Abstract
To establish α–tropomyosin (Tm)’s structure-function relationships in cooperative regulation of muscle contraction, thin filaments were reconstituted with a variety of Tm mutants (Δ2Tm, Δ3Tm, Δ6Tm, P2sTm, P3sTm, P2P3sTm, P1P5Tm, wtTm), and force and sliding velocity of the thin filament were studied using an in vitro motility assay. In the case of deletion mutants, Δ indicates which of the quasi-equivalent repeats in Tm was deleted. In the case of period (P) mutants, an Ala cluster was introduced into the indicated period to strengthen the Tm-actin interaction. In P1P5Tm, the N-terminal half of period 5 was substituted with that of period 1 to test the quasi-equivalence of these two Tm periods. The reconstitution included bovine cardiac troponin. Deletion studies revealed that period 3 is important for the positive cooperative effect of Tm on actin filament regulation, and that period 2 also contributes to this effect at low ionic strength, but to a lesser degree. Furthermore, Tm with one extra Ala cluster at period 2 (P2s) or period 3 (P3s) did not increase force or velocity, whereas Tm with two extra Ala clusters (P2P3s) increased both force and velocity, demonstrating interaction between these periods. Most mutants did not move in the absence of Ca2+. Notable exceptions were Δ6Tm and P1P5Tm, which moved near at the full velocity, but with reduced force, which indicate impaired relaxation. These results are consistent with the mechanism that the Tm-actin interaction cooperatively affects actin to result in generation of greater force and velocity.
Keywords: Force and velocity, actin, mutant, optical tweezers, in vitro motility
INTRODUCTION
The regulation of muscle contraction is highly cooperative. While both Ca2+ binding to troponin (Tn) and the actomyosin interaction regulate the thin filament, it is tropomyosin (Tm) that makes the switch from relaxation to full activation cooperative. The interdependence of myosin and Tm in binding to the actin filament, and also the role of Tm-actin in cooperative activation by myosin have long been known1; 2; 3; 4. More recently it was suggested that Tm has a positive cooperative effect on actin in the regulated thin filament, causing an increase in the contractile force in the fibers5; 6. Such an effect implies that strengthening one or more parameters of the Tm-actin interaction increases force. In the present work we identify regions of Tm that function with myosin and Tn in cooperative regulation of the thin filament.
Tm is a two-chained α-helical coiled-coil protein that binds along the length of the actin filament and regulates muscle contraction in cooperation with Tn and myosin. The Tm sequence contains a seven-fold, quasi-periodic repeat that is a product of gene duplications7; 8. Although these repeats were originally proposed to each serve as a binding partner for the seven actin monomers along its length7; 9; 10, and they have clear sequence homologies, each contributes to actin affinity and regulatory function in a different way. Specifically, the two ends of Tm (periods 1 and 7) and the first half of period 5 are essential for actin binding11; 12; 13; the middle of the molecule, in particular period 3, is involved in positive cooperative regulation of actin5; 12; 14; 15; 16; 17; and the second half of period 4, the entire length of period 5, and the first half of period 6 are involved in Ca2+-dependent regulatory function12; 18; 19.
In this report, specific contributions of Tm’s periodic domains to force and sliding velocity were analyzed for Tm mutants by in vitro motility assays. In mutants of one class, a specific period (period 2, 3 or 6) was deleted. In the second class of mutants, the quasi-equivalence of Tm’s periodic repeats with respect to cooperative regulation was tested by replacing N-terminal half of period 5 with that of period 1. Studies with this mutant provided the first experimental support for quasi-equivalent periodic binding sites; the mutant Tm binds actin, however, it has altered regulation of the acto-myosin ATPase with troponin18.
In addition to the periodic repeats of surface residues, there is a less regular repeat of small non-polar interface residues noted by McLachlan and Stewart10 and more recently called “Ala clusters20”, that are the basis for the third class of mutants. Interruption of the hydrophobic interface between the two chains with residues such as Ala locally destabilizes the coiled-coil interface and can be associated with bends or staggers in Tm20; 21 that may allow local flexibility or directional bending of the Tm’s supercoil to optimize binding to the actin subunits in the helical filament. Naturally-occurring Ala or other destabilizing interface residues at these sites are critical for actin binding22; 23; 24. These mutations locally destabilize the coiled coil and increase actin affinity or the binding cooperativity24, consistent with the proposal that Ala clusters enhance affinity of a local periodic repeat for actin. Here we studied the effect of introducing Ala clusters in periods 2 and 3 on regulatory function in three mutants.
Using period 2 and 3 deletion mutants, we found that the sliding velocity and force for the period 3 deletion mutant were similar to that with unregulated actin, and intermediate between unregulated actin and wildtype (wt) Tm for the period 2 deletion mutant. The period 6 deletion mutant was similar to wtTm. These results suggest that, at the ionic strength of the current experiments (50 mM), period 3 is most critical for cooperative activation of the thin filament. The mutant designed to increase actin affinity by introducing Ala clusters within proposed actin binding regions in both periods 2 and 3 had the greatest force and velocity, that were greater than those observed when wtTm was used, indicating cooperativity between periods 2 and 3. These results provide further evidence that this region of Tm has modulatory function. The results show that there is not a simple relationship between the effects of deletions or introduction of Ala clusters on Tm’s affinity for actin filaments and the cooperative regulation of actin-myosin interaction by Tm-Tn.
RESULTS
We evaluated the contributions of the periodic repeats of Tm, as well as the contributions of structural parameters of the individual periods, to regulatory functions using two assays: an in vitro motility assay (measures velocity), and an optical tweezers displacement assay (measures force). Computerized analysis of the thin-filament velocity revealed that the major peak had a Gaussian distribution and its peak centered at 3–12 μm/s. The typical velocity distributions are shown in Fig. 1. The velocity distribution was fitted to Eq. 1 to remove the background and to determine Vc (center velocity), σ (dispersion), and A (strength). Table 1 summarizes the velocity and force measurements of the wildtype (wt) and mutant Tms. Experiments in Figs. 1–4 were performed in the presence of 2 mM MgATP.
Fig. 1.
Distributions of filament velocity for thin filaments reconstituted with Tm mutants and for filaments lacking Tm (−Tm) together with Tn. The distributions were calculated at each 0.15 μm/s interval. Each distribution is fitted to Eq. 1 (continuous curves), with the total area under the Gaussian curve normalized to 100% μm/s. Values shown in Table 1 are the averages calculated from many distributions. A–D: In the presence of Ca2+ (pCa 4.73). Each distribution was calculated based on a video record of 3 sec in length. E: In the absence of Ca2+. Each distribution is based on the sum of 3 video records. The distribution of wtTm is scaled down to 1/6, else the values are too large to show in this panel (To obtain actual values, 6 should be multiplied).
Table 1.
Velocity and force, measured by in vitro motility assays
| +Ca2+ (activating solution, pCa 4.73) | −Ca2+ (relaxing solution) | |||
|---|---|---|---|---|
| Velocity μm/s | Force pN/μm | Velocity μm/s | Force pN/μm | |
| −Tm | 4.68±0.09 (15)** | 1.07±0.06 (38)** | ||
| Δ2Tm | 5.71±0.09 (29)** | 0.48±0.04 (58)** | 0.26±0.05 (4) | 0 |
| Δ3Tm | 4.90±0.29 (9)** | 0.37±0.03 (74)** | 0.36±0.03 (10) | 0 |
| Δ6Tm | 6.26±0.13 (22)** | 1.52±0.16 (48) | 6.47±0.12 (9)** | 0.66±0.05 (47)** |
| wtTm | 7.70±0.07 (16) | 1.63±0.06 (101) | 0.33±0.02 (10) | 0 |
| P2sTm | 6.73±0.19 (10)** | 1.46±0.08 (55)(*) | 0.32±0.02 (10) | 0 |
| P3sTm | 6.92±0.14 (12)** | 1.53±0.09 (56) | 0.39±0.02 (10) | 0 |
| P2P3sTm | 9.27±0.18 (11)** | 2.11±0.12 (85)** | 0.33±0.02 (10) | 0 |
| P1P5Tm | 6.92±0.23 (18)** | 2.06±0.12 (66)** | 5.41±0.22 (8)** | 0.28±0.03 (32)** |
For velocity measurements, velocity distribution such as shown in Fig. 1 was deduced from a video record consisting of 3 sec, and then the distribution was fitted to Eq. 1 to find Vc. Vc was then averaged for many records, and shown with ±SEM in this Table. For force measurements, the filament-attached beads are trapped by optical tweezers, the thin filaments and HMM interaction was performed, then force was measured (Fig. 2), histogram (Fig. 3) was created, and averaged. 0: force was not detectable. Experiments were performed in the presence of 2 mM MgATP at 24°C. In comparison, the velocity measured in the rigor solution (no ATP, no Ca) was 0.40±0.01 (N=6) for wtTm, and 0.45±0.04 (N=4) in the absence of Tm and Tn; Force was not detectable.
P≤0.01,
0.01<P≤0.05, and
0.05<P≤0.1 compared to wtTm (based on paired T-tests).
Fig. 4.
Correlation of force and velocity. A, Experiments were carried out in the presence of Ca2+ (○) or in its absence (×). Error bars represent SEM. The regression line (V=aF+b) used only the data with Ca2+, where a=1.84±0.99 μm2pN−1s−1, b=4.07±1.47 μm/s (best fits ±95% confidence limits), and r (regression coefficient)=0.80 are found. B, The regression line for the data with and without Ca2+, which includes shift mutants (P2s, P3s, and P2P3s), wt, and −Tm, is represented by a=4.38±0.07 μm2pN−1s−1, b=0.34±0.02 μm/s, and r=0.99. Inset: The area around the zero intercept (filled rectangle) is magnified by 20 times and plotted. These are the data in the absence of Ca2+. The data of wtTm and P2P3sTm coincided.
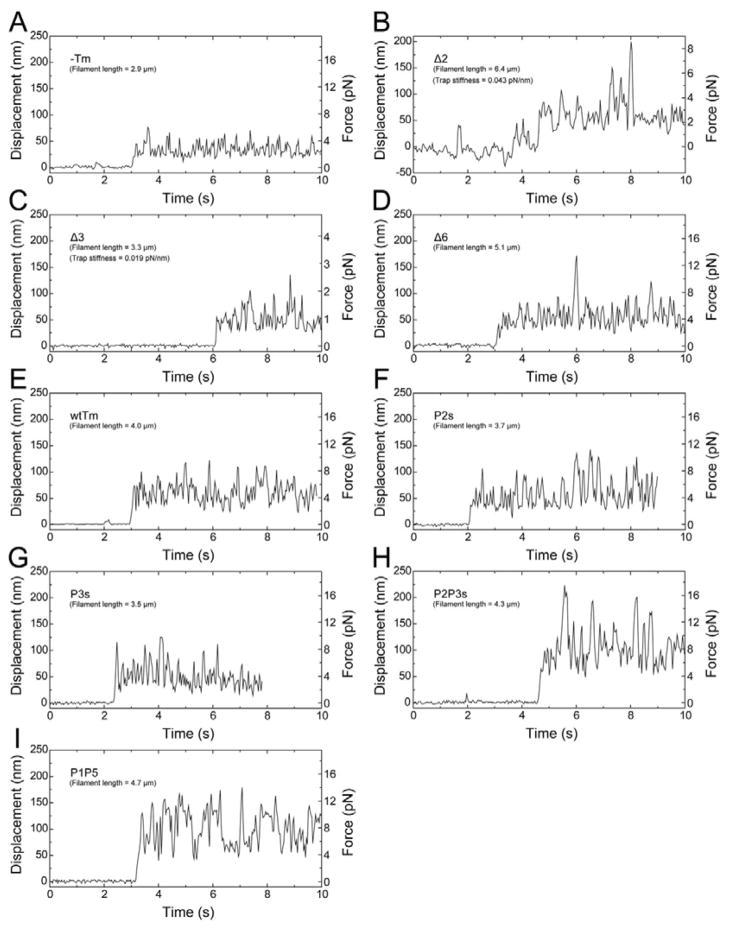
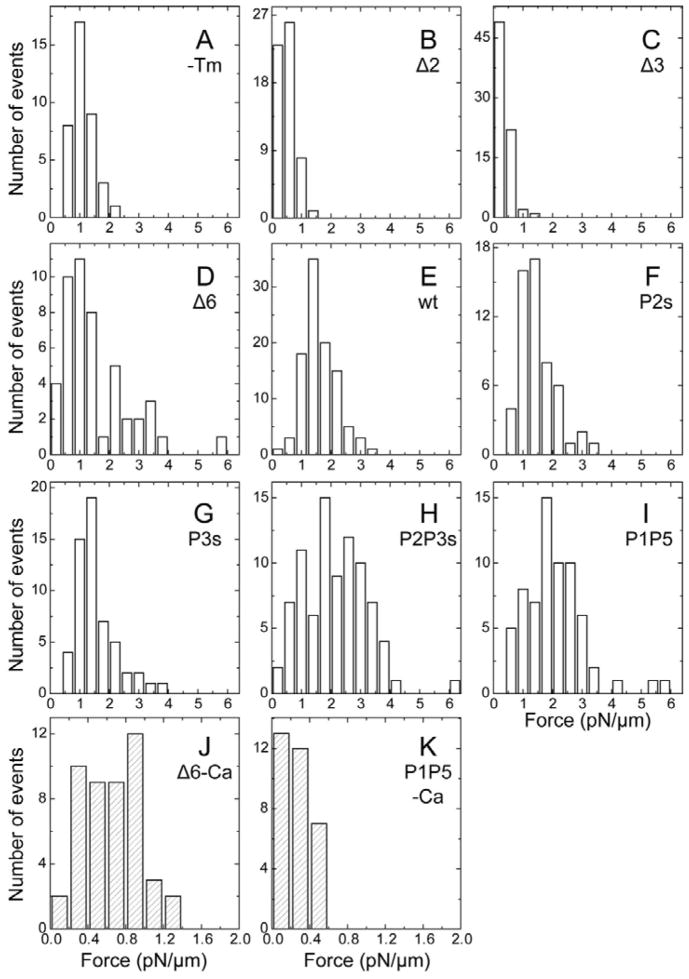
The time course of the interaction between the thin filament and HMM on the glass surface is shown in Fig. 2. The displacement of the bead to which the thin filament was attached was initially 0, as there was no interaction at the beginning of the experiment. Once the bead was lowered to the glass surface and the thin filament started interacting with HMM, the bead was suddenly displaced (Fig. 2, left ordinate). Force (right ordinate) was deduced by averaging this displacement and multiplying by the trap stiffness of the optical tweezers. Because measured force increases in proportion to the length of the thin filament25, the force/μm of the thin filament was obtained by dividing the force by the length of filament interacting with HMM, which was measured from the fluorescence image. For each mutant, a histogram of force values was created for every 0.4 pN/μm (Fig. 3A–I) or 0.2 pN/μm (Fig. 3J and K). Each histogram has an approximate Gaussian distribution.
Fig. 2.
Time course of the interaction between the thin filament and heavy meromyosin (HMM) on the glass surface in the activating solution. The bead to which the thin filament was attached was trapped by optical tweezers, and slowly lowered to the glass surface. Initially, there was no interaction between the thin filament and HMM, therefore, the displacement (left ordinate) of the bead is 0. When the thin filament started to interact with HMM, the bead was suddenly displaced. This displacement was averaged and multiplied by the trap stiffness of the optical tweezers to calculate force (right ordinates). The force value was then divided by the thin filament length (FA) interacting with HMM. A, in the absence of Tm and Tn. B–I, Reconstituted with mutant Tms (indicated) and wtTn. Optical trap stiffness was 0.078 pN/nm, except for B and C (indicated in each panel).
Fig. 3.
Histograms of force distribution. A, in the absence of Tm and Tn. B–K, Reconstituted with mutant Tms (indicated) and wtTn. A–I, Measured in the presence of Ca2+ (force interval: 0.4 pN/μm, pCa 4.73). J–K: Measured in the absence of Ca2+ (force interval: 0.2 pN/μm).
In the absence of regulatory proteins (−Tm), the averaged sliding velocity (Vc) of the actin filament was 4.68±0.09 μm/s (±SEM, Table 1; Fig. 1D —○—) and the force was 1.07±0.06 pN/μm (Table 1, Figs. 2A, 3A and 4). When the actin filament was reconstituted with wtTm and Tn and assayed in the presence of Ca2+ (Table 1, KFig. 1A —○—, Figs. 2E and 3E), the sliding velocity and the force significantly increased. Because both the velocity and the force increased by reconstitution of wtTm and Tn, we call this “positive cooperativity”, or simply cooperativity. The positive cooperativity of regulation is distinct from the cooperativity of Tm binding to actin where the actin affinity is measured by the app, and the cooperativity by the Hill coefficient, or the temperature and sharpness of dissociation of Tm from actin in DSC measurements. The recombinant wtTm used in these experiments has an unacetylated N-terminal Met. Its function with Tn is indistinguishable from that of N-acetylated Tm purified from striated muscle in regulation of the actomyosin or acto-myosin S1 ATPase11; 26, the myosin-S1 dependence of the acto-myosin S1 ATPase11, and in regulation of reconstituted myocardial fibers16.
Deletion mutants
The Hitchcock-DeGregori and Tobacman laboratories have studied a series of Tm deletion mutants to understand the requirements for and contribution of the periodic repeats to Tm’s actin binding and regulatory functions12; 14; 15; 27; 28; 29. Domain swapping and deletion are classic approaches for understanding protein design and function, and are naturally practiced in Tms by alternative exon splicing30; 31. There are caveats that there may be long range and/or other unanticipated consequences, as in any study that modifies protein structure. In the three mutants studied here, period 2, 3 and 6 (35 or 42 residues, according to the periodic repeats in the Phillips’ model, with an average of 38.7 residues9) were individually deleted to preserve both the coiled-coil heptad repeat and integral number of periods that are important for function28 (see Table 2 and Methods for the residues deleted). The mutant Tms fold normally and bind actin, albeit with reduced affinity (Kapp): previous studies showed that deletion of period 2, 3 or 6 reduces Kapp of Tm for F-actin by ~10-fold (Δ2Tm) to ~20-fold (Δ3Tm and Δ6Tm)12; 27; 28. The 35-residue deletions in Δ3Tm and Δ6Tm are in accordance with the Phillips 1986 model9. A Kapp (for actin) of a 42-residue deletion mutant of period 3 is indistinguishable from that of the Δ3Tm mutant used here28. The myosin S1-induced binding of Tm to actin was weaker in Δ3Tm and Δ6Tm; that of Δ2Tm was similar to wtTm. Regulation of the actomyosin ATPase was unaffected with Δ2Tm; activation in the presence of Ca2+ was impaired with Δ3Tm, whereas inhibition in the absence of Ca2+ was poor with Δ6Tm. Extensive analysis of these and other related mutants has shown that the specific region deleted determines function, not the 35 versus 42-residue deletion10; 12; 14; 15; 27; 28; 29. The possible functional consequences of the deletions on the orientation of the chains in the flexible intermolecular junction appear to be minor, presumably because of the variable length of the supercoil in the molecule32.
Table 2.
Description of tropomyosin mutants used in the study
| Tropomyosin1 | Description | Length | Ref. |
|---|---|---|---|
| wtTm | Recombinant rat striated α-Tm | 284 residues | 11 |
| Δ2Tm2,3 | Deletion of period 2, residues 47–88 | 242 residues | 27 |
| Δ3Tm2 | Deletion of period 3, residues 89–123 | 249 residues | 28 |
| Δ6Tm2,3 | Deletion of period 6, residues 208–242 | 249 residues | 12 |
| P2sTm4 | Y60A, L64A. Generation of an additional Ala cluster within proposed period 2 actin binding site | 284 residues | 24 |
| P3sTm4 | L106A, A120L. Generation of an additional Ala cluster within proposed period 3 actin binding site. | 284 residues | 24 |
| P2P3sTm4 | L60A, L64A, L106A, A120L. Generation of additional clusters within periods 2 and 3. | 284 residues | 24 |
| P1P55 | Replacement of residues 165–188 in period 5 with residues 4–27, the homologous residues in period 1. Duplication of period 1. | 284 residues | 18 |
Notes:
All proteins were expressed in E. coli. The N-terminal Met is unacetylated.
The seven periods are those defined by the model in Phillips 9. The model differs from that of McLachlan and Stewart 10 in that it considers the supercoiled coiled coil structure. The periods are all multiples of seven but differ in length, being 35 or 42 residues, with the average being 38 2/3 residues, as in McLachlan and Stewart. Period 2 has 42 residues in the Phillips model.
Periods 3 and 6 have 35 residues in the Phillips model. Deletions of 35 residues (89–123) or 42 residues (89–130) from this area had equivalent effects on the Kapp and cooperativity of binding to actin 28.
Periods 1 and 5 have Ala clusters within proposed actin recognition motifs. The shift mutants create or relocate Ala clusters in periods 2 and 3 in positions homologous to those in periods 1 and 5.
Residues 165–188 contain the motif Phillips 9 proposed is important for actin binding, and the rest of the Ala cluster. Singh and Hitchcock-DeGregori 23 provided support for the model and showed this region is critical for actin binding. Period 1 (residues 4–27) is the only other period to have an Ala cluster in the same relationship as in period 5.
The functional effects of the deletion mutants reported here are period specific. Notably, the velocity of filaments reconstituted with Δ3Tm (Table 1, Fig. 1B - - + - -) was similar to that with unregulated (−Tm) actin filaments (Table 1), much lower than that of filaments reconstituted with wtTm (Table 1). The force with Δ3Tm was also lower, only 23% of wtTm (Table 1, Figs. 2C and 3C, P<0.001). When Δ2Tm was used for reconstitution, the values for velocity (Fig. 1A ---×---) and force (Table 1, Figs. 2B and 3B) were both significantly larger than those for Δ3Tm (P<0.001 for velocity; P<0.05 for force). With regulated actin filaments reconstituted with Δ6Tm, the velocity (Table 1, Fig. 1B —○—) and especially the force (Table 1, Figs. 2D and 3D) were closer to wtTm levels.
While periods 2 and 3 both appear to contribute to Tm’s activating effect of the actin-myosin interaction, the contribution of period 3 is greater than that of period 2. These results are consistent with the importance of period 3 to activate actomyosin ATPase activity in the presence of Ca2+,12 and to induce positive cooperative effect on force generation in reconstituted skinned cardiac fibers5; 16.
Shift mutants
Whereas the deletion mutants are all shorter than wtTm, the “shift” mutants have the same length as wtTm: 284 residues per chain. We assayed two types of shift mutants to test hypotheses concerning the design of Tm’s periodic repeats for regulatory function. In the first type, P2sTm, P3sTm, and P2P3sTm, Ala clusters were introduced at the coiled-coil interface in the regions of the Tm periods proposed to be involved in actin recognition. The design mimics the relation of the Ala clusters to the actin binding site in period 5, where the Ala cluster (or similarly destabilized interface) is required for actin binding24. These mutations locally destabilize the coiled coil and increase actin affinity (P2sTm and P2P3sTm), or cooperativity of binding (P3sTm), as measured by cosedimentation or DSC24. Here, P2P3sTm developed significantly greater force (2.11±0.12 pN/μm) and velocity (9.27±0.18 μm/s) than wtTm (Table 1), indicating that this mutation results in greater cooperativity in the actomyosin interaction, consistent with the involvement of this region in cooperative regulation5; 12; 14; 15; 16; 24. On the other hand, the velocity and force with P2sTm and P3sTm were similar to those with wtTm (Fig. 4).
In the P1P5 mutant, residues 165–188 of period 5, the periodic repeat shown to be the most critical for actin binding12; 22, were replaced with the homologous region of period 1 (residues 4–27), which, like period 5, contains an Ala cluster. This mutant has reduced actin affinity, and the myosin S1-induced binding of Tm to actin was close to wtTm; The major difference was that inhibition of the regulated actomyosin ATPase in the absence of Ca2+ was impaired18. Thus, the function of period 1 appears to be quasi-equivalent to that of period 5 with respect to actin binding and myosin cooperativity, but not with respect to Ca2+-dependent regulatory function (see below). P1P5Tm developed significantly greater force (2.06±0.12 pN/μm), but somewhat less velocity (6.92±0.23 μm/s) than wtTm (Table 1). Both the force and velocity values of shift mutants were significantly greater than those obtained using deletion mutants Δ2Tm, Δ3Tm, and unregulated actin filaments (−Tm) (Fig. 4A).
To show how force and velocity are related, we plotted in Fig. 4A the force-velocity relationship of the data in Table 1. This figure demonstrates that force and velocity are linearly, though loosely, related with the regression coefficient of r=0.80 (data in the presence of Ca2+ (○) are used). Fig. 4B plots the data for wtTm, −Tm, and three shift mutants, but no deletion mutants. The data includes those in the presence of Ca2+ and in its absence (inset), and their regression line. This regression is even better (r=0.99) and extrapolates to near the origin, demonstrating that force and velocity are proportionately related in this class of mutants.
Regulation in the absence of Ca2+
When Ca2+ was depleted, the sliding velocity decreased to near zero (0.26–0.39 μm/s, Table 1) in most of the Tm mutants, indicating the perfect Ca2+ regulation. Similar velocity values were found under the rigor condition. When video records were examined, no filaments moved; relaxed filaments had a thermal fluctuation, and rigor filaments did not even fluctuate. Consequently, the small velocity values deduced (represented by Fig. 1E, wtTm) set the limit to the image analysis method used here. Under these conditions force was not detectable. Two notable exceptions were Δ6Tm and P1P5Tm. In the case of Δ6Tm, the force in the absence of Ca2+ was 43% of that in the presence of Ca2+ (Fig. 3J), and the filaments moved with the velocity indifferent to Ca2+ (Table 1). With P1P5Tm, the force was 14% (Fig. 3K) and the velocity was 78% of that in the presence of Ca2+ (Table 1), a value significantly different from 100%. In both mutants, the thin filament movement was more sluggish than in the presence of Ca2+, which is reflected by broader velocity distributions of Δ6Tm and P1P5Tm (Fig. 1E and Table 1).
The number of cross-bridges during rigor
For our study to be meaningful, it is important to establish the number of cross-bridges involved in the actomyosin interaction is approximately the same when various Tm mutants are used. To count the number of cross-bridges, the concentration of HMM is reduced to 5 μg/ml so that less HMM is adsorbed to the glass surface in flow cells. This is to reduce the number of cross-bridges that attach to the thin filament for ease of counting. Thin-filament attached beads are suspended in the rigor solution and injected to the flow cell. A bead with a thin filament was trapped by the optical tweezers, lined up with the glass surface, and lowered to the surface for the rigor interaction to take place (Fig. 5A; time 0 in Fig. 5D). Thereafter, the bead was elevated slowly, which displaced the bead (Fig. 5B; linear rising phase in Fig. 5D) because of the stretch of the thin filament. When a myosin cross-bridge detached from the thin filament (Fig. 5C), there was a sudden reduction in the displacement (vertical drops in Fig. 5D with down arrows). The number of such reductions corresponds to the number of cross-bridges. The results were divided by the length of the thin filament interacting with HMM-coated glass surface and plotted in Fig. 5E for mutant Tms and in the case of −Tm. This plot demonstrates that the number of cross-bridges averaged 1.92 μm−1; it was slightly larger for Δ3Tm and Δ6Tm, and slightly less in P2sTm. Other Tms including the case of −Tm are close to the average.
Fig. 5.
The method of counting the number of cross-bridges for interaction with a thin filament in the rigor solution. A filament-attached bead is trapped by the optical tweezers and lowered for the interaction to take place (A; time 0 sec in D). The bead is elevated slowly, which displaces the bead (B; linearly rising phase in D) because of the strain on the thin filament. When myosin (HMM) detaches from the thin filament (C; down arrows), there is a sudden reduction in the displacement (D). The number of such reductions is the number of cross-bridges. The results are plotted in E for mutant Tms and −Tm (N=13–22).
To determine the force per cross-bridge, the force values of Table 1 and Fig. 4 must be divided by the number of cross-bridges. Because the variation in the number of cross-bridges is small, this division does not alter the conclusions derived in this report.
DISCUSSION
Tropomyosin’s domains and cooperative activation
Our observation (Table 1, Fig. 4) that the increased force (~1.5×) and velocity (~1.6×) in the presence of Ca2+ when wtTm was reconstituted in the actin filament together with Tn illustrates the activating role of Tm on the actin-myosin interaction, which we call “positive cooperativity”. These results are consistent with our earlier results that ~1.5× force developed when regulatory proteins were added to actin filament reconstituted cardiac muscle fibers6; 16; 35. The cooperativity of the thin filament regulation by Tm, Tn and myosin has been previously demonstrated by using in vitro reconstituted systems3; 33; 34; 35; 36; 37; 38; 39, as well as in the skinned fiber systems6; 17.
The present work assigns period 3 of Tm a major role in the activation of the regulated thin filament in the presence of Ca2+, because wildtype levels of both force and velocity require period 3. This conclusion is consistent with the outcomes of previous studies on the thin filament-reconstituted myocardium (cardiac muscle fibers) 16, as well as with measurements of actomyosin ATPase and myosin S1-induced Tm binding 12; 28; 29. The conclusion is further consistent with findings from studies in which both periods 2 and 3 were deleted (Δ23Tm): Lu et al 5 for the thin-filament reconstituted myocardium; Landis et al 15 for in vitro motility assays; Siththanandan et al 40 for myofibril mechanics; and Hitchcock-DeGregori et al 29 for actomyosin ATPase assays.
Period 2 also contributes to positive cooperative regulation. With regulated filaments reconstituted with Δ2Tm, both the force and the velocity were intermediate between wtTm and Δ3Tm. In fiber studies the force value obtained for reconstitution with Δ2Tm was much closer to that with wtTm 16, whereas in the current in vitro motility assays both force and velocity values were closer to those with Δ3Tm. This difference may have arisen because of a difference in the experimental conditions. The fiber study was carried out at 200 mM ionic strength, whereas the in vitro motility assay was carried out at 50 mM ionic strength. While the hydrophobic interaction does not change with ionic strength, the ionic interaction is stronger at 50 mM than at 200 mM. The proposed actin binding site (heptad positions b, c and f) in period 2 has 8 ionic residues, 1 Gly, and no hydrophobic residues, whereas the corresponding region of period 3 has 5 ionic and 4 hydrophobic amino acid residues. Consequently, the deletion of period 2 would significantly reduce the cooperative effect at 50 mM ionic strength (in vitro system), whereas the hydrophobic residues in period 3 may explain how deletion of this period would affect the cooperative activation regardless of ionic strength (fiber 16 and in vitro systems; Table 1). These results further suggest that both the hydrophobic and ionic interactions have roles in regulatory function.
Tm-Tn interaction and imperfect regulation in some mutants
When Ca2+ was depleted, the force and velocity decreased to a near zero values in wtTm and the most mutant Tms (Table 1), indicating perfect Ca2+ regulation together with Tn. This observation further demonstrates that all the actin molecules are saturated with regulatory proteins. There are two exceptions to this. The velocity of reconstituted thin filaments with Δ6Tm or P1P5Tm was similar in the presence and absence of Ca2. However, force was significantly reduced in the absence of Ca2+ to 43% (Δ6Tm) or to 14% (P1P5Tm) (Table 1). From these observations we infer that the regulatory system with Δ6Tm or P1P5Tm could not perfectly turn off actomyosin interaction in the absence of Ca2+. The regions altered in these mutants (residues 165–188 and 208–242) are included in the Ca2+-sensitive, Tn binding region of Tm (reviewed in Brown and Cohen41). It is unlikely the poor regulatory function in the absence of Ca2+ results from incomplete saturation with Tm-Tn, because both mutants had a positive effect in the presence of Ca2+ where the affinity of Tm for the regulated actin filament is weaker than in the absence of Ca2+.
With regard to the quasi-equivalence of periods 1 and 5, regulatory function with P1P5 is primarily impaired in relaxation conditions, as discussed above. In the presence of Ca2+, cooperative activation with P1P5 is the closest to wt. We conclude that the design of period 5 of wtTm is optimal for cooperative binding and regulation, even though it is a constitutively-expressed exon, found in tissues that do not have Tn30. In human striated muscle αTm, the region is the site of several cardiomyopathy-causing mutations. 42; 43
Ala clusters in tropomyosin
Regional interruptions of the canonical coiled coil are essential for actin binding 23; 44. In wtTm, the only periodic actin binding motifs that contain destabilizing interface Ala (Ala clusters) are periods 1 (aa 18–22–25) and 5 (179–183–186). Mutations to introduce a third Ala cluster into either period 2 (60–64–67) (P2s) or period 3 (102–106–109) (P3s), or both periods (P2P3s) increased actin affinity or binding cooperativity and locally destabilized the coiled coil24. In the present study, P2s or P3s slightly decreased the velocity without a significant change in the force. However, when the Ala clusters were introduced to both periods (P2P3s), there were significant increases relative to wtTm in both force (1.3×) and velocity (1.2×). Our results demonstrate that the neighboring Ala clusters may work cooperatively in actin binding and activation of the regulated thin filament.
It has been reported that Ala clusters are associated with bends or staggers and a reduced interchain distance in crystal structures of Tm 20; 45; 46; 47. We have suggested that one function of Ala clusters is to increase actin affinity by allowing nearby binding sites to optimally interact with actin.44 The mechanism may operate via segmental flexibility caused by directional bending or staggers to enhance conformation of the Tm supercoil to the helical actin filament.21 This can be achieved by local disorder that may enable particular side chains to bind to sites on actin and be stabilized upon binding 23; 24, or by allowing a gradual (but not discrete) bending that confers a shape to Tm that is favorable for binding the actin helix 48; 49. It is likely that cooperative activation by myosin promotes these mechanisms by direct, weak interaction with Tm and/or by a change in the underlying actin structure that increases the affinity of myosin for actin.
It is interesting to observe that the force and the velocity are proportionally related in one class of Tms (shift mutants, wt, and −Tm: Fig. 4B). This implies that the velocity (a representative of actomyosin ATPase) and the force are directly coupled. This result is consistent with the formulation of Sato et al50, who showed that the unloaded sliding velocity is related to force by: (active force generated by each cross-bridge)/(molecular friction). That is, the molecular friction51 is the reciprocal of the slope of Fig. 4. The other classes of mutants showing the deviation from this proportional relationship may have a different mechanism of coupling between the sliding movement and the force generation. It may be that these mutants have a larger molecular friction (note less slope in Fig. 4A than in Fig. 4B), because of the shorter Tm’s molecular length introduced a higher Tn density (deletion mutants), or an extra bend that may have been introduced to the Tm molecule by the mutation (P1P5).
Conclusions
We conclude that (1) period 3, and to some extent period 2, is required for cooperative activation; (2) local destabilization of periods 2 and 3 by introduction of Ala clusters increases cooperative activation; (3) periods 5 and 6 are required for full relaxation, but not for cooperative activation. These conclusions are consistent with the hypothesis that the periodic repeats have specific functions. The regulated thin filament is a complex system; the different facets of regulatory function are not directly correlated with a single parameter such as the Kapp of Tm for the actin filament. Recent molecular models for the Tm-actin filament based on computational and functional analyses have proposed the contribution of ionic interactions that allow for weak but specific binding of Tm to actin49; 52. Weak ionic interactions have the character that would allow rapid, cooperative switching between states, although molecular models for Tm in the positions proposed by the steric blocking mechanism are not yet available. We suggest, here, that Tm’s periodic repeats are not quasi-equivalent with respect to regulatory functions. The involvement of internal periods in cooperative activation of the thin filament suggests that some of the signals are transmitted via the actin filament and/or by weak interactions with myosin, not only via the intermolecular junctions of tropomyosin molecules along the actin filament. Our study demonstrates the significance of Tm’s modulatory role in the actomyosin interaction.
MATERIALS AND METHODS
Proteins
Mutant and wt Tm proteins were made in rat striated-muscle αTm cDNA, and expressed in E. coli. This isoform is expressed in cardiac and skeletal muscles and in some studies is referred to as “cardiac” Tm. These proteins were purified in the Hitchcock-DeGregori laboratory, and the N-terminal Met is not acetylated.12; 16; 24 In previous studies with these Tms, actin binding was measured in the presence of skeletal muscle Tn12; 28 (and Ca2+), or a N-terminal fragment of cardiac TnT.18; 24 Both Tn and TnT increase the affinity of unacetylated Tm for actin.22; 26 Tn was extracted from bovine ventricle in the Kawai laboratory as described previously53. Actin and heavy meromyosin (HMM) were purified from rabbit white skeletal muscles in the Ishiwata laboratory as described previously.54; 55 All experimental procedures conformed to the “Guidelines for Proper Conduct of Animal Experiments” approved by the Science Council of Japan, and were performed according to the “Regulations for Animal Experimentation at Waseda University.
Actin filaments labeled with rhodamine-phalloidin were attached to polystyrene beads 1 μm in diameter via gelsolin and G-actin, using the carboxyl group attached to the beads.55 The bead-tailed actin filaments were added to the solution of Tm+Tn in a test tube, and the mixture was incubated for 30 min on ice to reconstitute the thin filaments.56 The annealing treatment usually used to disperse the filaments was not carried out because of the low concentration of proteins 57. The thin filament was visualized by fluorescence when illuminated by a green laser (532 nm).
The Tm mutants are listed in Table 2. In the Δ2Tm mutant, residues 47–88, which correspond to period 2 of the 7 quasi-equivalent repeats, were deleted.27 In the Δ3Tm mutant, residues 89–123 (period 3) were deleted.28 In the Δ6Tm mutant, residues 208–242 (period 6) were deleted. These mutants are described and their biochemical properties have been reported12. In P2sTm (period 2 shift Tm), mutations Y60A and L64A were introduced in period 2 to generate an Ala cluster20 at residues 60–64–67 (aa 67 is Ala in αTm). In P3sTm, mutations L106A and A120L were introduced into period 3 to generate another Ala cluster in residues 102–106–109; the A120L mutation was necessary to stabilize the coiled coil. In the P2P3sTm (period 2 and period 3 shift) mutant, both sets of mutations were introduced to yield two additional Ala clusters. In P1P5Tm, the first half of P5 (residues 165–188) was replaced with that of P1 (residues 4–27) to examine the interchangeability and the effect of the Ala cluster that is normally present in period 1 (residues 18–22–25). All constructs were cloned in pET11d58 and expressed in E. coli BL21 (DE3) cells as unacetylated proteins. The deletion mutants, P2sTm, P3sTm, P2P3sTm, and P1P5Tm mutants were described and their biochemical properties were previously reported.12; 18; 24; 27; 28
Solutions
Relaxing solution contained (mM:) 2 Na2ATP, 4 MgCl2, 1 EGTA, 25 KCl, 10 dithiothreitol (DTT), 25 Im-HCl (Im=imidazole), and 1 mg/ml BSA. Activating solution contained (mM) 2 Na2ATP, 1 CaEGTA (pCa 4.73), 4 MgCl2, 25 KCl, 10 DTT, 25 Im-HCl, and 1 mg/ml BSA. Rigor solution contained 4 mM MgCl2, 1 mM EGTA, 28 mM KCl, 10 mM DTT, 25 mM Im-HCl, and 1 mg/ml BSA. In addition, all solutions contained 25 mM glucose, 0.216 mg/ml glucose oxydase, and 0.036 mg/ml catalase to remove dissolved oxygen and minimize photobleaching of tetra methyl rhodamine (TMR)59. Experiments were carried out at 24±1°C in low ionic-strength (50 mM) solution in the absence of Ca2+, or in its presence with the pH adjusted to 7.40.
Flow cell (chamber)
Both surfaces of a large cover slip were coated with collodion dissolved in 3-methylbutyl acetate. Another cover slip of normal size was glued to the large cover slip using double-sided tape, to yield a flow-cell volume of ~20 μl. 20 μl of HMM solution (30 μg/ml) was applied from one side of the flow cell and left on the slide for 60 sec, so that the HMM could settle and attach to the collodion-coated glass surface. Another drop of HMM solution was then applied from the other side and to settle for 60 sec. Subsequently, 20 μl of BSA solution (5 mg/ml) was applied and allowed to settle for 5 min. Thereafter, 20 μl of the experimental solution containing thin filaments was applied. The two open sides were sealed with non-fluorescent enamel, and the flow cell was placed in the experimental setup. The thin filament was monitored using a fluorescence microscope, and video images were captured at the rate of 30 frames/s. For details, see our earlier reports.56; 60
The reconstitution of the thin filament was performed in Eppendorf pipettes in the presence of 0.25 μM F-actin, 0.25 μM Tm, and 0.25 μM Tn at 0°C at least for 30 min. This was diluted to 1/60 before injecting into the flow cell. We used 3–4 flow cells per one mutant, and 10–20 force and velocity measurements were made per each flow cell.
Velocity measurements
A video record consisting of 3 sec was analyzed using an image analysis program developed at the Fink laboratory at the University of Heidelberg56; 61 to deduce the velocity distribution [Dist(V)] of the thin filaments (Fig. 1) at every 0.15 μm/s interval. This program uses a structure-tensor based algorithm to identify a group of pixels moving together, follows its trajectory with time, and calculates the velocity of the pixel group. The velocity distribution was fitted to Eq. 1:
| (1) |
where V is the velocity, Vc is the center velocity, σ is the dispersion (~half width) of the velocity distribution, and A is the intensity of the distribution. The first term on the right side of Eq. 1 is the Gaussian distribution. B, C and D are parameters to fit the background to the quadratic form, and are needed primarily for instances in which some filaments are slowly moving (velocities of <2–3 μm/s) or immobile. For plotting the velocity distribution (Fig. 1), the area of the Gaussian was normalized to 100% μm/s by using Eq. 2.
| (2) |
This means that only the Gaussian portion of the distribution (first term in Eq. 1) was used for normalization according to Eq. 2, but the background portion (2nd–4th terms in Eq. 1) was not used for normalization. This is because the main peak and Vc were reproducible, whereas the background was not as much reproducible.
Force measurements
For the measurement of active force, a bead to which a single thin filament was attached was trapped with optical tweezers, and placed ~5 μm above the HMM-coated glass surface. The flow cell was moved to one direction to line up the thin filament with the flow cell. The bead was lowered until the thin filament interacted with the HMM. The position of the bead throughout the course of the interaction was monitored using a phase-contrast microscope. The force developed between HMM and the thin filament led to a displacement of the bead (Fig. 2), and the magnitude of this displacement was measured. Force was deduced after multiplication by the trap stiffness of the optical tweezers. For the calibration of the trap stiffness, a bead was trapped close to the glass surface, and the position of the bead was detected by a quadrant photodiode at 10 kHz. From the power spectrum of displacement of the trapped bead, we determined the trap stiffness to be 0.019 –0.078 pN/nm; the trap stiffness was adjusted at every preparation to maximize signal output by controlling the incident laser power.
Measurement of the number of cross-bridges in the rigor condition
We measured the number of bound cross-bridges in the rigor condition as reported by Nishizaka et al.62 and Kawai et al.56
Highlights.
The structure-function relationship of α–tropomyosin (Tm) was investigated.
Deletion (Δ) and shift mutants of Tm was used with in vitro motility assays.
Significance of Tm’s period 3 was demonstrated for cooperative activation of actin.
A large increase in cooperativity when Ala cluster was introduced to periods 2 and 3.
Δ6Tm impaired relaxation, because of uncoupling of C-TnT/TnIReg with period 5.
Acknowledgments
The authors would like to thank to Dr. Rainer Fink of Heidelberg University for the image analysis program used, and Dr. Frederick Wagner (Heidelberg University) for teaching MK how to use this program. We also thank Dr. Abhishek Singh, formerly of Robert Wood Johnson Medical School (now at the University of California, San Francisco) for the gift of P2s, P3s, P2P3s, and P1P5 Tms. This work was supported by Grants-in-Aid for Specially Promoted Research and Scientific Research (S) from the Ministry of Education, Sports, Culture, Science and Technology of Japan to SI; by grants NIH GM36326, NIH GM93065 and the UMDNJ Foundation to SEHD; and by grants from NIH HL70041 and AHA 0850184Z to MK. The work reported here and the conclusions drawn are solely the responsibility of the authors, and do not necessarily represent the official views of the awarding organizations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Yusuke Oguchi, Email: oguchi@gsc.riken.jp.
Junji Ishizuka, Email: junji.i@akane.waseda.jp.
Sarah E. Hitchcock-DeGregori, Email: hitchcoc@umdnj.edu.
Shin’ichi Ishiwata, Email: ishiwata@waseda.jp.
Masataka Kawai, Email: masataka-kawai@uiowa.edu.
References
- 1.Lehrer SS, Golitsina NL, Geeves MA. Actin-tropomyosin activation of myosin subfragment 1 ATPase and thin filament cooperativity. The role of tropomyosin flexibility and end-to-end interactions. Biochemistry. 1997;36:13449–54. doi: 10.1021/bi971568w. [DOI] [PubMed] [Google Scholar]
- 2.Bremel RD, Weber A. Cooperation within actin filament in vertebrate skeletal muscle. Nat New Biol. 1972;238:97–101. doi: 10.1038/newbio238097a0. [DOI] [PubMed] [Google Scholar]
- 3.VanBuren P, Palmiter KA, Warshaw DM. Tropomyosin directly modulates actomyosin mechanical performance at the level of a single actin filament. Proc Natl Acad Sci USA. 1999;96:12488–12493. doi: 10.1073/pnas.96.22.12488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eaton BL. Tropomyosin binding to F-actin induced by myosin heads. Science. 1976;192:1337–9. doi: 10.1126/science.131972. [DOI] [PubMed] [Google Scholar]
- 5.Lu X, Tobacman LS, Kawai M. Effects of tropomyosin internal deletion Δ23Tm on isometric tension and the cross-bridge kinetics in bovine myocardium. J Physiol. 2003;553:457–471. doi: 10.1113/jphysiol.2003.053694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita H, Sasaki D, Ishiwata S, Kawai M. Elementary steps of the cross-bridge cycle in bovine myocardium with and without regulatory proteins. Biophys J. 2002;82:915–928. doi: 10.1016/S0006-3495(02)75453-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parry DA. Analysis of the primary sequence of alpha-tropomyosin from rabbit skeletalmuscle. J Mol Biol. 1975;98:519–535. doi: 10.1016/s0022-2836(75)80084-2. [DOI] [PubMed] [Google Scholar]
- 8.Irimia M, Maeso I, Gunning PW, Garcia-Fernàndez J, Roy SW. Internal and external paralogy in the evolution of tropomyosin genes in metazoans. Mol Biol Evol. 2010;27:1504–1517. doi: 10.1093/molbev/msq018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phillips GN. Construction of an atomic model for tropomyosin and implications for interactions with actin. J Mol Biol. 1986;192:128–31. doi: 10.1016/0022-2836(86)90469-9. [DOI] [PubMed] [Google Scholar]
- 10.McLachlan AD, Stewart M. The 14-fold periodicity in alpha-tropomyosin and the interaction with actin. J Mol Biol. 1976;103:271–98. doi: 10.1016/0022-2836(76)90313-2. [DOI] [PubMed] [Google Scholar]
- 11.Cho YJ, Liu J, Hitchcock-DeGregori SE. The amino terminus of muscle tropomyosin is a major determinant for function. J Biol Chem. 1990;265:538–45. [PubMed] [Google Scholar]
- 12.Hitchcock-DeGregori SE, Song Y, Greenfield NJ. Functions of tropomyosin’s periodic repeats. Biochemistry. 2002;41:15036–44. doi: 10.1021/bi026519k. [DOI] [PubMed] [Google Scholar]
- 13.Moraczewska J, Hitchcock-DeGregori SE. Independent functions for the N- and C-termini in the overlap region of tropomyosin. Biochemistry. 2000;39:6891–7. doi: 10.1021/bi000242b. [DOI] [PubMed] [Google Scholar]
- 14.Landis CA, Bobkova A, Homsher E, Tobacman LS. The active state of the thin filament is destabilized by an internal deletion in tropomyosin. J Biol Chem. 1997;272:14051–6. doi: 10.1074/jbc.272.22.14051. [DOI] [PubMed] [Google Scholar]
- 15.Landis C, Back N, Homsher E, Tobacman LS. Effects of tropomyosin internal deletions on thin filament function. J Biol Chem. 1999;274:31279–85. doi: 10.1074/jbc.274.44.31279. [DOI] [PubMed] [Google Scholar]
- 16.Kawai M, Lu X, Hitchcock-Degregori SE, Stanton KJ, Wandling MW. Tropomyosin period 3 is essential for enhancement of isometric tension in thin filament-reconstituted bovine myocardium. J Biophys. 2009;2009:1–17. doi: 10.1155/2009/380967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawai M, Ishiwata S. Use of thin filament reconstituted muscle fibres to probe the mechanism of force generation. J Muscle Res Cell Motil. 2006;27:455–468. doi: 10.1007/s10974-006-9075-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh A, Hitchcock-DeGregori SE. Tropomyosin’s periods are quasi-equivalent for actin binding but have specific regulatory functions. Biochemistry. 2007;46:14917–27. doi: 10.1021/bi701570b. [DOI] [PubMed] [Google Scholar]
- 19.Sakuma A, Kimura-Sakiyama C, Onoue A, Shitaka Y, Kusakabe T, Miki M. The second half of the fourth period of tropomyosin is a key region for Ca2+-dependent regulation of striated muscle thin filaments. Biochemistry. 2006;45:9550–8. doi: 10.1021/bi060963w. [DOI] [PubMed] [Google Scholar]
- 20.Brown JH, Kim KH, Jun G, Greenfield NJ, Dominguez R, Volkmann N, Hitchcock-DeGregori SE, Cohen C. Deciphering the design of the tropomyosin molecule. Proc Natl Acad Sci U S A. 2001;98:8496–501. doi: 10.1073/pnas.131219198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown JH. How sequence directs bending in tropomyosin and other two-stranded alpha-helical coiled coils. Protein Sci. 2010;19:1366–75. doi: 10.1002/pro.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh A, Hitchcock-DeGregori SE. Local destabilization of the tropomyosin coiled coil gives the molecular flexibility required for actin binding. Biochemistry. 2003;42:14114–14121. doi: 10.1021/bi0348462. [DOI] [PubMed] [Google Scholar]
- 23.Singh A, Hitchcock-DeGregori SE. Dual requirement for flexibility and specificity for binding of the coiled-coil tropomyosin to its target, actin. Structure. 2006;14:43–50. doi: 10.1016/j.str.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 24.Singh A, Hitchcock-Degregori SE. A peek into tropomyosin binding and unfolding on the actin filament. PLoS One. 2009;4:e6336. doi: 10.1371/journal.pone.0006336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kishino A, Yanagida T. Force measurements by micromanipulation of a single actin filament by glass needles. Nature. 1988;334:74–76. doi: 10.1038/334074a0. [DOI] [PubMed] [Google Scholar]
- 26.Hitchcock-DeGregori SE, Heald RW. Altered actin and troponin binding of amino-terminal variants of chicken striated muscle alpha-tropomyosin expressed in Escherichia coli. J Biol Chem. 1987;262:9730–5. [PubMed] [Google Scholar]
- 27.Hitchcock-DeGregori SE, Varnell TA. Tropomyosin has discrete actin-binding sites with sevenfold and fourteenfold periodicities. J Mol Biol. 1990;214:885–96. doi: 10.1016/0022-2836(90)90343-K. [DOI] [PubMed] [Google Scholar]
- 28.Hitchcock-DeGregori SE, An Y. Integral repeats and a continuous coiled coil are required for binding of striated muscle tropomyosin to the regulated actin filament. J Biol Chem. 1996;271:3600–3. doi: 10.1074/jbc.271.7.3600. [DOI] [PubMed] [Google Scholar]
- 29.Hitchcock-DeGregori SE, Song Y, Moraczewska J. Importance of internal regions and the overall length of tropomyosin for actin binding and regulatory function. Biochemistry. 2001;40:2104–12. doi: 10.1021/bi002421z. [DOI] [PubMed] [Google Scholar]
- 30.Gunning P, O’Neill G, Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol Rev. 2008;88:1–35. doi: 10.1152/physrev.00001.2007. [DOI] [PubMed] [Google Scholar]
- 31.Wang CL, Coluccio LM. New insights into the regulation of the actin cytoskeleton by tropomyosin. Int Rev Cell Mol Biol. 2010;281:91–128. doi: 10.1016/S1937-6448(10)81003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greenfield NJ, Huang YJ, Swapna GV, Bhattacharya A, Rapp B, Singh A, Montelione GT, Hitchcock-DeGregori SE. Solution NMR structure of the junction between tropomyosin molecules: implications for actin binding and regulation. J Mol Biol. 2006;364:80–96. doi: 10.1016/j.jmb.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 33.Gordon AM, Chen Y, Liang B, LaMadri M, Luo Z, Chase PB. Skeletal muscle regulatory proteins enhance F-actin in vitro motility. Adv Exp Med Biol. 1998;453:187–196. doi: 10.1007/978-1-4684-6039-1_22. [DOI] [PubMed] [Google Scholar]
- 34.Bing W, Knott A, Marston SB. A simple method for measuring the relative force exerted by myosin on actin filaments in the in vitro motility assay: evidence that tropomyosin and troponin increase force in single thin filaments. Biochem J. 2000;350:693–699. [PMC free article] [PubMed] [Google Scholar]
- 35.Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 36.Gorga JA, Fishbaugher DE, VanBuren P. Activation of the calcium-regulated thin filament by myosin strong binding. Biophys J. 2003:2484–91. 2484–91. doi: 10.1016/s0006-3495(03)74671-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Homsher E, Lee DM, Morris C, Pavlov D, Tobacman LS. Regulation of force and unloaded sliding speed in single thin filaments: effects of regulatory proteins and calcium. J Physiol 524 Pt. 2000;1:233–43. doi: 10.1111/j.1469-7793.2000.00233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clemmens EW, Regnier M. Skeletal regulatory proteins enhance thin filament sliding speed and force by skeletal HMM. J Muscle Res Cell Motil. 2004;25:515–25. doi: 10.1007/s10974-004-3787-0. [DOI] [PubMed] [Google Scholar]
- 39.Clemmens EW, Entezari M, Martyn DA, Regnier M. Different effects of cardiac versus skeletal muscle regulatory proteins on in vitro measures of actin filament speed and force. J Physiol. 2005;566:737–46. doi: 10.1113/jphysiol.2005.084194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siththanandan VB, Tobacman LS, Van Gorder N, Homsher E. Mechanical and kinetic effects of shortened tropomyosin reconstituted into myofibrils. Pflugers Arch. 2009;458:761–76. doi: 10.1007/s00424-009-0653-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown JH, Cohen C. Regulation of muscle contraction by tropomyosin and troponin: how structure illuminates function. Adv Protein Chem. 2005;71:121–59. doi: 10.1016/S0065-3233(04)71004-9. [DOI] [PubMed] [Google Scholar]
- 42.Kee AJ, Hardeman EC. Tropomyosins in skeletal muscle diseases. Adv Exp Med Biol. 2008;644:143–57. doi: 10.1007/978-0-387-85766-4_12. [DOI] [PubMed] [Google Scholar]
- 43.Wieczorek DF, Jagatheesan G, Rajan S. The role of tropomyosin in heart disease. Adv Exp Med Biol. 2008;644:132–42. doi: 10.1007/978-0-387-85766-4_11. [DOI] [PubMed] [Google Scholar]
- 44.Hitchcock-DeGregori SE, Singh A. What makes tropomyosin an actin binding protein? A perspective. J Struct Biol. 2010;170:319–324. doi: 10.1016/j.jsb.2009.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown JH, Zhou Z, Reshetnikova L, Robinson H, Yammani RD, Tobacman LS, Cohen C. Structure of the mid-region of tropomyosin: bending and binding sites for actin. Proc Natl Acad Sci U S A. 2005;102:18878–83. doi: 10.1073/pnas.0509269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minakata S, Maeda K, Oda N, Wakabayashi K, Nitanai Y, Maeda Y. Two-crystal structures of tropomyosin C-terminal fragment 176–273: exposure of the hydrophobic core to the solvent destabilizes the tropomyosin molecule. Biophys J. 2008;95:710–9. doi: 10.1529/biophysj.107.126144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nitanai Y, Minakata S, Maeda K, Oda N, Maeda Y. Crystal structures of tropomyosin: flexible coiled-coil. Adv Exp Med Biol. 2007;592:137–51. doi: 10.1007/978-4-431-38453-3_13. [DOI] [PubMed] [Google Scholar]
- 48.Li XE, Holmes KC, Lehman W, Jung H, Fischer S. The shape and flexibility of tropomyosin coiled coils: implications for actin filament assembly and regulation. J Mol Biol. 395:327–39. doi: 10.1016/j.jmb.2009.10.060. [DOI] [PubMed] [Google Scholar]
- 49.Li XE, Lehman W, Fischer S. The relationship between curvature, flexibility and persistence length in the tropomyosin coiled-coil. J Struct Biol. 2010;170:313–318. doi: 10.1016/j.jsb.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sato K, Ohtaki M, Shimamoto Y, Ishiwata S. A theory on auto-oscillation and contraction in striated muscle. Prog Biophys Mol Biol. 105:199–207. doi: 10.1016/j.pbiomolbio.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 51.Tawada K, Sekimoto K. Protein friction exerted by motor enzymes through a weak-binding interaction. J Theor Biol. 1991;150:193–200. doi: 10.1016/s0022-5193(05)80331-5. [DOI] [PubMed] [Google Scholar]
- 52.Barua B, Pamula MC, Hitchcock-DeGregori SE. Evolutionarily conserved surface residues constitute actin binding sites of tropomyosin. Proc Natl Acad Sci U S A. 2011;108:10150–5. doi: 10.1073/pnas.1101221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Potter JD. Preparation of troponin and its subunits. Methods Enzymol. 1982;85(B):241–63. doi: 10.1016/0076-6879(82)85024-6. [DOI] [PubMed] [Google Scholar]
- 54.Fujita H, Yasuda K, Niitsu S, Funatsu T, Ishiwata S. Structural and functional reconstitution of thin filaments in the contractile apparatus of cardiac muscle. Biophys J. 1996;71:2307–2318. doi: 10.1016/S0006-3495(96)79465-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suzuki N, Miyata H, Ishiwata S, Kinosita K. Preparation of bead-tailed actin filaments: estimation of the torque produced by the sliding force in an in vitro motility assay. Biophys J. 1996;70:401–408. doi: 10.1016/S0006-3495(96)79583-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawai M, Kido T, Vogel M, Fink RH, Ishiwata S. Temperature change does not affect force between regulated actin filaments and heavy meromyosin in single-molecule experiments. J Physiol. 2006;574:877–87. doi: 10.1113/jphysiol.2006.111708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ishiwata S. A study on the F-actin-tropomyosin-troponin complex. I. Gel-filament transformation. Biochim Biophys Acta. 1973;303:77–89. doi: 10.1016/0005-2795(73)90150-5. [DOI] [PubMed] [Google Scholar]
- 58.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 59.Harada Y, Sakurada K, Aoki T, Thomas DD, Yanagida T. Mechanochemical coupling in actomyosin energy transduction studied by in vitro movement assay. J Mol Biol. 1990;216:49–68. doi: 10.1016/S0022-2836(05)80060-9. [DOI] [PubMed] [Google Scholar]
- 60.Kawai M, Kawaguchi K, Saito M, Ishiwata S. Temperature change does not affect force between single actin filaments and HMM from rabbit muscles. Biophys J. 2000;78:3112–3119. doi: 10.1016/S0006-3495(00)76848-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uttenweiler D, Veigel C, Steubing R, Goetz C, Mann S, Haubecker H, Jaehne B, Fink RHA. Motion determination in actin filament fluorescence images with a spatio-temporal orientation analysis method. Biophys J. 2000;78:2709–2715. doi: 10.1016/S0006-3495(00)76815-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nishizaka T, Seo R, Tadakuma H, Kinosita K, Jr, Ishiwata S. Characterization of single actomyosin rigor bonds: load dependence of lifetime and mechanical properties. Biophys J. 2000;79:962–74. doi: 10.1016/S0006-3495(00)76350-8. [DOI] [PMC free article] [PubMed] [Google Scholar]