Abstract
Numerous examples of phage display applied to soluble proteins demonstrate the power of the technique for protein engineering, affinity reagent discovery and structure-function studies. Recent reports have expanded phage display to include membrane proteins. The scope and limitations of membrane protein display remain undefined. Therefore, we report data from the phage display of representative types of membrane-associated proteins including plasma, nuclear, peripheral, single and multipass. The peripheral membrane protein neuromodulin displays robustly with packaging by conventional M13-KO7 helper phage. The monotopic membrane protein, Nogo-66 can also display on the phage surface, if packaged by the modified M13-KO7+ helper phage. The modified phage coat of KO7+ can better mimic the zwitterionic character of the plasma membrane. Four examples of putatively α-helical, integral membrane proteins failed to express as fusions to an anchoring phage coat protein, and therefore did not display on the phage surface. However, the β-barrel membrane proteins ShuA and MOMP, which pass through the membrane 22- and 16-times respectively, can display surprisingly well on the surfaces of both conventional and KO7+ phage. The results provide a guide for protein engineering and large-scale mutagenesis enabled by the phage display of membrane proteins.
Keywords: Phage display, membrane proteins, protein engineering, mutagenesis, β-barrel membrane proteins
Introduction
Reflecting their critical importance to cellular functions, membrane proteins comprise approximately 30% of the human proteome.1 Cellular processes relying on membrane proteins range from energy generation to cell signaling. This preeminence also accounts for the large number of membrane proteins targeted by >50% of FDA-approved therapeutics.2 Thus, understanding the relationship between membrane protein sequence and function could provide valuable insights in biology and drug development.
The properties of membrane proteins challenge attempts to produce sufficient quantities for biophysical studies. For example, the Membrane Proteins of Known 3D Structures (http://blanco.biomol.uci.edu/mpstruc) includes < 300 unique membrane protein structures, a low number limited by difficulties with expression, isolation, and purification of folded membrane proteins.3 Protein aggregation can result from the inherent hydrophobicity of membrane proteins. Typical methods to solubilize membrane proteins use strong detergents, which can destabilize the proteins.4 Furthermore, expression of membrane proteins in prokaryotes can fail due to, amongst other reasons, the lack of post-translational modifications required in eukaryotes.
Phage display provides an alternative to the expression of membrane proteins for in vitro binding and drug discovery studies.5 Phage display allows identification of functional residues in the displayed protein by high throughput mutagenesis. Alanine shotgun scanning of the displayed protein, for example, can identify residues contributing to protein binding interfaces.6,7 Furthermore, the technique can be used to reverse engineer soluble and functional variants of phage-displayed membrane proteins8 and also to identify membrane protein ligands.9
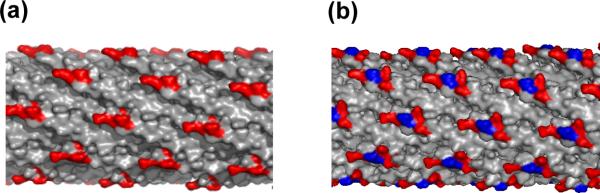
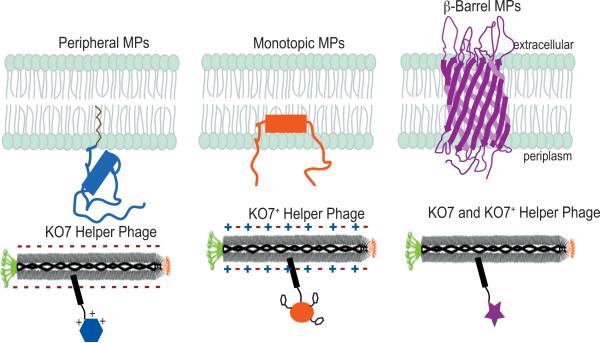
Essential to the phage display approach, the phenotype of the phage-displayed protein is linked to the encoded genotype, which is packaged within the phage particle.10 Proteins can be displayed on the phage surface using a phage vector or a plasmid-based phagemid vector.11 Unlike the natural Ff phage genome of a phage vector, the phagemid encodes the open reading frame with a single coat protein fused to the displayed protein. A helper phage provides the phage proteins required for virus packaging and assembly.12 This report applies two helper phage, the conventional M13-KO7 and the recently reported M13-KO7+ bacteriophage.13 KO7+ helper phage includes a tetrapeptide (AKAS) near the N-terminus of each copy of the major coat protein (g8p or P8). Originally developed to reduce background binding, this tetrapeptide insertion in the P8 of KO7+ (Fig. 1) has also been shown to allow display of two membrane proteins, caveolin-1 and HIV-gp4.14
Fig. 1.
A schematic diagram depicting the surfaces of KO7 and KO7+ helper phage. Positively and negatively charged side chain functionalities are highlighted in blue and red respectively. (a) The major coat protein, P8, of M13-KO7 includes three carboxylate bearing side chains at its N-terminus (PDB accession: 1ifd). (b) An insertion of a 4-mer peptide into each copy of P8 alters the surface charge of M13-KO7+. The resultant zwitterionic surface can better display monotopic membrane proteins.
The successful display of a functional protein on the phage surface requires protein translocation to the periplasm, subsequent folding into its functional conformation, and incorporation of the phage coat-fused protein during virus assembly. Although a large number of soluble proteins have successfully displayed on the phage surface,12,15 some proteins prove refractory to display.16 Failure in phage display can result from protein aggregation,17 or potentially incomplete translocation into the periplasm due to “stop transfer” signals in the protein sequence.18 Attempts to improve display levels include mutations to the anchoring P8 coat protein,19 co-expression of periplasmic chaperones,20 and translocation of the displayed peptides to the periplasm by SRP21 or Tat translocation pathways.18
Despite successful display of soluble proteins, only a handful of membrane-associated proteins have been displayed to date on phage.14 Here, we define the scope and current limitations in the display of membrane proteins on the phage surface. The proteins tested include plasma, nuclear, peripheral, single and multipass membrane-associated proteins (Table 1). The accumulation of successes and failures with phage-displayed, membrane-associated proteins provides a predictive model for the design of phage-based experiments to explore the structure and function of membrane proteins.
Table 1.
Phage Display of Membrane-Associated Proteins
| Membrane-Associated Protein | MW (kDa) | Membrane Topology | TM Secondary Structure | Display | Helper Phage |
|---|---|---|---|---|---|
| Nef (6) | 23 | Peripheral | +++ | KO7 | |
| Neuromodulin | 25 | Peripheral | ++ | KO7 | |
| Nogo-66 | 7.5 | Monotopic | +++ | KO7+ | |
| Caveolin-1(15) | 22 | Monotopic | ++ | KO7+ | |
| Emerin | 29 | Bitopic | α-helicala | - | KO7/KO7+ |
| MAN1 | 100 | Polytopic | α-helicala | - | KO7/KO7+ |
| NET25 | 57 | Polytopic | α-helicala | - | KO7/KO7+ |
| Matriptase-2 | 51.6 | Bitopic | α-helicala | - | KO7/KO7+ |
| ShuA | 72.5 | Polytopic | β-barrel | ++ | KO7/KO7+ |
| MOMP | 40 | Polytopic | β-barrela | +++ | KO7 |
- No display, ++ Moderate display levels, +++ High display levels.
Putative secondary structure.
Results and Discussion
Cloning membrane proteins for display
The genes encoding the targeted membrane proteins were subcloned into a conventional phagemid vector for phage display initially as fusions to the N-terminus of P8. Fusion to the N-terminus of the minor coat protein P3 (C-terminal domain) was also attempted for the display of the putatively α-helical transmembrane (TM) proteins. Each displayed protein included a FLAG antibody epitope fused to the N-terminus for quantification of the relative display levels. DNA sequencing verified successful cloning of an ORF encoding the following sequences from N-terminus to C-terminus: the periplasmic signal peptide targeting the Sec translocation pathway, the FLAG epitope, the membrane-associated protein, the linker (amino acid sequence of GGGSGSSS) and the anchoring phage coat protein. The phage were packaged with wild-type KO7 or the variant KO7+ helper phage, as discussed below.
A phage-based ELISA quantified the relative display levels of the membrane proteins. In the ELISA, microtiter plate-adsorbed, anti-FLAG antibody bound to the FLAG epitope of full-length proteins displayed on phage surfaces. The negative and positive controls for all ELISA experiments yielded similar expected results. Negative controls resulted in little or no detectable binding; one negative control examined binding by the displayed protein to the blocking agent (BSA, NFM or ovalbumin), and a second control tested binding to the anti-FLAG antibody by phage lacking a displayed protein (KO7 or KO7+ helper phage). Phage-displayed Polymerase I and transcript release factor (PTRF), provided the positive control for successful display, as the protein displayed consistently well on the phage surfaces provided by both KO7 and KO7+ helper phage.
Phage Display of Peripheral Membrane Proteins
Peripheral membrane proteins loosely and sometimes reversibly associate with the plasma membrane, through interactions with the membrane or other membrane-associated proteins.22 Post-translational modifications, including acylation,23 prenylation,24 and glycosylphophatidylinositol (GPI) anchors,25 can covalently anchor the protein to the membrane, though protein expression in E. coli precludes this possibility. Successful phage display of at least one peripheral membrane protein has been reported previously. For example, the HIV-1 protein Nef, responsible for down-regulation of CD4, displays robustly on the phage surface.5 Display of Nef was reported before the discovery of KO7+ for phage display of membrane proteins. Therefore, KO7 and KO7+ helper phage were compared for the packaging of the peripheral membrane protein, neuromodulin.
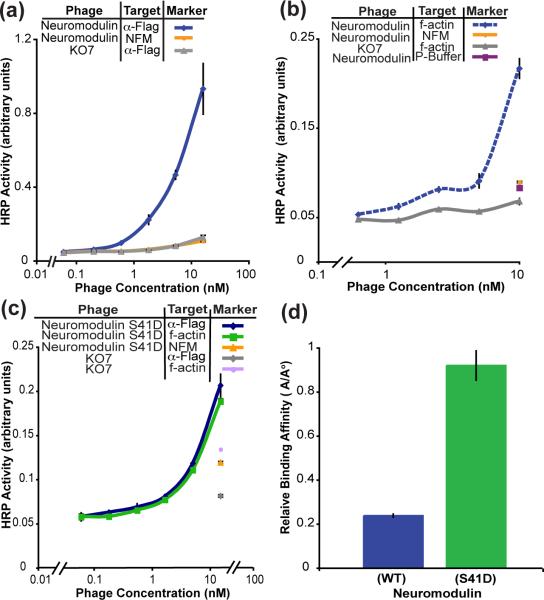
Neuromodulin (GAP-43), a neurite growth stimulating protein, associates with the membrane through palmitoylation and electrostatic interactions between the positively charged residues in the membrane binding domain at the N-terminus and the headgroups of the lipid bilayer.26 For display on the phage surface, the sites of palmitoylation, cysteine residues C3 and C4, were mutated to alanine. The neuromodulin-displayed phage was packaged by either KO7 or KO7+ helper phage. An ELISA to measure display of the FLAG epitope at the N-terminus of neuromodulin demonstrates successful display with packaging by KO7, but not KO7+, helper phage (Fig. 2a and Fig. S1, respectively). The display of neuromodulin through KO7 packaging is consistent with the display of the peripheral membrane protein HIV-1 Nef also using KO7 helper phage.
Fig. 2.
Phage display of functional neuromodulin. (a) In this phage-based ELISA, the levels of neuromodulin displayed were assayed through immobilized anti-FLAG antibody binding to a FLAG epitope fused to the N-terminus of the phage-displayed protein. Successful display of neuromodulin on the phage surface required the helper phage KO7. The negative controls for all ELISAs in this report include membrane protein-displayed phage binding to the blocking agent BSA, NFM or ovalbumin, and the helper phage KO7 or KO7+ binding to the anti-FLAG antibody or the target protein. (b) The phage-displayed neuromodulin remained functional as demonstrated through binding to the known binding partner, f-actin. Binding to Polymerization Buffer (P-Buffer) provided an additional negative control. (c) The displayed protein variant, neuromodulin S41D also binds to immobilized f-actin as expected. (d) The relative binding affinity of the neuromodulin(S41D) for f-actin is four fold higher than the affinity of the wild type neuromodulin-f-actin interaction. Throughout the report, error bars indicate standard error (n = 3), except where specified.
KO7 helper phage has three carboxylate bearing sidechains near the surface-exposed N-terminus of each P8 to create a forest of negatively charged residues (Fig. 1a) Such sidechains could facilitate electrostatic interactions with the basic residues of the membrane binding domain of neuromodulin. However, the more positively charged surface of KO7+ phage could negatively affect such interactions, and thus interfere with the display of this peripheral membrane protein. The experimental results with two peripheral membrane proteins demonstrate that proteins with lipid binding motifs can be readily packaged with conventional KO7 helper phage.
Phage-displayed neuromodulin also retains its functional conformation on the phage surface. To demonstrate the functionality of displayed neuromodulin, a binding assay with a known binding partner was performed. F-actin binds to unphosphorylated neuromodulin with a dissociation constant (KD) of ≈1.2 μM.27 An ELISA with immobilized f-actin and phage-displayed wild-type neuromodulin demonstrated this interaction, thus confirming the folding of the displayed protein (Fig. 2b). Although neuromodulin binds to f-actin, phosphorylation at Ser41 increases the binding affinity for f-actin about seven-fold (KD = 161 nM).27 To further demonstrate functional folding of the phage-displayed neuromodulin, a constitutively active neuromodulin variant was generated through a S41D substitution in the phage-displayed protein. Introduction of the negative charge at this phosphorylation site can mimic phosphorylated neuromodulin.28 An ELISA quantified display levels of neuromodulin (S41D) and binding to f-actin (Fig. 2c). As observed with the wild-type protein, neuromodulin (S41D) binds well to f-actin, though the display levels of the S41D variant were lower than for the wild-type protein. The reduced display levels could result from decreased expression levels of the S41D variant of neuromodulin.
With the successful phage display of neuromodulin (wild-type and the S41D variant), the relative binding affinities of the two phage-displayed proteins were calculated through a single ELISA, which quantifies both display and binding levels. As shown previously, the ratio of the protein display levels (Ao) to binding levels (A) has a linear correlation to the KD values for the interactions.29 The linear correlation requires the concentration of the target protein (f-actin) to exceed the concentration of the displayed protein, which is a reasonable assumption for a phage-displayed protein. The constitutively active neuromodulin (S41D) bound to f-actin with 4-fold higher affinity than wild-type, a measurement within the range of previous measurements made without phage display27 (Fig. 2d). The ability to bind to f-actin with high affinity and predictable outcomes from mutagenesis suggests that this peripheral protein is displayed in its functional conformation. Furthermore, the phage-displayed neuromodulin could be amenable to further mutational analysis for structure-function studies.
Phage Display of Monotopic Membrane Proteins
Monotopic membrane proteins penetrate only one leaflet of the plasma membrane, and do not traverse the bilayer.30 Such proteins associate tightly with the membrane either through amphipathic helices31 or through electrostatic and hydrophobic interactions.32 Reported structures of monotopic membrane proteins suggest that both basic residues and hydrophobic amino acids are involved in the anchoring of the membrane protein. Monotopic membrane proteins include both “shallow inserters” that interact with the surface of the lipid bilayer and others that can penetrate deeper into the bilayer, aided by a hydrophobic stretch and key basic residues.32
Phage display of two monotopic membrane proteins demonstrates the capabilities of KO7+ helper phage to permit display of previously inaccessible membrane proteins. As reported previously, but not examined, successful phage display of the monotopic membrane proteins requires packaging by KO7+ helper phage.14 At the physiological pH, the AKAS peptide inserted into each copy of the P8 phage coat protein provides a positively charge lysine sidechain surrounded by three negatively charged carboxylate bearing sidechains. Extended to ≈2700 copies of P8 per phage, the protein displayed on the KO7+ surface will be surrounded by both positive and negatively charged sidechains (Fig. 1b). In this model, the AKAS insertion within P8 of KO7+ allows the phage surface to better mimic the zwitterionic character of the phospholipid head groups found at the plasma membrane interface. The ε-amine of the lysine sidechain, for example, could substitute for the primary and quaternary amines of the phospholipid headgroups.
Our laboratory has previously reported successful display of functional full-length caveolin-1 on the phage surface.14 Successful display of functional caveolin-1 on the phage surface allowed engineering of a soluble, but functional, caveolin variant.8 Interestingly, the extensive mutagenesis of caveolin on the phage surface required solubilizing mutations targeting the aromatic residues in the intramembrane domain. Such aromatic sidechains could form cation-π interactions with primary and quaternary amines of the phospholipid head groups. The cation-π interactions provide a mechanistic hypothesis for the efficacy of KO7+ in monotopic membrane protein display.
A second example of monotopic protein display, Nogo-66, consists of the 66-residues of the extracellular domain of the neuronal protein Nogo. As shown by circular dichroism and NMR spectra, Nogo-66 requires dodecylphosphocholine micelles to fold.33 The structure of Nogo-66 determined by NMR shows a monotopic membrane topology. The aromatic residues of Nogo-66 contribute to the lipid binding through potential cation-π interactions with the quaternary amines of phosphotidylcholine. Given the requirement for protein-lipid interactions to assist folding, Nogo-66 represents a particularly challenging test for display by M13 phages, which lack an enveloping phospholipid.
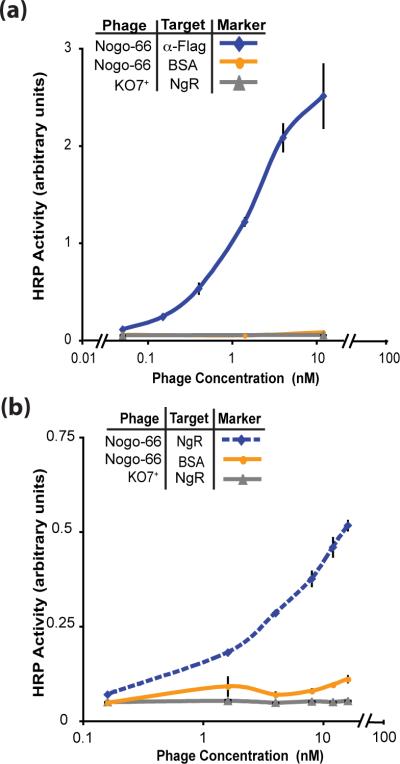
Using KO7+ as the helper phage, folded and functional Nogo-66 successfully displayed on the phage surface (Fig. 3a). In this case, KO7+ contributes two essential functions to allow successful phage display. First, KO7+ mimics phospholipid headgroups, which are required for the folding of the protein. Binding to the Nogo receptor (NgR) by phage displayed Nogo-66 (Fig. 3b) demonstrates functional folding of the displayed protein. Second, packaging with KO7+ helper phage reduced background binding to NgR, which can result from interactions between the conventional KO7 helper phage surface and high pI target proteins (the pI of NgR is 8.4). Successful Nogo-66 display reinforces the hypothesis that the extra positive charge included with each copy of KO7+ P8 can mimic the necessary zwitterionic character inherent to phospholipid headgroups.
Fig. 3.
Functional Nogo-66 displayed on the phage surface. (a) Display of Nogo-66 with KO7+ helper phage is assessed by ELISA with anti-FLAG antibody immobilized on the microtiter plate. (b) The phage-displayed Nogo-66 remained functional, as demonstrated through binding to its known receptor, NgR, immobilized on the microtiter plate.
Phage Display of α-Helical TM proteins
The α-helical TM proteins comprise >70% of the membrane proteins in the Membrane Protein Data Bank.34 This class of TM proteins can traverse the lipid bilayer in a single pass (bitopic) or cross two or more times in polytopic TM proteins. Residing in cellular and nuclear membranes, α-helical TM proteins can form monomers or oligomers. Through their extramembrane domains, such proteins can perform a myriad of functions ranging from enzymatic activity to signal transduction.35
To examine different types of α-helical membrane proteins, four membrane proteins were subcloned into a vector for phage display. The tested proteins include single and multipass TM proteins, and also cytoplasmic and nuclear membrane proteins (Table 1). Sub-cloned as fusions to the major coat protein, P8, the phage displaying the α-helical membrane proteins were propagated using either KO7 or KO7+ helper phage. Attempts to display these TM proteins also involved fusion to the minor coat protein, P3, and also expression in different strains of E. coli bacteria (Table S2 in the Supporting Information). As demonstrated by ELISA experiments, no combination of helper phage, coat protein and cell-line allowed successful display of the targeted α-helical integral membrane proteins (Fig. S2 and S3).
To understand the basis for this inability to display α-helical TM proteins, Western blots targeting the FLAG epitope at the N-terminus of each protein examined protein expression levels during phage assembly. The phage coat proteins, including P3 and P8 fused to the displayed proteins, must insert into the bacterial membrane before assembly into the filamentous phage. Thus, the analysis of fusion protein levels focused on the cell pellets where such membrane proteins should localize. None of the α-helical TM proteins were detectable by Western blot (Fig. S4). The positive control for the Western blot, PTRF fused to P8 was readily detected in the cell pellets. The absence of the expected fusion proteins in the immuno-blot demonstrates the α-helical TM proteins, emerin, MAN1, NET25 and matriptase-2, fail to express sufficiently well for phage-based applications. Furthermore this inability to express sufficient quantities of the fusion protein results in the lack of display observed for α-helical TM proteins. In addition, the P8- and P3-fused proteins could fail to translocate to the periplasm, resulting in degradation of the fusion protein in the cytosol. Despite these results, the display of α-helical TM proteins might be successful if the fusion protein can be expressed and targeted to the periplasm.
Phage Display of β-Barrel TM proteins
β-barrel membrane proteins are observed only in the outer membranes of Gram negative bacteria, mitochondria and chloroplasts. The proteins can form pores for the transport of nutrients, and can also possess enzymatic activity.36 Ranging from 8 to 22 β-strands, β-integral membrane proteins are polytopic. Both monomeric and oligomeric β-barrel TM proteins have been described. As described below, two β-barrel proteins, Shigella heme uptake A (ShuA), an outer membrane heme transporter from Shigella dysenteriae and the major outer membrane protein (MOMP), a porin from Chlamydia muridarum Nigg, displayed well on the phage surface.
ShuA belongs to the family of TonB-dependent transporters.37 The C-terminal domain of ShuA forms a 22-stranded barrel structure. The N-terminal domain provides a plug partitioning the extracellular milieu from the periplasm.38Shigella dysenteriae, a gram negative pathogen, uses TonB-dependent transporters to acquire heme from the host's heme-binding protein, hemoglobin.37
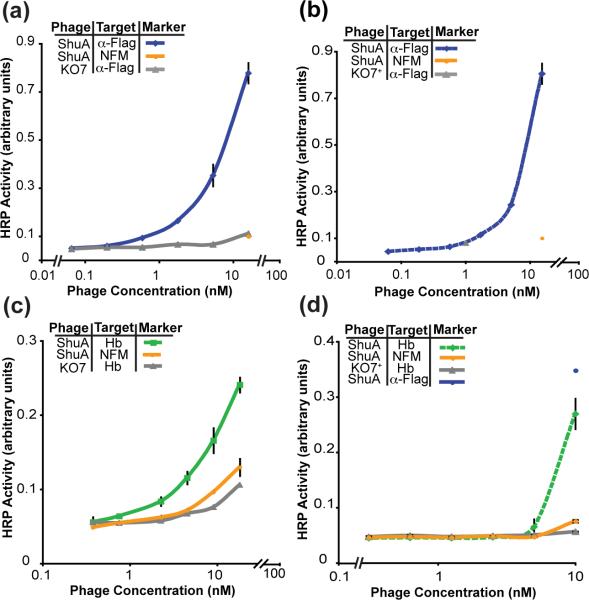
ShuA fused to P8 displayed robustly on the phage surface with packaging by both KO7 and KO7+ helper phage (Fig. 4a and 4b). Improved display of ShuA resulted from phage propagation at 30 °C. The slower growth at this reduced temperature could allow for increased expression of the functional fusion protein, and could assist in the assembly and packaging of phage particles. Furthermore, binding to immobilized hemoglobin A demonstrated that phage-displayed ShuA remains functionally folded (Fig. 4c and 4d). To our knowledge, ShuA is the first example of a functional, full length, β-barrel integral membrane protein successfully displayed on the phage surface.
Fig. 4.
Functional display of full-length ShuA packaged with (a) KO7 helper phage and (b) KO7+ helper phage. Display of ShuA with KO7 and KO7+ helper phage is assessed by ELISA with anti-FLAG antibody immobilized on the plate. (c) and (d) Binding between phage-displayed ShuA on KO7 and KO7+ and its binding partner hemoglobin (Hb) is assessed by ELISA with Hb immobilized on the plate.
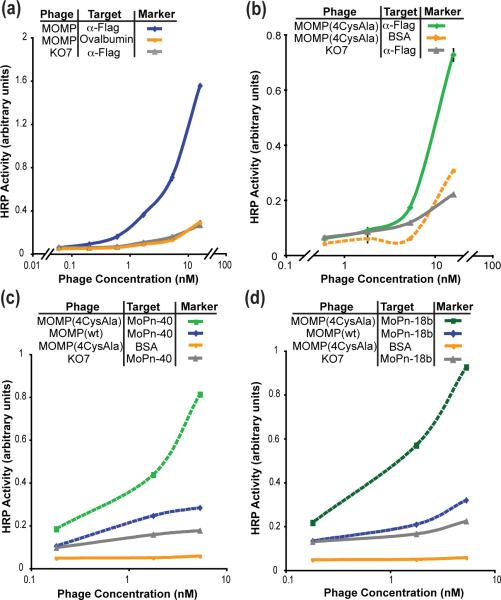
To demonstrate the generality of the approach, a second putative β-barrel membrane protein was successfully displayed. A cysteine-rich outer membrane protein, MOMP of the Chlamydia mouse pneumonitis (MoPn serovar) is expected to form a 16-strand β-barrel secondary structure. Functioning as a porin, MOMP likely forms a homotrimer.39 To allow formation of a trimer on the phage surface, the phagemid ORF included the gene encoding MOMP, followed by an amber stop codon, before the linker and the phage coat protein (P3). In an amber suppressor strain of E. coli, the amber stop is recognized as a glutamine residue with 10-30% efficiency40 resulting in the production of both free and P3-fused MOMP monomers. The two MOMP constructs can form trimers in the periplasm prior to phage assembly.
An ELISA to determine display levels with immobilized anti-FLAG antibody demonstrated that MOMP displayed at high levels (Fig. 5a). The natural binding partners to MOMP remain unidentified, and we, therefore, demonstrate functionality of the phage-displayed protein through binding to two monoclonal antibodies with well-defined epitopes. The first anti-MOMP antibody MoPn-40 binds a linear epitope in one of the MOMP variable domains, VD1 and the second MoPn-18b recognizes a structural epitope of the homotrimer.39 The wild-type MOMP displayed well, yet bound poorly to the two antibodies (Fig. 5c and 5d).
Fig. 5.
Display of the putative β-barrel membrane protein, MOMP. (a) Successful display of MOMP as a fusion to the P3 coat protein is demonstrated by ELISA with anti-FLAG antibody immobilized on the microtiter plate (n=2). (b) Successful display of MOMP (4CysAla) as a fusion to the P3 coat protein is assessed by ELISA with anti-FLAG antibody immobilized on the microtiter plate (n=2). (c) The MOMP variant with four free cysteines mutated to alanine binds to the MoPn-40 antibody, which recognizes a linear epitope (n=1). (d) The MOMP variant also binds to the conformational antibody MoPn-18b which recognizes the trimeric form of MOMP (n=1).
Modifications to the assay conditions and the gene encoding MOMP allow MOMP-antibody binding assays. Treatment of the phage-displayed wild-type MOMP with 2M urea and the reducing agent tris (2 carboxymethyl) phosphine (TCEP) (10 mM) produced phage-displayed MOMP that could be recognized by the MoPn-40, which requires a linear epitope of MOMP (Fig. S5). The results confirm successful display of MOMP, and implicate difficulties with disulfide bonding as a complicating variable. To improve folding, the four free cysteines of MOMP, C51, C136, C226, and C351, were mutated to alanine. The replacement of the free cysteines resulted in a displayed and folded MOMP (Fig. 5b, 5c, 5d). The MOMP variant protein, MOMP (4CysAla) bound to both MOMP antibodies, including the antibody MoPn-18b, which recognizes a structural epitope of the trimeric MOMP (Fig. 5d).
Several factors unique to β-barrel membrane protein translocation and folding could contribute to the successful display of folded β-barrel membrane proteins. First, β-barrel transmembrane domains are less hydrophobic than α-helical TM proteins. Furthermore, the β-barrel TM proteins use alternate folding and membrane insertion pathways, are targeted to the periplasm, and remain unfolded with periplasmic chaperones preventing their aggregation prior to insertion in the outer membrane.41,42 The moderate hydrophobicity and the lack of aggregation allow the transmembrane regions of such proteins to insert spontaneously into the lipid bilayer. Such features complement the requirements for successful phage display, which involves targeting of the displayed protein to the periplasm. Furthermore the decreased hydrophobicity could assist with the removal of the fusion protein during phage assembly.
Conclusions
The results presented here expand the palette of phage-displayed proteins from solely soluble proteins to include membrane-associated proteins (Fig. 6). The successful display of HIV-1 Nef5 and neuromodulin as fusions to the P8 coat protein on the KO7 phage surface demonstrate that the conventional helper phage, M13-KO7, can be used for display of peripheral membrane proteins. Monotopic membrane proteins, caveolin-114 and Nogo-66, also displayed robustly and consistently as fusions to P8, when packaged with the modified helper phage, KO7+. The β-barrel membrane protein, MOMP displayed as a fusion to P3 and likely formed a trimer as evident by the binding assay with a conformational antibody; a second β-barrel membrane protein, ShuA also displayed successfully as a folded and functional protein. In addition, protein size is not a limiting factor in membrane protein phage display, as the examples of successful display range from 7 to 73 kDa. Though phage display proved ineffective for the α-helical TM proteins, emerin, MAN1, matriptase-2, and NET25, successful display of peripheral, monotopic and β-barrel membrane proteins illustrates the efficacy of the approach. For the three classes of TM proteins displayed on phage, we report at least two examples of each class, which highlight functional folding through binding to known ligands and receptors. Given the tremendous potential of phage-based mutagenesis for protein engineering and structure-function studies, extending the technique to TM proteins should provide new possibilities for exploring the world of membrane proteins.
Fig. 6.
A guide to the phage display of membrane proteins. As demonstrated here, peripheral membrane proteins can require the negatively charged surface of wild-type, KO7 helper phage. Monotopic membrane proteins benefit from the zwitterionic surface of KO7+ helper phage. Unlike the requirements of peripheral and monotopic membrane proteins, β-barrels can be displayed through packaging with either helper phage.
Materials and Methods
Phage-display Vector Design
The genes encoding the membrane proteins, emerin, MAN1, matriptase-2, MoPn MOMP, NET25, neuromodulin, mouse Nogo-66, and ShuA were sub-cloned into the pM1165a phagemid between the ORF encoding a periplasmic secretion signal peptide and P8. The pM1165a phagemid has been previously reported43 and is based on pS1607 phagemid.44 For display of the membrane proteins as fusions to P3, the modified pS1602 phagemid45 was used. A FLAG epitope peptide (amino acid sequence DYKDDDDK) fused to the N-terminus of the proteins provided an antibody epitope for monitoring protein display levels. Throughout this report, successful cloning and mutagenesis was verified by DNA sequencing (Genewiz, Inc.).
Site-Directed Mutagenesis of Neuromodulin and MOMP
Sequences of the mutagenic oligonucleotides appear in Supporting Information. Using the Stratagene Quikchange site-directed mutagenesis protocol, and oligonucleotides NeuromodulinS41D-Fwd and NeuromodulinS41D-Rev, the neuromodulin S41D gene was generated. The C51A, C136A, C226A and C351A mutations were introduced into the MOMP gene using oligonucleotide-directed mutagenesis.46 MOMP (4CysAla) was also subcloned into the phagemid vector to encode a fusion to the P3 coat protein.
Purification of membrane protein-displayed phage
As listed in the Supporting Information, different strains of E. coli were found to improve expression and display on the phage surface (Table S2). K91, XL-1 or SS320 cells were transformed with a phagemid encoding each of the membrane proteins and cells were grown at 30 or 37 °C in 2 ml 2YT medium supplemented with 50 μg/ml carbenicillin until the culture reached log-phase growth. KO7 or KO7+ helper phage (1010 phage /ml) were added, and the culture was shaken at 37 °C for ½ h before transferring to 70 ml 2YT supplemented with carbenicillin and kanamycin (25 μg/ml). The culture was incubated overnight for 21 or 16 h at 30 or 37 °C respectively. Cells were removed by centrifugation (10000 × g, 10 min), and the phage precipitated from the supernatant by addition of 1/5th volume of 20% polyethylene glycol 8000 in 2.5 M NaCl. The phage were recovered by centrifugation (10000 × g for 20 min and 5000 × g for 5 min), resuspended in 1X PBS and 0.05% Tween-20 and isolated by centrifugation (10 min at 12000 × g). The precipitation of the phage was repeated as described above with the addition of 20% PEG 8000 in 2.5M NaCl, followed by centrifugation and resuspension in PBS. Phage concentrations were estimated spectrophotometrically (OD268 = 8.31 nM = 5 × 1012 phage/ml).
Phage-based ELISAs
A phage-based ELISA was used to assess relative display and binding levels of the membrane proteins. Specific wells of a 96-well Nunc maxisorb plate were coated with anti-FLAG antibody (Stratagene, 100 μl/well, 1:1000 dilution in 50 mM Na2CO3, pH 9.6), or the target protein (10μg/ml) and incubated for 2 h at room temperature (RT) or overnight at 4 °C. For ELISAs targeting f-actin, the actin protein (Sigma Aldrich) was allowed to polymerize in Polymerization buffer (2 mM MgCl2, 100 mM KCl, 0.2 mM DTT, 0.2 mM ATP, 2mM Tris pH 7.6) for 1 h at RT before coating the plate 27. After removal of the coating solution, the wells were blocked for 30 min with BSA, ovalbumin or non fat milk (0.2%) in 50 mM PBS, pH 7.2. Control wells, which were not coated with the anti-FLAG antibody or target protein, were coated with the blocking buffer. The wells were rinsed three times with wash buffer (0.05% Tween-20 in PBS). Separately, serial dilutions of the membrane protein displaying phage and KO7+ phage (negative control) were made in dilution buffer (0.1% BSA with 0.05% Tween-20 in PBS). The phage solution was added to the corresponding wells of target-coated ELISA plates and shaken on an orbital shaker for 1 h at RT. The wells were washed five times with wash buffer, and then incubated with HRP-conjugated anti-M13 antibody (GE Healthcare, 100 μl/well; 1:5000 in the phage dilution buffer) for 30 min. The wells were washed five times with wash buffer and twice with PBS. The bound reactants were detected by incubating with the substrate solution (100 μl/well; 2 mg/ml o-phenylenediamine dihydrochloride and 0.02% w/v H2O2, in citric acid buffer, pH 5). Following an appropriate incubation time, the absorbance was measured spectrophotometrically at 450 nm using a microtiter plate reader (μQuant, Bio-Tek).
Western Blot of α-Helical TM proteins
After the protocol described above for phage propagation and isolation, samples of the E. coli cell pellets expressing the α-helical TM proteins, emerin, MAN1, matriptase-2, Net25 and the control, PTRF were electrophoretically separated by SDS-PAGE before transferring to a nitrocellulose membrane. The membrane was blocked with 5% non fat milk in PBS for an hour. The primary antibody was mouse anti-FLAG antibody (Stratagene, 1:1000 dilution), and the secondary antibody was horseradish peroxidase conjugated anti-mouse IgG antibody (Sigma, 1:1000 dilution). To visualize the immune blot, the membrane was incubated for 10 min with the addition of a detection reagent (4-chloro-1-naphthol with 3, 3’-diaminobenzidine, tetrahydrochloride).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Larry Gerace for the NET25 vector, Frank Torti for matriptase-2 encoding plasmid, Luis de la Maza for the MOMP vector and the MoPn-40 and MoPn-18b antibodies, Angela Wilks, Thomas L. Poulos and Sarvind Tripathi for the Shu A vector, Sarvind Tripathi for the hemoglobin protein, and Sachdev Sidhu for providing the modified pS1602 phagemid. We also thank Dr. Sudipta Majumdar for technical assistance and Davoud Mozhdehi for assistance with a figure. This work was supported by the National Institute of Health, National Institute of General Medical Sciences (RO1-GM078528-01 to G.A.W) and the Roman Reed Research Fund (RR05-155 to M.J.C.)
Abbreviations used
- SRP
signal recognition particle
- TM
transmembrane protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers: NCBI accession numbers for emerin, MAN1(LEMD3), matriptase-2 (TMPRSS6), MOMP, Net25(LEMD2), neuromodulin(GAP43), nogo-A (reticulon-4) and ShuA are BC000738, BC167864, BC039082, NP_296436, NP_851853, BC007936, NP_918943, AAC27809, respectively.
References
- 1.Stevens TJ, Arkin IT. Do more complex organisms have a greater proportion of membrane proteins in their genomes? Protein. 2000;39:417–420. doi: 10.1002/(sici)1097-0134(20000601)39:4<417::aid-prot140>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 2.Russell RB, Eggleston DS. New roles for structure in biology and drug discovery. Nat. Struct. Biol. 2000;7(Suppl):928–930. doi: 10.1038/80691. [DOI] [PubMed] [Google Scholar]
- 3.White SH. The progress of membrane protein structure determination. Protein Sci. 2004;13:1948–1949. doi: 10.1110/ps.04712004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Popot JL. Amphipols, nanodiscs, and fluorinated surfactants: three nonconventional approaches to studying membrane proteins in aqueous solutions. Annu. Rev. Biochem. 2010;79:737–775. doi: 10.1146/annurev.biochem.052208.114057. [DOI] [PubMed] [Google Scholar]
- 5.Olszewski A, Sato K, Aron ZD, Cohen F, Harris A, McDougall BR, Robinson WE, Jr., Overman LE, Weiss GA. Guanidine alkaloid analogs as inhibitors of HIV-1 Nef interactions with p53, actin, and p56lck. Proc. Natl. Acad. Sci. USA. 2004;101:14079–14084. doi: 10.1073/pnas.0406040101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrison KL, Weiss GA. Combinatorial alanine-scanning. Curr. Opin. Chem. Biol. 2001;5:302–307. doi: 10.1016/s1367-5931(00)00206-4. [DOI] [PubMed] [Google Scholar]
- 7.Weiss GA, Watanabe CK, Zhong A, Goddard A, Sidhu SS. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc. Natl. Acad. Sci. USA. 2000;97:8950–8954. doi: 10.1073/pnas.160252097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hajduczki A, Majumdar S, Fricke M, Brown IA, Weiss GA. Solubilization of a Membrane Protein by Combinatorial Supercharging. ACS Chem. Biol. 2011;6:301–307. doi: 10.1021/cb1001729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majumdar S, Hajduczki A, Vithayathil R, Olsen T, Spitler R, Mendez A, Thompson T, Weiss GA. In vitro evolution of ligands to the membrane protein Caveolin. J. Am. Chem. Soc. 2011;133:9855–9862. doi: 10.1021/ja201792q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 11.Wells JA, Lowman HB. Rapid evolution of peptide and protein binding properties in vitro. Curr. Opin. Struct. Biol. 1992;2:597–604. doi: 10.1016/0958-1669(92)90163-d. [DOI] [PubMed] [Google Scholar]
- 12.Kehoe JW, Kay BK. Filamentous phage display in the new millennium. Chem. Rev. 2005;105:4056–4072. doi: 10.1021/cr000261r. [DOI] [PubMed] [Google Scholar]
- 13.Lamboy JA, Tam PY, Lee LS, Jackson PJ, Avrantinis SK, Lee HJ, Corn RM, Weiss GA. Chemical and genetic wrappers for improved phage and RNA display. Chembiochem. 2008;9:2846–2852. doi: 10.1002/cbic.200800366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majumdar S, Hajduczki A, Mendez AS, Weiss GA. Phage display of functional, full-length human and viral membrane proteins. Bioorg. Med. Chem. Lett. 2008;18:5937–5940. doi: 10.1016/j.bmcl.2008.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clackson T, Wells JA. In vitro selection from protein and peptide libraries. Trends Biotechnol. 1994;12:173–184. doi: 10.1016/0167-7799(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 16.Wilson DR, Finlay BB. Phage display: applications, innovations, and issues in phage and host biology. Can. J. Microbiol. 1998;44:313–329. [PubMed] [Google Scholar]
- 17.Takahashi TT, Austin RJ, Roberts RW. mRNA display: ligand discovery, interaction analysis and beyond. Trends Biochem. Sci. 2003;28:159–165. doi: 10.1016/S0968-0004(03)00036-7. [DOI] [PubMed] [Google Scholar]
- 18.Paschke M, Hohne W. A twin-arginine translocation (Tat)-mediated phage display system. Gene. 2005;350:79–88. doi: 10.1016/j.gene.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Weiss GA, Wells JA, Sidhu SS. Mutational analysis of the major coat protein of M13 identifies residues that control protein display. Protein Sci. 2000;9:647–654. doi: 10.1110/ps.9.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bothmann H, Pluckthun A. Selection for a periplasmic factor improving phage display and functional periplasmic expression. Nat. Biotechnol. 1998;16:376–380. doi: 10.1038/nbt0498-376. [DOI] [PubMed] [Google Scholar]
- 21.Steiner D, Forrer P, Stumpp MT, Pluckthun A. Signal sequences directing cotranslational translocation expand the range of proteins amenable to phage display. Nat. Biotechnol. 2006;24:823–831. doi: 10.1038/nbt1218. [DOI] [PubMed] [Google Scholar]
- 22.Singer SJ, Nicolson GL. The fluid mosaic model of the structure of cell membranes. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 23.Resh MD. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 24.Jackson JH, Cochrane CG, Bourne JR, Solski PA, Buss JE, Der CJ. Farnesol modification of Kirsten-ras exon 4B protein is essential for transformation. Proc. Natl. Acad. Sci. USA. 1990;87:3042–3046. doi: 10.1073/pnas.87.8.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferguson MA, Williams AF. Cell-surface anchoring of proteins via glycosyl-phosphatidylinositol structures. Annu. Rev. Biochem. 1988;57:285–320. doi: 10.1146/annurev.bi.57.070188.001441. [DOI] [PubMed] [Google Scholar]
- 26.Liang X, Lu Y, Neubert TA, Resh MD. Mass spectrometric analysis of GAP-43/neuromodulin reveals the presence of a variety of fatty acylated species. J. Biol. Chem. 2002;277:33032–33040. doi: 10.1074/jbc.M204607200. [DOI] [PubMed] [Google Scholar]
- 27.He Q, Dent EW, Meiri KF. Modulation of actin filament behavior by GAP-43 (neuromodulin) is dependent on the phosphorylation status of serine 41, the protein kinase C site. J. Neurosci. 1997;17:3515–3524. doi: 10.1523/JNEUROSCI.17-10-03515.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chapman ER, Au D, Alexander KA, Nicolson TA, Storm DR. Characterization of the calmodulin binding domain of neuromodulin. Functional significance of serine 41 and phenylalanine 42. J. Biol. Chem. 1991;266:207–213. [PubMed] [Google Scholar]
- 29.Rossenu S, Leyman S, Dewitte D, Peelaers D, Jonckheere V, Van Troys M, Vandekerckhove J, Ampe C. A phage display-based method for determination of relative affinities of mutants. Application of the actin-binding motifs in thymosin beta 4 and the villin headpiece. J. Biol. Chem. 2003;278:16642–16650. doi: 10.1074/jbc.M208311200. [DOI] [PubMed] [Google Scholar]
- 30.Blobel G. Intracellular protein topogenesis. Proc. Natl. Acad. Sci. USA. 1980;77:1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sapay N, Montserret R, Chipot C, Brass V, Moradpour D, Deleage G, Penin F. NMR structure and molecular dynamics of the in-plane membrane anchor of nonstructural protein 5A from bovine viral diarrhea virus. Biochemistry. 2006;45:2221–2233. doi: 10.1021/bi0517685. [DOI] [PubMed] [Google Scholar]
- 32.Balali-Mood K, Bond PJ, Sansom MS. Interaction of monotopic membrane enzymes with a lipid bilayer: a coarse-grained MD simulation study. Biochemistry. 2009;48:2135–2145. doi: 10.1021/bi8017398. [DOI] [PubMed] [Google Scholar]
- 33.Vasudevan SV, Schulz J, Zhou C, Cocco MJ. Protein folding at the membrane interface, the structure of Nogo-66 requires interactions with a phosphocholine surface. Proc. Natl. Acad. Sci. USA. 2010;107:6847–6851. doi: 10.1073/pnas.0911817107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raman P, Cherezov V, Caffrey M. The Membrane Protein Data Bank. Cell. Mol. Life Sci. 2006;63:36–51. doi: 10.1007/s00018-005-5350-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eisenberg D. Three-dimensional structure of membrane and surface proteins. Annu. Rev. Biochem. 1984;53:595–623. doi: 10.1146/annurev.bi.53.070184.003115. [DOI] [PubMed] [Google Scholar]
- 36.Schulz GE. beta-Barrel membrane proteins. Curr. Opin. Struct. Biol. 2000;10:443–447. doi: 10.1016/s0959-440x(00)00120-2. [DOI] [PubMed] [Google Scholar]
- 37.Burkhard KA, Wilks A. Characterization of the outer membrane receptor ShuA from the heme uptake system of Shigella dysenteriae. Substrate specificity and identification of the heme protein ligands. J. Biol. Chem. 2007;282:15126–15136. doi: 10.1074/jbc.M611121200. [DOI] [PubMed] [Google Scholar]
- 38.Cobessi D, Meksem A, Brillet K. Structure of the heme/hemoglobin outer membrane receptor ShuA from Shigella dysenteriae: heme binding by an induced fit mechanism. Proteins. 78:286–294. doi: 10.1002/prot.22539. [DOI] [PubMed] [Google Scholar]
- 39.Sun G, Pal S, Sarcon AK, Kim S, Sugawara E, Nikaido H, Cocco MJ, Peterson EM, de la Maza LM. Structural and functional analyses of the major outer membrane protein of Chlamydia trachomatis. J. Bacteriol. 2007;189:6222–6235. doi: 10.1128/JB.00552-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelley RF, Totpal K, Lindstrom SH, Mathieu M, Billeci K, Deforge L, Pai R, Hymowitz SG, Ashkenazi A. Receptor-selective mutants of apoptosis-inducing ligand 2/tumor necrosis factor-related apoptosis-inducing ligand reveal a greater contribution of death receptor (DR) 5 than DR4 to apoptosis signaling. J. Biol. Chem. 2005;280:2205–2212. doi: 10.1074/jbc.M410660200. [DOI] [PubMed] [Google Scholar]
- 41.Bulieris PV, Behrens S, Holst O, Kleinschmidt JH. Folding and insertion of the outer membrane protein OmpA is assisted by the chaperone Skp and by lipopolysaccharide. J. Biol. Chem. 2003;278:9092–9099. doi: 10.1074/jbc.M211177200. [DOI] [PubMed] [Google Scholar]
- 42.Lazar SW, Kolter R. SurA assists the folding of Escherichia coli outer membrane proteins. J. Bacteriol. 1996;178:1770–1773. doi: 10.1128/jb.178.6.1770-1773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murase K, Morrison KL, Tam PY, Stafford RL, Jurnak F, Weiss GA. EF-Tu binding peptides identified, dissected, and affinity optimized by phage display. Chem. Biol. 2003;10:161–168. doi: 10.1016/s1074-5521(03)00025-5. [DOI] [PubMed] [Google Scholar]
- 44.Sidhu SS, Weiss GA, Wells JA. High copy display of large proteins on phage for functional selections. J. Mol. Biol. 2000;296:487–495. doi: 10.1006/jmbi.1999.3465. [DOI] [PubMed] [Google Scholar]
- 45.Lee CV, Sidhu SS, Fuh G. Bivalent antibody phage display mimics natural immunoglobulin. J. Immunol. Methods. 2004;284:119–132. doi: 10.1016/j.jim.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 46.Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.