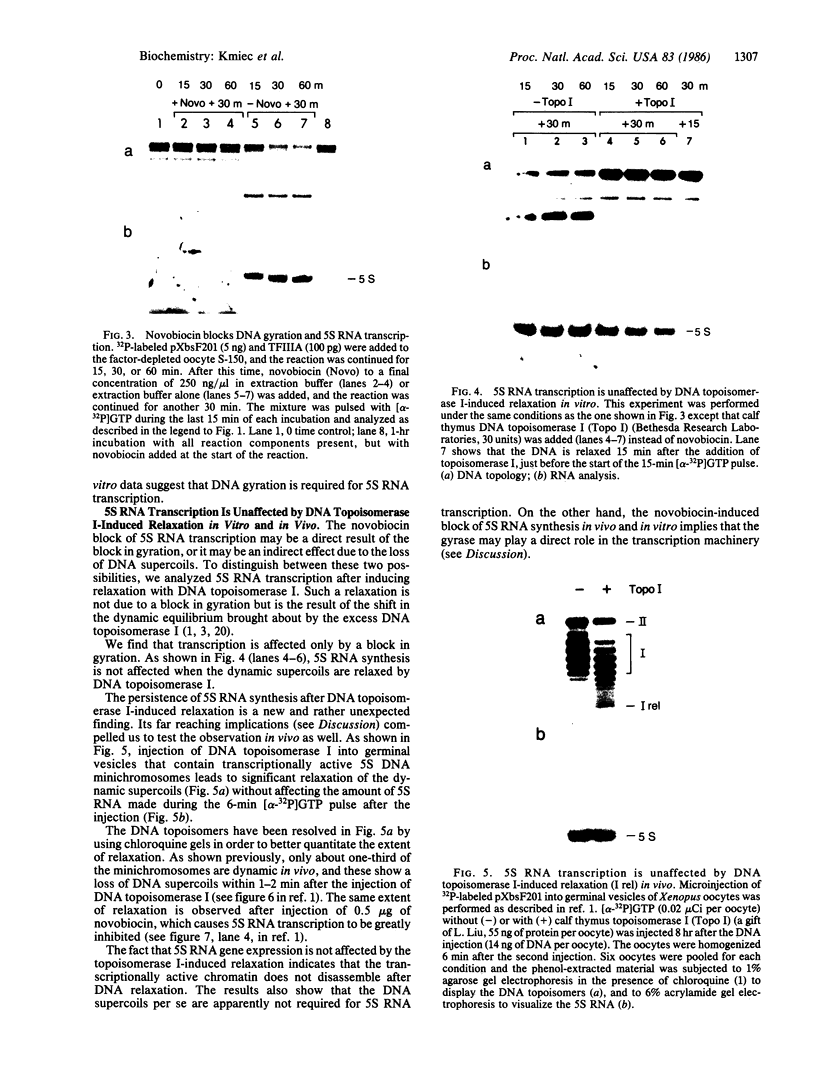


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benedetti P., Baldi M. I., Mattoccia E., Tocchini-Valentini G. P. Purification and characterization of Xenopus laevis topoisomerase II. EMBO J. 1983;2(8):1303–1308. doi: 10.1002/j.1460-2075.1983.tb01585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieker J. J., Martin P. L., Roeder R. G. Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell. 1985 Jan;40(1):119–127. doi: 10.1016/0092-8674(85)90315-0. [DOI] [PubMed] [Google Scholar]
- Bieker J. J., Roeder R. G. Physical properties and DNA-binding stoichiometry of a 5 S gene-specific transcription factor. J Biol Chem. 1984 May 25;259(10):6158–6164. [PubMed] [Google Scholar]
- Bogenhagen D. F., Wormington W. M., Brown D. D. Stable transcription complexes of Xenopus 5S RNA genes: a means to maintain the differentiated state. Cell. 1982 Feb;28(2):413–421. doi: 10.1016/0092-8674(82)90359-2. [DOI] [PubMed] [Google Scholar]
- Cozzarelli N. R., Gerrard S. P., Schlissel M., Brown D. D., Bogenhagen D. F. Purified RNA polymerase III accurately and efficiently terminates transcription of 5S RNA genes. Cell. 1983 Oct;34(3):829–835. doi: 10.1016/0092-8674(83)90540-8. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Martin P. L., Shastry B. S., Roeder R. G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Gargiulo G., Razvi F., Worcel A. Assembly of transcriptionally active chromatin in Xenopus oocytes requires specific DNA binding factors. Cell. 1984 Sep;38(2):511–521. doi: 10.1016/0092-8674(84)90506-3. [DOI] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glikin G. C., Ruberti I., Worcel A. Chromatin assembly in Xenopus oocytes: in vitro studies. Cell. 1984 May;37(1):33–41. doi: 10.1016/0092-8674(84)90298-8. [DOI] [PubMed] [Google Scholar]
- Han S., Udvardy A., Schedl P. Novobiocin blocks the Drosophila heat shock response. J Mol Biol. 1985 May 5;183(1):13–29. doi: 10.1016/0022-2836(85)90277-3. [DOI] [PubMed] [Google Scholar]
- Hsieh T., Brutlag D. ATP-dependent DNA topoisonmerase from D. melanogaster reversibly catenates duplex DNA rings. Cell. 1980 Aug;21(1):115–125. doi: 10.1016/0092-8674(80)90119-1. [DOI] [PubMed] [Google Scholar]
- Kmiec E. B., Worcel A. The positive transcription factor of the 5S RNA gene induces a 5S DNA-specific gyration in Xenopus oocyte extracts. Cell. 1985 Jul;41(3):945–953. doi: 10.1016/s0092-8674(85)80075-1. [DOI] [PubMed] [Google Scholar]
- Luchnik A. N., Bakayev V. V., Zbarsky I. B., Georgiev G. P. Elastic torsional strain in DNA within a fraction of SV40 minichromosomes: relation to transcriptionally active chromatin. EMBO J. 1982;1(11):1353–1358. doi: 10.1002/j.1460-2075.1982.tb01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. G., Liu L. F., Englund P. T. A homogeneous type II DNA topoisomerase from HeLa cell nuclei. J Biol Chem. 1981 Sep 10;256(17):9334–9339. [PubMed] [Google Scholar]
- Razvi F., Gargiulo G., Worcel A. A simple procedure for parallel sequence analysis of both strands of 5'-labeled DNA. Gene. 1983 Aug;23(2):175–183. doi: 10.1016/0378-1119(83)90049-5. [DOI] [PubMed] [Google Scholar]
- Roeder R. G. Multiple forms of deoxyribonucleic acid-dependent ribonucleic acid polymerase in Xenopus laevis. Isolation and partial characterization. J Biol Chem. 1974 Jan 10;249(1):241–248. [PubMed] [Google Scholar]
- Ryoji M., Worcel A. Chromatin assembly in Xenopus oocytes: in vivo studies. Cell. 1984 May;37(1):21–32. doi: 10.1016/0092-8674(84)90297-6. [DOI] [PubMed] [Google Scholar]
- Ryoji M., Worcel A. Structure of the two distinct types of minichromosomes that are assembled on DNA injected in Xenopus oocytes. Cell. 1985 Apr;40(4):923–932. doi: 10.1016/0092-8674(85)90352-6. [DOI] [PubMed] [Google Scholar]
- Shlomai J., Zadok A. Reversible decatenation of kinetoplast DNA by a DNA topoisomerase from trypanosomatids. Nucleic Acids Res. 1983 Jun 25;11(12):4019–4034. doi: 10.1093/nar/11.12.4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. R., Jackson I. J., Brown D. D. Domains of the positive transcription factor specific for the Xenopus 5S RNA gene. Cell. 1984 Jun;37(2):645–652. doi: 10.1016/0092-8674(84)90396-9. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Wormington W. M., Schlissel M., Brown D. D. Developmental regulation of Xenopus 5S RNA genes. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):879–884. doi: 10.1101/sqb.1983.047.01.101. [DOI] [PubMed] [Google Scholar]











