Abstract
Background
Despite overwhelming evidence that major depression is highly heritable, recent studies have localized only a single depression-related locus reaching genome-wide significance and have yet to identify a causal gene. Focusing on family-based studies of quantitative intermediate phenotypes or endophenotypes, in tandem with studies of unrelated individuals using categorical diagnoses, should improve the likelihood of identifying major depression genes. However, there is currently no empirically-derived statistically rigorous method for selecting optimal endophentypes for mental illnesses. Here we describe the Endophenotype Ranking Value (ERV), a new objective index of the genetic utility of endophenotypes for any heritable illness.
Methods
Applying ERV analysis to a high-dimensional set of over 11,000 traits drawn from behavioral/neurocognitive, neuroanatomic, and transcriptomic phenotypic domains, we identified a set of objective endophenotypes for recurrent major depression in a sample of Mexican American individiauls (n=1122) from large randomly-selected extended pedigrees.
Results
Top-ranked endophenotypes included the Beck Depression Inventory, bilateral ventral diencephalon volume and expression levels of the RNF123 transcript. To illustrate the utility of endophentypes in this context, each of these traits were utlized along with disease status in bivariate linkage analysis. A genome-wide significant quantitative trait locus was localized on chromsome 4p15 (LOD=3.5) exhibiting pleiotropic effects on both the endophenotype (lymphocyte-derived expression levels of the RNF123 gene) and disease risk.
Conclusions
The wider use of quantitative endophentpyes, combined with unbiased methods for selecting among these measures, should spur new insights into the biological mechanisms that influence mental illnesses like major depression.
Keywords: major depression, recurrent major depression, endophenotype, endophenotype ranking, linkage, family studies
Introduction
Major depression is a clinically heterogeneous and common mental illness (1) with lifetime prevalence rates approaching 20% (2, 3). Despite overwhelming evidence that major depression is heritable (4), recent case-control association studies have failed to identify a locus reaching genome-wide significance (5–10), leading some to conclude that common genetic variants with substantial odds ratios are unlikely to exist for the disease (7). In contrast, recent family-based linkage studies of major depression identified a significant quantitative trait locus (QTL) on chromosome 3p25-26 (LOD=4.0) in a large sample of affected sibling pairs (11). This effect replicated in a smaller sample of individuals ascertained for heavy smoking (12). However, the causal gene(s) for this QTL remain to be identified. Given our slow pace of discovery, new approaches may be necessary to improve understanding of specific causal genes influencing risk of mental illness. One possible approach to speed gene localization/identification is the use of informative quantitative intermediate phenotypes or endophenotypes in families (13, 14). Such an approach has strategic benefits (e.g., simultaneous identification of endophenotypes, increased power to identify genes, increased power to detect rare functional variants) over the more common paradigm that has focused upon collections of unrelated individuals and relied solely upon categorical diagnoses. Endophenotype exploitation should improve the likelihood of identifying major depression genes (13, 15–17).
Although the application of allied phenotypes has been successful in other complex illnesses (14, 18–20), difficulties choosing appropriate endophenotypes for mental disorders have limited their use in psychiatry, where relatively less is know about the biological mechanisms that predispose illness than in other areas of medicine. While the endophenotype concept is widely espoused in psychiatric genetics (14, 16, 21), a formal or standardized approach for the identification of endophenotypes is lacking. Most studies employ purely phenotypic correlations between disease risk and a quantitative risk factor to define putative endophenotypes. However, the endophenotype concept fundamentally depends upon the existence of joint genetic determination of both endophenotype and disease risk (13, 14). This obligatory pleiotropy is most efficiently tested using family-based observations to assess both the heritability of the endophenotype and its genetic correlation with disease liability. To facilitate the identification of optimal endophenotypes, we developed the Endophenotype Ranking Value (ERV), a novel objective index of the genetic utility of endophenotypes for an illness. The ERV provides an unbiased and empirically derived method for choosing appropriate endophenotypes in a manner that balances the strength of the genetic signal for the endophenotype and the strength of its relation to the disorder of interest. It is defined using the square-root of the heritability of the illness (hi2), the square-root of the heritability of the endophenotype (he2), and their genetic correlation (ρg), and is expressed in the following formula:
ERV values vary between 0 and 1, where higher values indicate that the endophenotype and the illness are more strongly influenced by shared genetic factors. This method necessitates that endophenotypes be heritable and have some level of pleiotropy with the studied illness, reducing the heterogeneity of the disease and focusing on the proportion of shared genetic factors influencing both the endophenotype and the illness. An advantage of the ERV approach is that very large numbers of potential endophenotypes can be efficiently assessed prior to conducting molecular genetic analyses, analogous to high-throughput screening methods developed for drug discovery. Furthermore, the ERV approach is applicable to any heritable disease and any set of potentially relevant traits.
Applying ERV analysis to a high dimensional set of traits we identified a set of significant endophenotypes for recurrent major depression (rMDD). We focus on recurrent depression to reduce the clinical heterogeneity of the disorder and potentially increase the genetic control over the illness (3, 6). We perform an automated high dimensional search for endophenotypes via the ranking of 37 behavioral/neurocognitive, 85 neuroanatomic and 11,337 lymphocyte-based transcriptional candidate endophenotypes for rMDD using data acquired from 1,122 Mexican-American individuals from large randomly ascertained extended pedigrees who participated in the "Genetics of Brain Structure and Function" study. Finally, we employed the top-ranked endophentypes in bivariate linkage analysis, localizing a significant QTL exhibiting pleiotropic effects on both endophenotype and disease risk.
Materials and Methods
Participants
1,122 Mexican-American individuals from extended pedigrees (71 families, average size 14.9 [1–87] people) were included in the analysis. Participants were 64% female and ranged in age from 18 to 97 (mean±SD 47.11±14.2) years. Individuals in this cohort have actively participated in research for over 18 years and were randomly selected from the community with the constraints that they are of Mexican-American ancestry, part of a large family and live within the San Antonio region (see (22) for recruitment details). No other inclusion or exclusion criteria were imposed in the initial study. However, individuals were excluded from scanning for MRI contraindications. In addition, individuals were excluded from scanning and neurocognitive evaluation for history of neurological illnesses, stroke or other major neurological event. Reported pedigree relationships were empirically verified with autosomal markers and intra-familial relationships were edited if necessary (see Table 1 for familial relationships). All participants provided written informed consent on forms approved by the IRBs at the University of Texas Health Science Center San Antonio (UTHSCSA) and at Yale University.
Table 1. Pair-Wise Pedigree Relationships.
| Relationship | Number of Pairs |
|---|---|
| Parent-offspring | 689 |
| Siblings | 784 |
| Grandparent-grandchild | 122 |
| Avuncular | 1248 |
| Half siblings | 135 |
| 1st cousins | 1602 |
| 3rd degree | 2128 |
| 4th degree | 2235 |
| 5th degree | 1341 |
| 6th degree | 584 |
| 7th degree | 309 |
| 8th degree | 36 |
Diagnostic Assessment
All participants received the Mini-International Neuropsychiatric Interview (23), a semi-structured interview augmented to include items on lifetime diagnostic history. Masters and doctorate level research staff, with established reliability (κ>=0.85) for affective disorders, conducted all interviews. All subjects with possible psychopathology were discussed in case conferences that included licensed psychologists or psychiatrists. Lifetime consensus diagnoses were determined based on available medical records, the MINI interview and the interviewer’s narrative. Recurrent major depression was defined as two or more distinct episodes of depression meeting DSM-IV criteria.
Behavioral and Neurocognitive Assessment
Each participant received a 90-min neuropsychological evaluation consisting of standard and computerized measures (24, 25). Thirty-five neurocognitive variables were derived from 17 separate neuropsychological tests, including measures of attention/concentration, executive processing, working memory, declarative memory, language processing, intelligence and emotional processing. In addition, participants completed two questionnaires indexing depressive mood: the Beck Depression Inventory II (BDI (26)) and the neuroticism questions of the Eysenck Personality Questionnaire (27).
Image Acquisition and Processing
MRI data were acquired on a 3T Siemens Trio scanner with an 8-channel head coil in the Research Imaging Institute, UTHSCSA. 800 µm isotropic anatomic images were acquired for each subject using a retrospective motion-corrected protocol (28). This protocol included the acquisition of seven full-resolution volumes using a T1-weighted, 3D TurboFlash sequence with the following scan parameters: TE/TR/TI=3.04/2100/785 ms, flip angle=13°. Surface-based image analyses were conducted with FreeSurfer (29, 30) following standardized protocols (31). T1-weighted images were segmented into gray matter thickness measures for 53 cortical regions and 21 subcortical volumes (averaged across both hemispheres).
T2-weighted imaging data were acquired using a 1mm isotropic, turbo-spin-echo FLAIR sequence with the following parameters: TR/TE/TI/Flip angle/ETL=5s/353ms/1.8s/180°/221. White-matter hyperintensities were manually delineated in 3D-space using in-house software by experienced neuroanatomists with high (r2>0.90) test-retest reproducibility (32).
Diffusion tensor imaging (DTI) data acquisition used a single-shot single spin-echo, echo-planar imaging sequence with a spatial resolution of 1.7×1.7×3.0mm (TR/TE=8000/87ms, FOV=200mm, 55 directions, b=0 and 800 s/mm2). Fractional anisotropy values were estimated for each subject on thirteen tracts using Tract-Based Spatial Statistics software (TBSS) (33). DTI images provided fractional anisotropy indices for 13 white-matter tracts (averaged across both hemispheres).
Transcriptional Profiling
Transcriptional profiling followed procedures described by Göring and colleagues (34). Total RNA was isolated from lymphocytes and hybridized to Illumina Sentrix Human Whole Genome (WG-6) Series 1 BeadChips. These BeadChips simultaneously probe ~48,000 transcripts, representing more than 25,000 annotated human genes. Although we previously identified 20,413 quantitative transcripts in lymphocytes, we only examined those with heritabilities greater than or equal to 0.20 (n=11,337) in the current analysis.
Genotyping
DNA extracted from lymphocytes was used in polymerase chain reactions (PCRs) for the amplification of individual DNA at 432 dinucleotide repeat microsatellite loci (STRs), spaced approximately 10 centiMorgan (cM) intervals apart across the 22 autosomes, with fluorescently labeled primers from the MapPairs Human Screening set, versions 6 and 8 (Research Genetics, Huntsville, AL). PCRs were performed separately according to manufacturer specifications in Applied Biosystems 9700 thermocyclers (Applied Biosystems, Foster City, CA). For each individual, the products of separate PRCs were pooled using the Robbins Hydra-96 Microdispenser, and a labeled size standard was added to each pool. The pooled PCR products were loaded into an ABI PRISM 377 or 3100 Genetic Analyzer for laser based automated genotyping. The STRs and standards were detected and quantified, and genotypes were scored using the Genotyper software package (Applied Biosystems).
Quantitative Genetic Analyses
All analyses were conducted with SOLAR (35), which employs maximum likelihood variance decomposition methods to determine the relative importance of genetic and environmental influences by modeling the covariance among family members as a function of genetic proximity (see Supplement for detail on variance components methods).
The Endophenotype Ranking Value (ERV) represents the standardized genetic covariance between the endophenotype (denoted by the subscript, e) and illness (denoted by the subscript, i), and is defined as ERVie = |√hi2√he2ρg|. Heritability (h2) represents the portion of the phenotypic variance accounted for by additive genetic variance (h2 = σ2g/σ2p). Genetic correlation represents the common genetic covariance between two traits, or pleiotropy (36). Bivariate quantitative genetic analysis was used to estimate the genetic (ρg) and environmental (ρe) correlations between each potential endophenotype and rMDD. The phenotypic correlation (ρp), which quantifies the overall relationship between the two traits, can be derived from the genetic and environmental correlations as ρp = ρg√(h2eh2i) + ρe√((1−h2e)(1−h2i)). These parameters are estimated by jointly utilizing all available pedigree information with a multivariate normal threshold model for combined dichotomous/continuous traits (37, 38). The significance of the ERV was tested by comparing the ln likelihood for the restricted null model (with ρg constrained to equal 0) against the ln likelihood for the alternative model in which the ρg parameter is estimated. The resultant likelihood ratio test is asymptotically distributed as a chi-square with a single degree of freedom. The corresponding p-value is identical to that used for genetic correlation. Prior to analysis, endophenotypes were normalized using an inverse Gaussian transformation. Age, sex, age×sex, age2, and age2×sex were included as covariates whose effects were simultaneously estimated in all analyses.
Bivariate Linkage Analysis
Bivariate linkage analysis exploits the genetic covariance between the endophenotype and the illness to improve the power to localize QTLs and to detect QTL-specific pleiotropic effects (37). After addressing (by blanking, recalling, or retyping) mistyping errors identified using Simwalk II (39), genotype data were used to compute maximum likelihood estimates of allele frequencies. Matrices of empirical estimates of identity-by-descent (IBD) allele sharing at points throughout the genome for every relative pair were computed using the Loki package (40). We used high-resolution chromosomal maps provided by deCODE genetics (41). For the localization of QTLs, we performed both univariate and bivariate variance components linkage analyses by employing the models for combined analysis of quantitative and dichotomous phenotypes described by Williams and colleagues (37, 38). Once a genome-wide significant localization was made, formal single degree of freedom likelihood ratio tests for pleiotropy were performed to test the specific hypothesis that a QTL at that location influenced a given endophenotype/rMDD risk (36).
Results
Heritability of Recurrent Major Depression
Two hundred and fifteen individuals met criteria for lifetime rMDD (19% of the sample; 73% female). Eighty-six individuals were clinically depressed at the time of the assessment. The estimated heritability for lifetime rMDD was h2=0.463 (standard error ±0.12), p=4.0×10−6. We previously demonstrated that this heritability estimate is not significantly influenced by common environmental factors as indexed by shared household (22). Additionally, there was no evidence for dominance (p=0.14) or additive×additive epistasis (p=0.18), suggesting that the heritability estimate reflects additive genetic factors.
Potential Behavioral/Neurocognitive Endophenotypes
The 10 top-ranked behavioral/cognitive endophenotypes are presented in Table 2. The BDI was the highest ranked endophenotype in this class. Although the BDI was developed as an index of mood state, the heritability of this measure was h2=0.254 (±0.07), p=5.6×10−5, demonstrating that 25% of the variability on this measure is due to additive genetic factors. The genetic correlation between the BDI and the neuroticism questions from the Eysenck, the second best ranked endophenotype in this domain, was ρG=0.870 (±0.09), p=3.3×10−4, suggesting significant pleiotropy and potential redundancy between these two measures. Top-ranked cognitive measures include indices of working and declarative memory, attention and emotion recognition.
Table 2. 10 Top-Ranked Endophenotypes per Domain for Recurrent Major Depression.
10 top ranked endophenotypes for recurrent major depression in the categories of behavioral/cognitive, neuroimaging and RNA transcripts (see Supplement for the complete rankings). Genetic correlations are between the respective endophenotypes and lifetime affection status. Endophenotype heritability estimates were estimated as part of bivariate models.
| Endophenotypes | ERV | P-value | Genetic Correlation (ρg) |
Heritability (h2) |
|---|---|---|---|---|
| Behavioral/Neurocognitive | ||||
| Beck Depression Inventory II | 0.263 | 1.9×10−5 | 0.825 | 0.253 |
| EPQ Neuroticism | 0.238 | 1.7×10−4 | 0.739 | 0.228 |
| Declarative Memory (CVLT Recognition) | 0.136 | 5.4×10−2 | −0.338 | 0.338 |
| Working Memory (Digit Span Forward) | 0.142 | 5.6×10−2 | −0.295 | 0.490 |
| Working Memory (Letter-Number | 0.135 | 6.3×10−2 | −0.267 | 0.541 |
| Penn Facial Memory (Immediate) | 0.127 | 6.9×10−2 | −0.319 | 0.344 |
| Penn Facial Memory (Delayed) | 0.134 | 8.1×10−2 | −0.295 | 0.439 |
| Attention (CPT hits) | 0.119 | 8.3×10−2 | −0.387 | 0.202 |
| Attention (Trails A) | 0.121 | 9.6×10−2 | 0.303 | 0.340 |
| Penn Emotion Recognition | 0.117 | 1.0×10−1 | −0.288 | 0.347 |
| Neuroimaging | ||||
| Ventral Diencephalon Volume | 0.240 | 3.9×10−3 | −0.425 | 0.694 |
| Parietal Hyperintensity Volume | 0.282 | 7.8×10−3 | 0.569 | 0.573 |
| Hippocampus Volume | 0.204 | 1.2×10−2 | −0.347 | 0.771 |
| Pallidum Volume | 0.203 | 1.3×10−2 | −0.396 | 0.562 |
| Cerebellar White Matter Volume | 0.218 | 1.3×10−2 | −0.443 | 0.524 |
| Frontal Hyperintensity Volume | 0.255 | 1.3×10−2 | 0.483 | 0.635 |
| CorticoSpinal Tract (FA) | 0.208 | 2.1×10−2 | −0.900 | 0.101 |
| Subcortical Hyperintensity Volume | 0.213 | 4.1×10−2 | 0.473 | 0.459 |
| Superior Parietal Gyrus Thickness | 0.178 | 4.5×10−2 | 0.363 | 0.480 |
| Thalamus Proper Volume | 0.172 | 4.8×10−2 | −0.294 | 0.739 |
| Transcriptional | ||||
| RNF123 (3p24) | 0.323 | 5.2×10−6 | −0.943 | 0.209 |
| PDXK (21q22) | 0.331 | 1.1×10−5 | −0.689 | 0.489 |
| ZFP64 (20q13) | 0.352 | 2.0×10−5 | −0.711 | 0.470 |
| RWDD2A (6q14) | 0.260 | 2.3×10−5 | 0.666 | 0.337 |
| B4GALT7 (5q35) | 0.276 | 3.6×10−5 | −0.732 | 0.309 |
| MARK2 (11q12) | 0.180 | 3.9×10−5 | −0.399 | 0.412 |
| GADD45A (1p31) | 0.344 | 4.0×10−5 | 0.729 | 0.432 |
| PIGN (18q21) | 0.274 | 7.9×10−5 | 0.646 | 0.399 |
| HTT (4p16) | 0.225 | 7.9×10−5 | −0.546 | 0.358 |
| ABHD12 (20p11) | 0.269 | 1.1×10−4 | 0.755 | 0.272 |
Potential Neuroimaging Endophenotypes
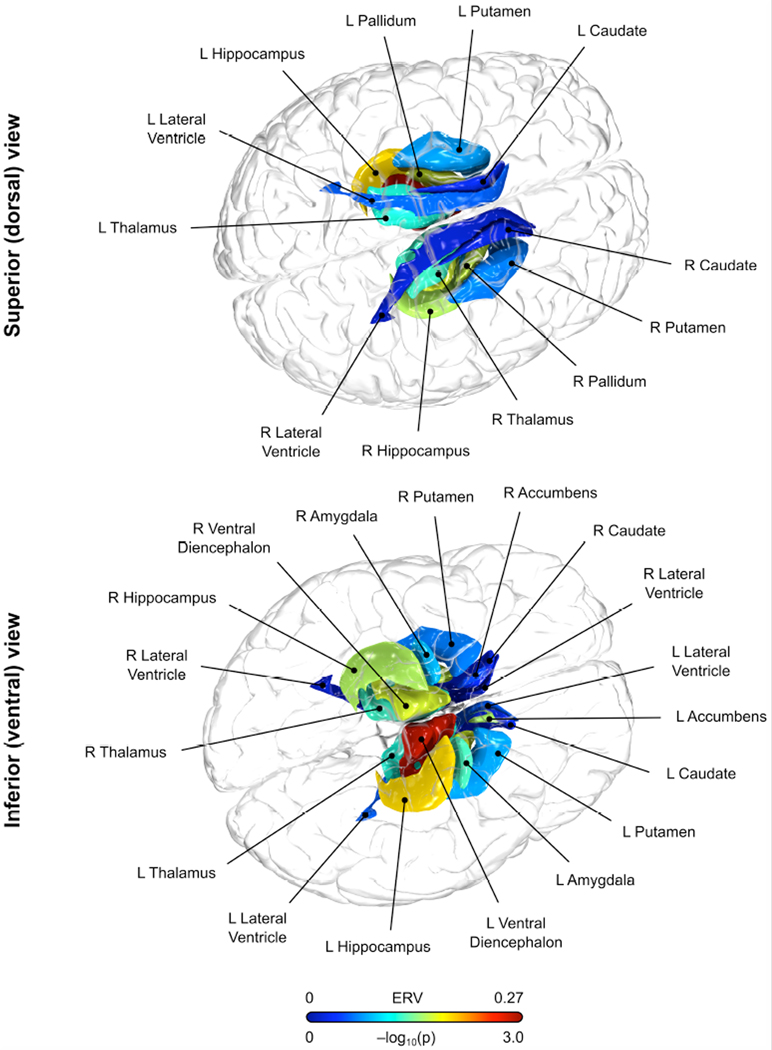
The top ranked brain region was bilateral ventral diencephalon volume (Table 2), a region primarily comprised of the hypothalamus. As part of the hypothalamic-pituitary-adrenal (HPA) axis, the hypothalamus mediates neuroendocrine and neurovegetative functions and has been consistently implicated in the neurobiology of depression (42). HPA axis activity is regulated in part by the hippocampus and amygdala (42), both regions with reasonably high ERV ranking (3rd and 13th ranked, respectively). Our results suggest that the genetic factors that influence the structure of these subcortical regions (Figure 1) also confer risk for rMDD. Additionally, white-matter hyperintensity measures, which are associated with aging, cerebrovascular dysfunction, smoking and a host of other depression-related pathologies (43), were highly ranked endophenotypes for rMDD. This result is consistent with and extends the vascular depression hypothesis (44), by suggesting common genetic factors increase risk for rMDD and white-matter hyperintensities.
Figure 1. ERV statistics for subcortical brain regions and recurrent major depression.
Volume measurements of subcortical nuclei were found to share genetic variance with liability for recurrent major depression in extended pedigrees selected without regard to phenotype. ERV statistics, which provide an unbiased and empirically derived method for choosing appropriate endophenotypes, were strongest for the ventral diencephalon volume, a region primarily comprised of the hypothalamus. For anatomical reference, in this image the cortex is shown as a semi-transparent structure.
Potential Transcriptional Endophenotypes
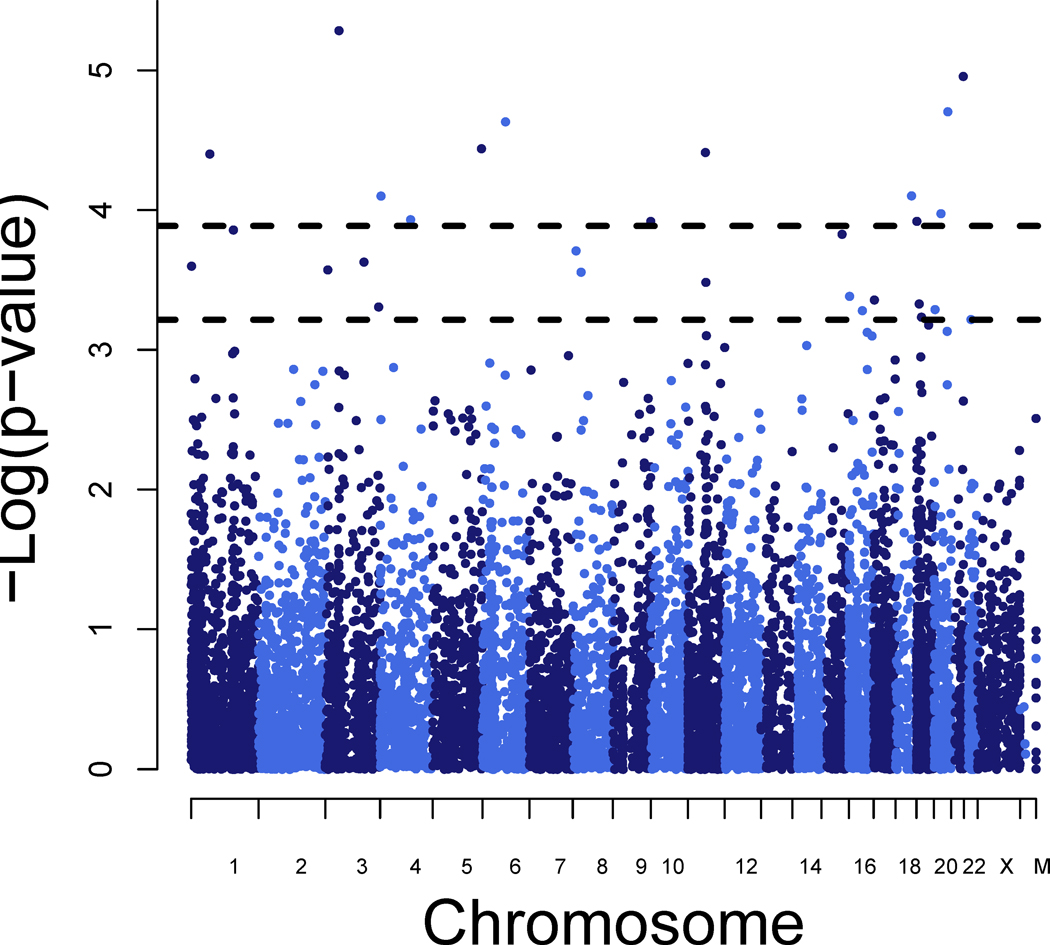
ERV analyses on 11,337 are presented in Figure 2 and top ranked transcriptional endophenotypes for rMDD are presented in Table 2. The top-ranking transcript RNF123, is a member of the E3 ubiquitin-protein ligase family, which have diverse functions including protein degradation and modulation of protein assembly, structure, function and localization (45, 46). Other members of the ubiquitin-proteosome system were previously implicated in anxiety, depression and vulnerability to seizures (47, 48). PDXK, an essential cofactor in the intermediate metabolism of amino acids and neurotransmitters, including serotonin and dopamine (49), confers risk for Parkinson disease (50) and epilepsy (49). Additionally, MARK2 and ABHD12 have previously been implicated in neuronal migration (51), degeneration (52) and regulation of endocannabinoid signaling pathways (53), respectively. Although other identified transcripts are less obvious candidates for rMDD risk, they may represent novel genes whose functions are not fully understood and may extend to depression phenotypes.
Figure 2. Manhattan plot depicting whole transcriptomic search for expression-based endophenotypes for recurrent major depression.
ERV values were calculated for 11,337 detected lymphocyte-based transcripts and recurrent major depression. Dashed lines reflect cutoff points for FDR < 0.10 (13 transcripts) and FDR < 0.25 (29 transcripts).
Genome-Wide Bivariate Linkage Analyses Using rMDD and Top-Ranked Endophenotypes
We performed a genome-wide linkage-based search for pleiotropic quantitative trait loci influencing disease risk and the top-ranked endophenotype from each class: BDI, bilateral ventral diencephalon volume and the RNF123 transcript. First, standard univariate linkage analyses were performed. Two traits exhibited genome-wide or near genome-wide significance QTLs. The best univariate score for rMDD was found on chromosome 4 at 47 cM (LOD=2.98, nominal p=0.00011). While not reaching traditional genome-wide significance, this results points to a potential disease-related QTL at chromosomal location 4p15. The bilateral ventral diencephalon exhibited an unequivocal genome-wide significant peak on chromosome 7 at 131 cM (LOD=3.40, nominal p=3.8×10−5). Neither BDI nor RNF123 expression levels showed strong evidence for causal QTLs in univariate analysis. Suggestive evidence for a QTL influencing BDI was found on chromosome 17 at 98 cM (LOD=2.57, nominal p=0.0003). We found little evidence for a QTL influencing quantitative RNF123 gene expression levels, with the single best univariate QTL location found on chromosome 6 at 53 cM (LOD=1.81).
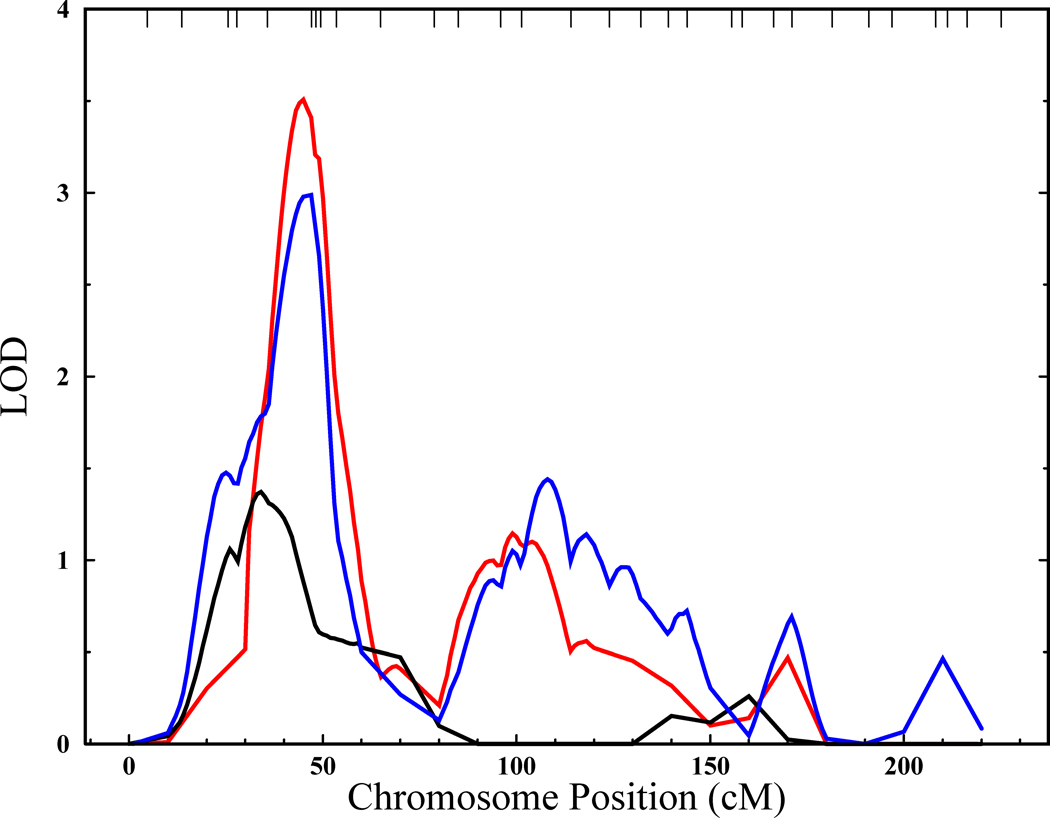
Bivariate linkage analyses were performed to determine if QTL localization could be enhanced via simultaneous analysis with rMDD affection status. The most dramatic improvement in localization was seen for rMDD and RNF123 transcription levels. The bivariate analysis of this endophenotype/disease combination substantially improved the evidence for a QTL located at 4p15 seen in the univariate rMDD results. Figure 3 shows the QTL localization results for the bivariate analysis and the two related univariate analyses. The peak bivariate LOD (scaled to a standard single degree of freedom LOD) for was 3.51 (nominal p=3.8×10−5) at 45 cM, a marked improvement over that seen for rMDD alone. No other rMDD/endophenotype combination provided genome-wide evidence for QTLs.
Figure 3. Detection of a QTL influencing recurrent major depression and RNF123 expression levels on chromosome 4.
Multipoint LOD functions for chromosome 4 in 1,122 individuals from large extended pedigrees from the Genetics of Brain Structure and Function study. The black line represents the univariate linkage analysis for RNF123 expression levels alone. The blue line represents the univariate linkage analysis for recurrent major depression alone. The red line represents the bivariate linkage analysis for recurrent major depression and RNF123 and reaches genome-wide significance (LOD=3.5) at 45 cM (chromosomal band 4p15). The vertical axis is in LOD score units, and the horizontal axis is in units of genetic distance (centi-Morgans, cM) from the p arm telomere.
Table 3 shows the results of likelihood ratio statistic-based formal tests of pleiotropy at the chromosome 4:45cM location obtained from the bivariate analysis of RNF123/rMDD. The marginal results from univariate analysis (technically co-incident linkage (36)) and the strict test of pleiotropy that can be performed using the bivariate linkage model. The chromosome 4 locus significantly influences rMDD (p=4.7×10−5), RNF123 (p=0.0010), and diencephalon volume (p=0.0290), and shows a trend for BDI (p=0.1170). The fact that this QTL influences both risk of rMDD and two of our three best endophenotypes provides additional validation for endophenotype identification, with evidence for rMDD increasing by nearly an order of magnitude. These results strongly support a QTL influencing rMDD and related endophenotypes at chromosome 4p15.
Table 3. Tests of Pleiotropy at the Chromosome 4:45cM Quantitative Trait Locus.
| Trait | Pleiotropy P-value from Bivariate Model |
Co-Incident Linkage P-value from Univariate Model |
|---|---|---|
| Recurrent MDD | 4.7×10−5 | 1.1×10−4 |
| RNF123 expression | 0.0010 | 0.0219 |
| Ventral Diencephalon Volume | 0.0290 | 0.0266 |
| BDI score | 0.1170 | 0.5000 |
Given evidence for a QTL influencing diencephalon volume on chromosome 7, we tested for pleiotropic effects. As expected, these tests revealed a major effect on diencephalon (pleiotropy p-value=1.6×10−5) and rMDD liability (pleiotropy p-value=0.0437). Both of these results are substantially improved over their univariate analogues and only with bivariate analysis do we detect a significant influence of this QTL on rMDD liability. The other two leading endophenotypes show no pleiotropic effects at this QTL.
Discussion
Our results demonstrate the utility of the ERV approach for formally identifying endophenotypes in high dimensional data and provide a novel genome-wide significant QTL for recurrent major depression. Bivariate genetic analyses including a quantitative endophenotype and disease risk significantly improved QTL detection over that observed utilizing diagnosis alone. These results may reflect the improved statistical sensitivity of quantitative over qualitative traits or that endophenotypes index a somewhat less heterogeneous aspect of the pathophysiology associated with mental illnesses (54). In either case, quantitative endophenotypes can significantly improve the potential to localize loci for complex disorders like rMDD, where multiple genes with varying effects and incomplete penetrance are thought to interact with environmental factors to determine illness susceptibility.
The present experiment demonstrates the utility of gene expression measures in peripheral tissues for psychiatric phenotypes. Transcripts can be considered endophenotypes that, while removed from the phenomenology-based diagnosis, are close to gene action, and in the case of primary cis-regulation, provide evidence for a gene’s involvement in the illness. Although brain tissue is ideal for gene expression studies in psychiatry, difficulty obtaining this tissue in genetically informative samples necessitates the use of a surrogate marker and lymphocytes appear to be good surrogates for detection of mental disease-relevant genes (55, 56). The lymphocyte measures used in the present experiment were collected 12–15 years prior to the current assessments, minimizing the potential that these traits were influenced by acute variation in mood or medication usage (57). It is notable that the top-ranked transcriptional endophenotype for rMDD was ranked higher than any of the behavioral/cognitive or neuroimaging traits, including BDI, suggesting that transcripts may provide an important new set of markers for disease risk.
Our single strongest ERV result was observed for quantitative mRNA levels of the RNF123 gene with risk for rMDD. This gene (also known as KPC1) encodes ring finger protein 123 which is likely involved in the regulation of neurite outgrowth via its modulation of the degradation of the cyclin-dependent kinase inhibitor p27(Kip1) (58, 59). p27(Kip1) is involved in increased hippocampal neuronal differentiation via a glucocorticoid receptor function that is observed upon administration of the antidepressant, sertraline (60). Thus, RNF123 appears to be a novel candidate involved in hippocampal neurogenesis of significant relevance to depression risk. We observed a significant negative genetic correlation between RNF123 expression level and disease risk consistent with evidence that RNF123 inhibits p27(Kip1) and depression amelioration. Thus, RNF123 represents a potential drug target for depression.
The dominant paradigm in psychiatric genetic studies focuses on a specific disease itself. However, as with most disease states, this endpoint is relatively distant from the causal anatomic or physiological disruption. In contrast, we supplement disease status with quantitative endophenotypes, selected through an empirically derived process, to identify and characterize genes that influence rMDD. Since these endophenotypes vary within the normal population, it is possible to localize genes influencing them in samples ascertained without regard to a specific phenotype (illness). The endophenotype and normal variation strategy has been successfully applied to the study of other complex diseases such as heart disease (18, 61, 62) obesity (19, 63, 64), diabetes (65, 66), hypertension (20, 67), and osteoporosis (68, 69). However, this strategy has not been effectively applied in the search for mental illness genes.
There is debate regarding the definition of a “good” endophenotype or even if endophenotypes will benefit the search for mental illness genes. We propose that endophenotypes that are heritable and genetically correlated with disease liability can facilitate gene identification. Although both disease and endophenotype must be heritable for the ERV approach, there is no requirement that the endophenotype exhibit higher heritability than the disease itself. Higher heritability estimates do not imply a simpler genetic architecture or improve the potential to localize genes (25). A quantitative endophenotype with a low but significant heritability estimate that is genetically correlated with disease still allows one to rank individuals along a continuous liability distribution (70), increasing power to identify genes. The ERV index includes no assumption about the genetic architecture of an endophenotype. While endophenotypes that are closer to gene action are desirable, this is not a requisite of an endophenotype as information about the genetic simplicity of a particular endophenotype is generally not available or easily quantified. A putative endophenotype with a high ERV value will reflect the genetic component of disease liability better than one with a low ERV. Therefore, even quantitative endophenotypes with complex genetic architectures (involving many genes) can offer major advantages in genetic dissection of disease liability. Indeed, the gold standard endophenotype for heart disease, LDL-C levels, is a complex quantitative trait that is not particularly close to gene action (given that it does not represent a single protein) that was successfully used to find cardiovascular disease risk genes (71, 72).
The present experiment establishes the value of randomly selected families in the search for common psychiatric illnesses genes. While we highlight the optimality of large families for the assessment of heritability, genetic correlations, and ERV calculations, we note that modern high density typing now allows empirical assessment of deep kinship between “unrelated” individuals that could in principle be used to estimate these parameters (albeit very inefficiently due to the remoteness of relationships). Thus, very large previously collected data sets of unrelateds may be of some future value in ERV estimation.
While we demonstrate the utlitiy of the ERV approach, the current experiment has several limitatons. For example, not all potential candiadate ednophentypes for affective disorders were included (17), as this is impractial in large-scale genetic studies. In addition, verification of endophenotypes in independent samples is warrented. However, when the goal of simultaneous evaluation of disease liability and endophenotype is focused on gene discovery, it may be folly to wait for such replication rather than immediately pursuing an independent discovery avenue like deep sequencing of a gene whose expression level is genetically correlated with disease liability. The formally testing and rigorously defining endophenotypes for a given disease should speed the identification of risk genes and improve our understanding of the underlying pathobiological processes. Endophenotypes identified by emperical approaches like the ERV will likely outperform non-objective ”expert”-derived putative endophenotypes.
The endophenotype strategy has the potential to significantly improve our understanding of the genetic architecture of psychiatric illnesses (14). However, choosing optimal endophenotypes for brain-related illness is difficult when relying upon theoretical factors alone. The ERV approach provides an unbiased method for selecting endophenotypes that is applicable to any heritable disease and should facilitate the use of endophentypes in the search for genes influencing brain-related illnesses. Objective formal identification of endophenotypes using the ERV procedure led to improved power to localization causal QTLs influencing risk of major depression and the identification of a novel potential player in depression risk focused on the RNF123 gene, its products, and its pathway.
Supplementary Material
Acknowledgments
We thank the study participants, our research staffs and Irving Gottesman for 50 years of championing endophenotypes in psychiatric genetics. Irv is the true source of the ERV. Financial support for this study was provided by the National Institute of Mental Health grants MH0708143 (PI: DC Glahn), MH078111 (PI: J Blangero), and MH083824 (PI: DC Glahn). Theoretical development of the ERV and its implementation in SOLAR is supported by National Institute of Mental Health grant MH59490 (PI: J Blangero). This investigation was conducted in part in facilities constructed with support from Research Facilities Improvement Program Grant Numbers C06 RR13556 and C06 RR017515 from the National Center for Research Resources, National Institutes of Health. We wish to acknowledge the Azar and Shepperd families and ChemGenex Pharmaceuticals for supporting the transcriptional profiling, sequencing, genotyping and statistical analysis. The supercomputing facilities used for this work at the AT&T Genomics Computing Center were supported in part by a gift from the SBC Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interests. All authors reported no biomedical financial interests or potential conflicts of interest.
References
- 1.Belmaker R, Agam G. Major depressive disorder. N Engl J Med. 2008;358:55–68. doi: 10.1056/NEJMra073096. [DOI] [PubMed] [Google Scholar]
- 2.Kessler R, Berglund P, Demler O, Jin R, Koretz D, Merikangas K, et al. The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R) JAMA. 2003;289:3095–3105. doi: 10.1001/jama.289.23.3095. [DOI] [PubMed] [Google Scholar]
- 3.McGuffin P, Katz R, Watkins S, Rutherford J. A hospital-based twin register of the heritability of DSM-IV unipolar depression. Arch Gen Psychiatry. 1996;53:129–136. doi: 10.1001/archpsyc.1996.01830020047006. [DOI] [PubMed] [Google Scholar]
- 4.Sullivan P, Neale M, Kendler K. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry. 2000;157:1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- 5.Sullivan P, de Geus E, Willemsen G, James M, Smit J, Zandbelt T, et al. Genome-wide association for major depressive disorder: a possible role for the presynaptic protein piccolo. Mol Psychiatry. 2009;14:359–375. doi: 10.1038/mp.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi J, Potash J, Knowles J, Weissman M, Coryell W, Scheftner W, et al. Genome-wide association study of recurrent early-onset major depressive disorder. Mol Psychiatry. 2010 doi: 10.1038/mp.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muglia P, Tozzi F, Galwey N, Francks C, Upmanyu R, Kong X, et al. Genome-wide association study of recurrent major depressive disorder in two European case-control cohorts. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.131. [DOI] [PubMed] [Google Scholar]
- 8.Uher R, Perroud N, Ng M, Hauser J, Henigsberg N, Maier W, et al. Genome-Wide Pharmacogenetics of Antidepressant Response in the GENDEP Project. Am J Psychiatry. 2010 doi: 10.1176/appi.ajp.2009.09070932. [DOI] [PubMed] [Google Scholar]
- 9.Lewis C, Ng M, Butler A, Cohen-Woods S, Uher R, Pirlo K, et al. Genome-wide association study of major recurrent depression in the U.K. population. Am J Psychiatry. 2010;167:949–957. doi: 10.1176/appi.ajp.2010.09091380. [DOI] [PubMed] [Google Scholar]
- 10.Lewis CM, Ng MY, Butler AW, Cohen-Woods S, Uher R, Pirlo K, et al. Genome-wide association study of major recurrent depression in the U.K. population. Am J Psychiatry. 2010;167:949–957. doi: 10.1176/appi.ajp.2010.09091380. [DOI] [PubMed] [Google Scholar]
- 11.Breen G, Webb BT, Butler AW, van den Oord EJ, Tozzi F, Craddock N, et al. A Genome-Wide Significant Linkage for Severe Depression on Chromosome 3: The Depression Network Study. Am J Psychiatry. 2011 doi: 10.1176/appi.ajp.2011.10091342. [DOI] [PubMed] [Google Scholar]
- 12.Pergadia ML, Glowinski AL, Wray NR, Agrawal A, Saccone SF, Loukola A, et al. A 3p26–3p25 Genetic Linkage Finding for DSM-IV Major Depression in Heavy Smoking Families. Am J Psychiatry. 2011 doi: 10.1176/appi.ajp.2011.10091319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blangero J, Williams JT, Almasy L. Novel family-based approaches to genetic risk in thrombosis. J Thromb Haemost. 2003;1:1391–1397. doi: 10.1046/j.1538-7836.2003.00310.x. [DOI] [PubMed] [Google Scholar]
- 14.Gottesman II, Gould TD. The Endophenotype Concept in Psychiatry: Etymology and Strategic Intentions. American Journal of Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 15.Merikangas KR, Chakravarti A, Moldin SO, Araj H, Blangero JC, Burmeister M, et al. Future of genetics of mood disorders research. Biological Psychiatry. 2002;52(6):457–477. doi: 10.1016/s0006-3223(02)01471-3. [DOI] [PubMed] [Google Scholar]
- 16.Insel T, Cuthbert B. Endophenotypes: bridging genomic complexity and disorder heterogeneity. Biol Psychiatry. 2009;66:988–989. doi: 10.1016/j.biopsych.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 17.Hasler G, Drevets WC, Manji HK, Charney DS. Discovering endophenotypes for major depression. Neuropsychopharmacology. 2004;29:1765–1781. doi: 10.1038/sj.npp.1300506. [DOI] [PubMed] [Google Scholar]
- 18.Kathiresan S, Willer C, Peloso G, Demissie S, Musunuru K, Schadt E, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Comuzzie A, Hixson J, Almasy L, Mitchell B, Mahaney M, Dyer T, et al. A major quantitative trait locus determining serum leptin levels and fat mass is located on human chromosome 2. Nature Genetics. 1997;15:273–276. doi: 10.1038/ng0397-273. [DOI] [PubMed] [Google Scholar]
- 20.Cho Y, Go M, Kim Y, Heo J, Oh J, Ban H, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41:527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 21.Ritsner M. Handbook of Neuropsychiatric Biomarkers, Endophenotypes and Genes: Neuroanatomical and Neuroimaging Endophenotypes and Biomarkers. Dordrecht: Springer; 2009. [Google Scholar]
- 22.Olvera RL, Bearden CE, Velligan DI, Almasy L, Carless MA, Curran JE, et al. Common genetic influences on depression, alcohol, and substance use disorders in Mexican-American families. Am J Med Genet B Neuropsychiatr Genet. 2011 doi: 10.1002/ajmg.b.31196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheehan DV, Lecrubier Y, Sheehan KH, Amorim P, Janavs J, Weiller E, et al. The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry. 1998;59 Suppl 20:22–33. quiz 34–57. [PubMed] [Google Scholar]
- 24.Glahn D, Almasy L, Blangero J, Burk G, Estrada J, Peralta J, et al. Adjudicating neurocognitive endophenotypes for schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:242–249. doi: 10.1002/ajmg.b.30446. [DOI] [PubMed] [Google Scholar]
- 25.Glahn D, Almasy L, Barguil M, Hare E, Peralta J, Kent JJ, et al. Neurocognitive endophenotypes for bipolar disorder identified in multiplex multigenerational families. Arch Gen Psychiatry. 2010;67:168–177. doi: 10.1001/archgenpsychiatry.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beck AT, Steer RA, GK B. Manual for the Beck Depression Inventory-II. San Antonio, TX: Psychological Corporation; 1996. [Google Scholar]
- 27.Eysenck HJ, SBG E. Manual of the Eysenck Personality Questionnaire. San Diego, CA: Educational and Industrial Testing Service; 1975. [Google Scholar]
- 28.Kochunov P, Lancaster J, Glahn D, Purdy D, Laird A, Gao F, et al. Retrospective motion correction protocol for high-resolution anatomical MRI. Hum Brain Mapp. 2006;27:957–962. doi: 10.1002/hbm.20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 30.Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 31.Winkler A, Kochunov P, Blangero J, Almasy L, Zilles K, Fox P, et al. Cortical Thickness or Grey Matter Volume? The Importance of Selecting the Phenotype for Imaging Genetics Studies. Neuroimage. 2009 doi: 10.1016/j.neuroimage.2009.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kochunov P, Glahn D, Winkler A, Duggirala R, Olvera R, Cole S, et al. Analysis of genetic variability and whole genome linkage of whole-brain, subcortical, and ependymal hyperintense white matter volume. Stroke. 2009;40:3685–3690. doi: 10.1161/STROKEAHA.109.565390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith SM, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols TE, Mackay CE, et al. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. NeuroImage. 2006;31:1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 34.Goring HH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 35.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. American Journal of Human Genetics. 1998;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Almasy L, Dyer TD, Blangero J. Bivariate quantitative trait linkage analysis: pleiotropy versus co-incident linkages. Genet Epidemiol. 1997;14:953–958. doi: 10.1002/(SICI)1098-2272(1997)14:6<953::AID-GEPI65>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 37.Williams JT, Van Eerdewegh P, Almasy L, Blangero J. Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. I. Likelihood formulation and simulation results. Am J Hum Genet. 1999;65:1134–1147. doi: 10.1086/302570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams JT, Begleiter H, Porjesz B, Edenberg HJ, Foroud T, Reich T, et al. Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. II. Alcoholism and event-related potentials. Am J Hum Genet. 1999;65:1148–1160. doi: 10.1086/302571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sobel E, Papp JC, Lange K. Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet. 2002;70:496–508. doi: 10.1086/338920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heath SC. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet. 1997;61:748–760. doi: 10.1086/515506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 42.Nestler E, Barrot M, DiLeone R, Eisch A, Gold S, Monteggia L. Neurobiology of depression. Neuron. 2002;34:13–25. doi: 10.1016/s0896-6273(02)00653-0. [DOI] [PubMed] [Google Scholar]
- 43.Fazekas F, Kleinert R, Offenbacher H, Schmidt R, Kleinert G, Payer F, et al. Pathologic correlates of incidental MRI white matter signal hyperintensities. Neurology. 1993;43:1683–1689. doi: 10.1212/wnl.43.9.1683. [DOI] [PubMed] [Google Scholar]
- 44.Alexopoulos G, Meyers B, Young R, Campbell S, Silbersweig D, Charlson M. 'Vascular depression' hypothesis. Arch Gen Psychiatry. 1997;54:915–922. doi: 10.1001/archpsyc.1997.01830220033006. [DOI] [PubMed] [Google Scholar]
- 45.Doolittle M, Ben-Zeev O, Bassilian S, Whitelegge J, Péterfy M, Wong H. Hepatic lipase maturation: a partial proteome of interacting factors. J Lipid Res. 2009;50:1173–1184. doi: 10.1194/jlr.M800603-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deshaies R, Joazeiro C. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 47.Kim S, Zhang S, Choi K, Reister R, Do C, Baykiz A, et al. An E3 ubiquitin ligase, Really Interesting New Gene (RING) Finger 41, is a candidate gene for anxiety-like behavior and beta-carboline-induced seizures. Biol Psychiatry. 2009;65:425–431. doi: 10.1016/j.biopsych.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishioka G, Yamada M, Kudo K, Takahashi K, Kiuchi Y, Higuchi T, et al. Induction of kf-1 after repeated electroconvulsive treatment and chronic antidepressant treatment in rat frontal cortex and hippocampus. J Neural Transm. 2003;110:277–285. doi: 10.1007/s00702-002-0779-3. [DOI] [PubMed] [Google Scholar]
- 49.Gachon F, Fonjallaz P, Damiola F, Gos P, Kodama T, Zakany J, et al. The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev. 2004;18:1397–1412. doi: 10.1101/gad.301404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elstner M, Morris C, Heim K, Lichtner P, Bender A, Mehta D, et al. Single-cell expression profiling of dopaminergic neurons combined with association analysis identifies pyridoxal kinase as Parkinson's disease gene. Ann Neurol. 2009;66:792–798. doi: 10.1002/ana.21780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sapir T, Sapoznik S, Levy T, Finkelshtein D, Shmueli A, Timm T, et al. Accurate balance of the polarity kinase MARK2/Par-1 is required for proper cortical neuronal migration. J Neurosci. 2008;28:5710–5720. doi: 10.1523/JNEUROSCI.0911-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishimura I, Yang Y, Lu B. PAR-1 kinase plays an initiator role in a temporally ordered phosphorylation process that confers tau toxicity in Drosophila. Cell. 2004;116:671–682. doi: 10.1016/s0092-8674(04)00170-9. [DOI] [PubMed] [Google Scholar]
- 53.Blankman J, Simon G, Cravatt B. A comprehensive profile of brain enzymes that hydrolyze the endocannabinoid 2-arachidonoylglycerol. Chem Biol. 2007;14:1347–1356. doi: 10.1016/j.chembiol.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blangero J. Localization and identification of human quantitative trait loci: king harvest has surely come. Curr Opin Genet Dev. 2004;14:233–240. doi: 10.1016/j.gde.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 55.Tsuang MT, Nossova N, Yager T, Tsuang MM, Guo SC, Shyu KG, et al. Assessing the validity of blood-based gene expression profiles for the classification of schizophrenia and bipolar disorder: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;133:1–5. doi: 10.1002/ajmg.b.30161. [DOI] [PubMed] [Google Scholar]
- 56.Borovecki F, Lovrecic L, Zhou J, Jeong H, Then F, Rosas HD, et al. Genome-wide expression profiling of human blood reveals biomarkers for Huntington's disease. Proc Natl Acad Sci U S A. 2005;102:11023–11028. doi: 10.1073/pnas.0504921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsankova N, Berton O, Renthal W, Kumar A, Neve R, Nestler E. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 58.Zheng YL, Li BS, Rudrabhatla P, Shukla V, Amin ND, Maric D, et al. Phosphorylation of p27Kip1 at Thr187 by cyclin-dependent kinase 5 modulates neural stem cell differentiation. Mol Biol Cell. 2010;21:3601–3614. doi: 10.1091/mbc.E10-01-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao J, Zhang S, Wu X, Huan W, Liu Z, Wei H, et al. KPC1 expression and essential role after acute spinal cord injury in adult rat. Neurochem Res. 2011;36:549–558. doi: 10.1007/s11064-010-0377-y. [DOI] [PubMed] [Google Scholar]
- 60.Anacker C, Zunszain PA, Cattaneo A, Carvalho LA, Garabedian MJ, Thuret S, et al. Antidepressants increase human hippocampal neurogenesis by activating the glucocorticoid receptor. Mol Psychiatry. 2011;16:738–750. doi: 10.1038/mp.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Almasy L, Hixson JE, Rainwater DL, Cole S, Williams JT, Mahaney MC, et al. Human pedigree-based quantitative-trait-locus mapping: localization of two genes influencing HDL-cholesterol metabolism. American Journal of Human Genetics. 1999;64(6):1686–1693. doi: 10.1086/302425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ayra R, Blangero J, Williams K, Almasy L, Dyer T, Leach R, et al. Factors of Insulin Resistance Syndrome (IRS)-Related Phenotypes are Linked to Genetic Locations on Chromosomes 6 and 7 in Nondiabetic Mexican Americans. Diabetes Care. 2002;51:841–847. doi: 10.2337/diabetes.51.3.841. [DOI] [PubMed] [Google Scholar]
- 63.Cai G, Cole SA, Freeland-Graves JH, MacCluer JW, Blangero J, Comuzzie AG. Genome-wide scans reveal quantitative trait Loci on 8p and 13q related to insulin action and glucose metabolism: the San Antonio Family Heart Study. Diabetes. 2004;53:1369–1374. doi: 10.2337/diabetes.53.5.1369. [DOI] [PubMed] [Google Scholar]
- 64.Willer C, Speliotes E, Loos R, Li S, Lindgren C, Heid I, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cai G, Cole SA, Bastarrachea-Sosa RA, Maccluer JW, Blangero J, Comuzzie AG. Quantitative trait locus determining dietary macronutrient intakes is located on human chromosome 2p22. Am J Clin Nutr. 2004;80:1410–1414. doi: 10.1093/ajcn/80.5.1410. [DOI] [PubMed] [Google Scholar]
- 66.Mitchell BD, Cole SA, Hsueh WC, Comuzzie AG, Blangero J, MacCluer JW, et al. Linkage of serum insulin concentrations to chromosome 3p in Mexican Americans. Diabetes. 2000;49:513–516. doi: 10.2337/diabetes.49.3.513. [DOI] [PubMed] [Google Scholar]
- 67.Kammerer CM, Gouin N, Samollow PB, VandeBerg JF, Hixson JE, Cole SA, et al. Two quantitative trait loci affect ACE activities in Mexican-Americans. Hypertension. 2004;43:466–470. doi: 10.1161/01.HYP.0000111830.36999.94. [DOI] [PubMed] [Google Scholar]
- 68.Kammerer CM, Schneider JL, Cole SA, Hixson JE, Samollow PB, O'Connell JR, et al. Quantitative trait loci on chromosomes 2p, 4p, and 13q influence bone mineral density of the forearm and hip in Mexican Americans. J Bone Miner Res. 2003;18:2245–2252. doi: 10.1359/jbmr.2003.18.12.2245. [DOI] [PubMed] [Google Scholar]
- 69.Kiel D, Demissie S, Dupuis J, Lunetta K, Murabito J, Karasik D. Genome-wide association with bone mass and geometry in the Framingham Heart Study. BMC Med Genet. 2007;8 Suppl 1:S14. doi: 10.1186/1471-2350-8-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Williams J, Blangero J. Power of variance component linkage analysis-II. Discrete traits. Ann Hum Genet. 2004;68:620–632. doi: 10.1046/j.1529-8817.2004.00128.x. [DOI] [PubMed] [Google Scholar]
- 71.Brown MS, Goldstein JL. Familial hypercholesterolemia: A genetic defect in the low-density lipoprotein receptor. N Engl J Med. 1976;294:1386–1390. doi: 10.1056/NEJM197606172942509. [DOI] [PubMed] [Google Scholar]
- 72.Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.