Abstract
Telomerase is a ribonucleoprotein (RNP) enzyme that maintains the ends of linear eukaryotic chromosomes and whose activation is a hallmark of 90% of all cancers. This RNP minimally contains a reverse transcriptase protein subunit (TERT) that catalyzes telomeric DNA synthesis and an RNA subunit (TER) that has templating, architectural and protein-scaffolding roles. Telomerase is unique among polymerases in that it synthesizes multiple copies of the template on the 3′ end of a primer following a single binding event, a process known as repeat addition processivity (RAP). Using biochemical assays and single-molecule Förster resonance energy transfer (smFRET) experiments on Tetrahymena thermophila telomerase, we now directly demonstrate that TER contributes to template positioning within the active site and to the template translocation required for RAP. We propose that the single-stranded RNA elements flanking the template act as a molecular accordion, undergoing reciprocal extension and compaction during telomerase translocation.
The ribonucleoprotein particle telomerase catalyzes the addition of telomeric DNA repeats to the ends of linear chromosomes by using the reverse transcriptase motifs within telomerase reverse transcriptase (TERT) and the template contained within telomerase RNA (TER)1,2. Telomerase can use its internal template RNA sequence to template several consecutive rounds of telomeric DNA repeat addition, a phenomenon known as repeat addition processivity (RAP); this feature of telomerase is unique among polymerases. Because inappropriate activation of telomerase contributes to malignant transformation, understanding the molecular mechanism of the enzyme’s unique ability to recycle an internal template is central to understanding telomerase function on basic and clinical levels.
While TER sequences vary widely, their RNA secondary and tertiary structural elements are largely conserved3–5. Most TERs contain a template sequence flanked on the 5′ side by a template boundary element (TBE) and on the 3′ side by a stretch of single-stranded RNA followed by a pseudoknot6–13, as exemplified in the exceptionally streamlined Tetrahymena thermophila TER (Fig. 1a). Experiments with circularly permuted RNAs showed that covalent connectivity between the template and the sequences flanking it, the TBE and the template recognition element (TRE), is required for efficient translocation and may be involved in template positioning in Tetrahymena14,15. In addition, nuclease protection experiments demonstrated the TRE and template are susceptible to both single- and double-stranded RNA nucleases, suggesting this region may alternate between a helical or stacked conformation and a flexible unstacked conformation16.
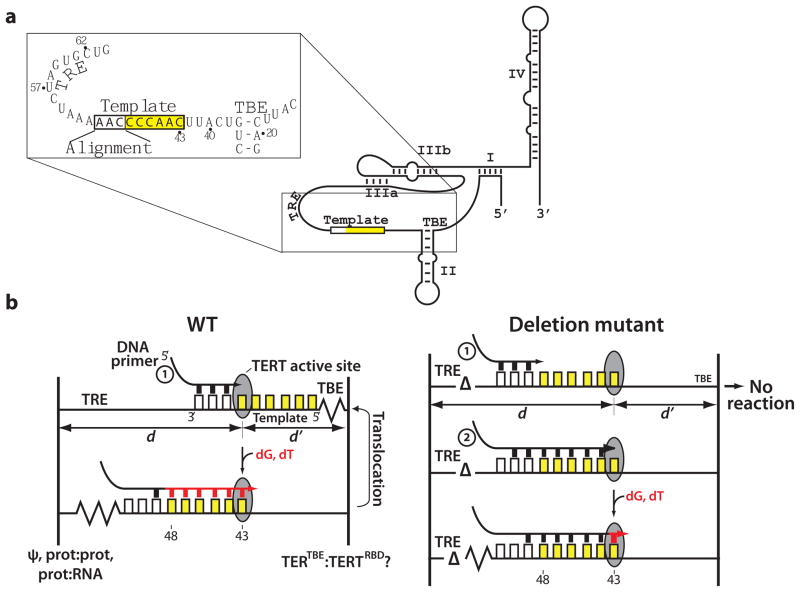
Figure 1.
The RNA accordion model for primer positioning and translocation. (a) Secondary structure of T. thermophila TER, with the inset showing the sequence of the template, the TBE and the TRE. The template is boxed; the alignment region is white and the sequence that is reverse transcribed is yellow; (b) Accordion model for template progression and translocation of telomerase. Left, in wild-type (WT) TER, primer extension is accompanied by compaction of the TRE and extension of the TBE. Right, deletion of nucleotides in the TRE (delta symbol) prevents bound primer 1 from reaching the active site, giving No reaction. Primer 2, which pairs near the 5′ end of the template, is able to undergo one round of extension. The active site of TERT is represented as a grey circle. Distances d and d′ represent our assumption that the position of the active site remains constant relative to an anchored TRE and TBE, respectively. The template sequence is represented as boxes, colored as in panel a. Primers are in black, newly added nucleotides in red. A maximum of seven base pairs forms between the primer and template30,31.
To explain these previous observations, we developed a detailed model for the contribution of TER to template progression through the TERT active site and the subsequent translocation required for RAP (Fig. 1b, left). Telomerase has a single active site for nucleotide addition2, so addition of nucleotides to the 3′ end of the primer must necessarily be accompanied by movement of the template through the active site. Given evidence for a site of strong protein-RNA interaction 5′ of the template (the TBE)17,18 and the existence of a conserved RNA structure 3′ of the template (the pseudoknot)5,13, it is likely that RNA elements on both sides of the template occupy fixed positions relative to the active site (distances d and d′ in Fig. 1b). We hypothesize that the single-stranded regions of RNA on either side of the template undergo reciprocal compression and expansion in an accordion-like manner to accommodate movement of the template RNA during the catalytic cycle. Here we provide evidence for the RNA positioning predicted by the accordion model, which may also have implications for the energetics of template translocation.
RESULTS
Deleting TRE nucleotides reduces RAP in a predictable manner
One prediction of the accordion model is that deletions in the TRE could prevent the 3′ template nucleotides from reaching the active site, so that primers paired in the alignment region (nucleotides 49–51) would give no reaction (No reaction, Fig. 1b, right). Primers with sequences that aligned within the template might be able to reach the active site, especially with smaller TRE deletions, but after one round of extension they would pair in the alignment region and undergo no further reaction. In general, the activities of different primers should reveal which template nucleotides are able to reach the active site in each mutant telomerase.
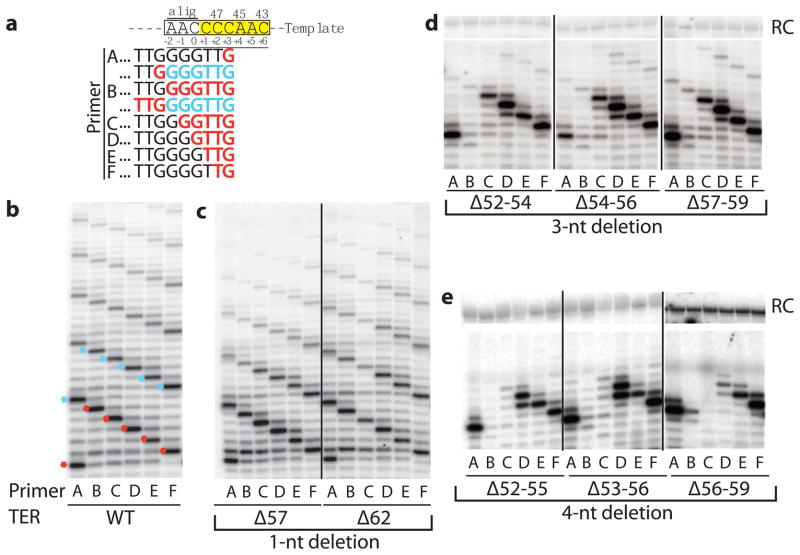
To test this hypothesis, we generated a series of TER molecules containing deletions in the TRE. Each mutant TER was assembled into a telomerase RNP and tested in telomerase activity assays with primers representing each of the six permutations of telomeric sequence (Fig. 2a); wild-type telomerase utilizes all primers efficiently (Fig. 2b). Single-nucleotide TRE deletions were tolerated by telomerase (Fig. 2c and Supplementary Fig. 1a). For all double-nucleotide deletion mutants, RAP was reduced compared to the activity of wild-type (WT) telomerase (Supplementary Fig. 1b). Deleting three sequential nucleotides had a dramatic effect, allowing robust addition of one repeat but very little RAP (Fig. 2d and Supplementary Fig. 1c). Primer B, which anneals to the alignment region of the template, was essentially inactive even for the first round of extension, and primer C showed reduced activity. This primer usage profile was independent of the location of the deletion within the TRE, suggesting that sequence is not as important as length. A similar observation was made for mutants with four sequential nucleotides deleted, but now primer C was also inactive (Fig. 2e), suggesting that in all four-nucleotide deletion mutants, template nucleotide 47 can no longer reach the active site. Deletion of six nucleotides (Δ54–59) continued the trend, preventing TER nucleotide 46 from being used as template. The largest deletion tested (Δ52–59) was only able to efficiently use primer A, which base pairs with the penultimate nucleotide of the template (Fig. 3). This eight-nucleotide deletion presumably acts as a molecular ruler, measuring the minimum distance between the pseudoknot and C43 in the active site.
Figure 2.
Deletion mutants in the TRE have predictable primer usage profiles. (a) The 3′ ends (black) of the six primers of permuted telomeric sequence aligned with the template. All primers were 18 nucleotides (three repeats of permuted telomeric sequence). Nucleotides added during the first round of synthesis are red.
Nucleotides added during the second round are shown for primers A and B in blue. Templating position is indicated below the template sequence, and is defined by the first nucleotide that can be copied8; (b–e) Telomerase activity assays for WT (b) and a selection of single (c), triple (d) and quadruple (e) deletion mutants within the TRE of TER with each of the indicated primers. Red and blue asterisks in the WT panel indicate the first and second rounds of RAP, respectively. RC, recovery control.
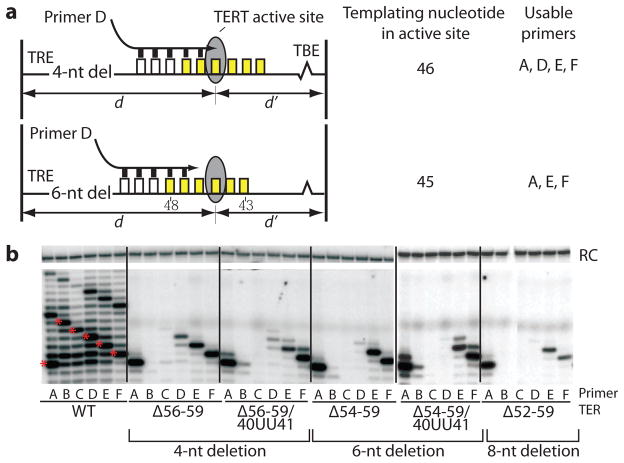
Figure 3.
The accordion model predicts primer usage for TRE deletion mutants. (a) Prediction of the interaction of primer D ([GTTGGG]3) with a four-nucleotide deletion mutant (top) and a six-nucleotide deletion mutant (bottom). (b) Telomerase activity assays on deletion and deletion-insertion combination mutants. Asterisks within the WT lanes indicate the first repeat for each primer. The exact nucleotides deleted and inserted are indicated for each panel. RC, recovery control.
Insertions in the TBE reinforce template misalignment
A second prediction of the accordion model is that flexibility in the single-stranded RNA portion of the TBE, which is 5′ of the template, is also required for template progression. Insertions of nucleotides adjacent to the TBE are predicted to reinforce the misalignment of the template relative to the active site in the context of TRE deletion mutants. We modified two TRE deletion mutants (Δ56–59, a four-nucleotide deletion, and Δ54–59, a six-nucleotide deletion) by inserting two uridines between nucleotides 40 and 41 of the TBE, just 5′ of the template (Fig. 3). While the four-nucleotide TRE deletion had only a modest ability to use primer D, the combination of the insertion of 2Us in the TBE and the four-nucleotide TRE deletion exacerbated this effect, resulting in a primer usage profile similar to that of the six-nucleotide TRE deletion mutant. Similarly, the six-nucleotide deletion mutant containing an insertion of two uridines 5′ of the template had a weakened ability to use primer E, matching the primer usage profile of the eight-nucleotide deletion mutant.
Discontinuities flanking the RNA template eliminate RAP
Previous experiments with circularly permuted (cp) telomerase RNAs demonstrated that discontinuities on either side of the template decreased RAP15. We interpret these results as showing that coordinated structural rearrangements on both sides of the template contribute to template movement. The previous experiments were performed in the absence of p65, the telomerase-specific RNA folding protein19 that is essential in vivo20 and has the ability to rescue many severely destabilizing TERT and TER mutations in vitro16. If nicked RNAs were defective simply because of structural destabilization, they might likewise be suppressed in telomerase containing p65.
We therefore tested the activities of cpRNAs assembled into telomerase RNPs in both the presence and absence of p65 (Supplementary Fig. 2). Because the TBE is necessary for TERT binding17,21 and proper telomerase function8, we tested four circularly permuted RNAs (cpRNA) within this region, and one in the TRE. Telomerase RNPs containing the cpRNAs had essentially full first-round activity, but thereafter RAP was severely reduced, with nicks in the TBE affecting the reaction similarly to the nick at position 59 in the TRE. This limited RAP even in the presence of p65 provides further support for the requirement for direct connectivity of the RNA on both sides of the template.
Single-molecule FRET reveals RNA movement as predicted
Taken together, these biochemical experiments demonstrate that specific deletions and insertions, as well as the introduction of nicks, in the RNA elements flanking the template evoke catalytic defects predicted by the accordion model. However, these biochemical assays cannot directly probe RNA structure; therefore single-molecule Förster resonance energy transfer (smFRET) was employed to monitor RNA conformational changes within individual telomerase-DNA primer complexes at successive steps in the telomerase catalytic cycle (Fig. 4a and Supplementary Fig. 3)19. Ideally, smFRET experiments performed in the presence of dTTP and dGTP could permit real-time observation of TER structural rearrangements throughout the telomerase catalytic cycle. To date, however, FRET-based monitoring of telomerase activity has remained elusive. Therefore, to simplify interpretation of our data, independent smFRET experiments were performed with primers successively extended on the 3′ end in the absence of dGTP and dTTP. In this way, the primers used in separate experiments emulate the different conformational states that would be sampled during telomerase-catalyzed DNA synthesis.
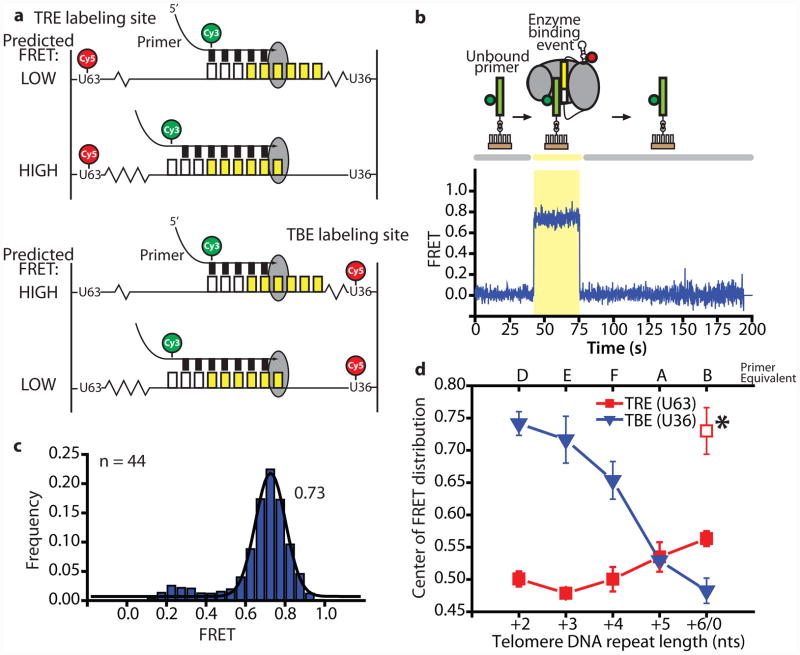
Figure 4.
Single-molecule FRET measurements of RNA compression and extension at successive steps of telomere repeat synthesis. (a) The accordion model predicts RNA extension (low FRET) and compression (high FRET) in the TRE for short and long telomere repeat primers, respectively (top panel). The opposite trend is predicted for the TBE region (bottom panel). In addition to the TBE, it is possible that part of the template itself compacts; (b) Representative single molecule trace of a primer-telomerase binding event, detected by the onset of FRET (shaded area); (c) Representative FRET histogram of 44 binding events of a GT-rich primer with the same 3′ terminal sequence as primer D (Figure 2 and Supplemental Fig. 4) bound by TBE (U36)-labeled telomerase. A Gaussian fit determines the center of the distribution to be 0.73 FRET; (d) Mean values for the center of each FRET distribution plotted as a function of the length of the 3′ extension of the primer. Complete primer sequences shown in Supplementary Fig. 4. TBE labeling site (U36) data, blue triangles. TRE labeling site (U63) data, red squares. Error bars are the standard deviation of triplicate experiments (number of binding events in each experiment varied from n = 23 to 106). Asterisk represents an alternative FRET state. Primer B with U63-labeled enzyme demonstrated a bimodal distribution, centered at FRET=0.57 and 0.73 (Supplementary Figs. 4 and 6). The points are plotted separately and the line is drawn to the 0.57 FRET state on the basis of continuity with previous primers.
Surface-immobilized telomeric DNA primers coupled to a donor dye (Cy3) were monitored in the presence of reconstituted telomerase enzymes harboring an acceptor dye (Cy5) at U63 in the TRE (Fig. 4a, top panel) or at U36 in the TBE (Fig. 4a, bottom panel). Importantly, these modified DNA primers and telomerase RNAs support near wild-type levels of telomerase activity22 (Supplementary Fig. 3). To ensure telomerase enzymes bound the RNA template in a single register, each primer used in our smFRET experiments consisted of a 5′-(TG)8 stretch followed by a single copy of one of the telomere repeat permutations (Supplementary Fig. 4)23. Since FRET efficiency is highly sensitive to the distance between the dyes, FRET provides a direct readout of the intervening RNA structure with high FRET and low FRET corresponding to compressed and extended RNA, respectively.
In this experiment, the binding of a Cy3-labeled DNA primer within the active site of a Cy5-labeled telomerase enzyme is observed in real time as the onset of energy transfer between the two FRET probes (Fig. 4b). For each experiment, histograms corresponding to the frequency of observed FRET values were compiled and fit with Gaussian functions to determine the center of the distribution (Fig. 4c and Supplementary Fig. 4).
Measurements from the bound primer to the TBE show a dramatic decrease in FRET as the template-complementary sequence at the primer 3′ end is extended (Fig. 4d). Measurements of the distance from the primer to the TRE show a smaller but appreciable rise in FRET with the same series of primers. These reciprocal changes are just as predicted by the accordion model. The larger FRET change measured across the TBE region than across the TRE has several possible explanations. First, the compression in the TBE may be greater in magnitude than the expansion in the TRE. A second possibility is that the U63 labeling position may not be fixed in space throughout the catalytic cycle and further expansion may occur 3′ of the TRE. Lastly, the observed asymmetry could be explained if the donor dye movement is out of plane with respect to the position of the acceptor dye. In other words, in three-dimensional space the relatively small FRET change observed for the TRE may in fact correspond to a significant rearrangement of the RNA.
Many of the smFRET distributions generated in our experiments are broader than would be expected for a single FRET distribution. In particular, we observed a gradual broadening of the FRET distributions as the length of the DNA primer was increased (Supplementary Fig. 4, primers F, A, and B). Analysis of smFRET trajectories revealed the broadening to be derived from anti-correlated donor and acceptor fluctuations in individual traces over a narrow range of FRET values (~0.15 FRET) (Supplementary Fig. 5 and 6). Since the physical basis for these FRET states is not known at present, we opted to treat the majority of the data by fitting to a single Gaussian function to extract the average FRET value for each primer.
One exception to this approach was for data obtained with the TRE (U63) label and the primer corresponding to a complete telomeric repeat (primer B). In this experiment, we observed a substantial number of binding events exhibiting a long-lived 0.73 FRET state (Fig. 4d, asterisk; Supplementary Fig. 6, Primer B) which is likely due to an alternative primer conformation. These data were therefore fit with two Gaussian functions (centered at 0.57 and 0.73) in order to avoid the artificially large apparent increase in FRET resulting from a single-Gaussian fit. Notably, transient binding events corresponding to a similar high FRET state were also observed for the shorter primers (D, E, F, A) (Supplementary Fig. 7, TRE (U63)-labeled enzyme), but their short durations resulted in a negligible contribution to the FRET histograms. Given that primer B is the only primer that shows this stable alternative high FRET state and that primer B represents the substrate for translocation during RAP, it is possible that the high FRET state represents a post-translocated state of the enzyme that is preferentially stabilized for telomeric primers terminating in a complete telomere repeat sequence. Importantly, our conclusion that there is increased FRET between the TRE (U63) and the most extended primer holds if we consider either of the two individual FRET states (0.57 and 0.73) or the average of the two.
DISCUSSION
Because TERT is a reverse transcriptase related to retroviral and retrotransposon reverse transcriptases, it is thought to use a similar metal-ion-catalyzed mechanism for nucleotide addition2,24,25. In contrast, the mechanism by which it achieves its unique activities -- template translocation and repeat addition processivity --remains unknown. Here, the predictable effects of nucleotide deletions on primer use and measurements of sequential primer position using single-molecule FRET both support an RNA accordion model for RAP. We conclude that the RNA elements flanking the template serve to position the template for nucleotide addition and for RAP.
We have considered alternative models that could explain our data. The requirement for connectivity of the RNA on the 5′ side of the template might be a consequence of its interaction with the RNA-binding domain of TERT, which might be disrupted by nicking. This is difficult to address in the absence of a detailed structure of the telomerase RNP. In addition, a small RNA stem-loop might form within the TRE during telomere synthesis, and this stem-loop could melt for translocation. This alternative model has been tested with deletion and point mutations predicted to stabilize or destabilize the putative hairpin. If hairpin formation and melting contributed to translocation, one would expect a large effect on RAP in the context of these mutants, similar to the effect on RAP observed with nicked RNAs or with RNAs containing deletions of three or more nucleotides in the TRE. However, in the presence of mutants that either stabilized or destabilized the putative hairpin, only a modest impact on RAP was observed; RAP decreased by less than fifty percent as compared to WT for all mutants tested (Supplementary Fig. 8).
While our data do not directly address the energetics of translocation, the disruption of RAP by RNA nicking observed by Collins15 and in our Supplementary Fig. 2 suggests that the RNA has an active role in template movement. More specifically, upon synthesis of a complete telomere DNA repeat, the TRE region of the RNA may exist in a high-energy state. For example, this single-stranded region of RNA could become compressed, or energetically unfavorable TER-TERT interactions could be produced. The relaxation of this high-energy state would then drive translocation.
A precedent exists for polymerase translocation powered by energy stored within nucleic acid. Polymerases move along their templates without external energy; the energy provided by nucleotide incorporation and pyrophosphate release can be converted directly into directional movement or it can be stored within scrunched nucleic acid that can later be manifested as translocation. In RNA polymerases, for example, the DNA template bunches near the active site during RNA synthesis initiation, resulting in a higher energy state that likely provides the energy for promoter clearance26–28. The conformational changes observed within telomerase TER are reminiscent of this mechanism. Here, however, rather than the primer DNA storing energy, perhaps the RNA downstream of the template bunches up as the template is copied, potentially storing energy during repeat synthesis, and releasing it to drive template recycling. The molecular basis for compaction could also involve base stacking16 and RNA-TERT protein interactions.
In addition, it is possible that a ratchet might be involved in translocation; the pawl for this ratchet would eliminate backtracking of the template until the end is reached29. A ratchet mechanism is compatible with RNA conformational changes being necessary for translocation, and thus could represent an elaboration of the accordion model. Further understanding of the translocation mechanism awaits detailed structural information concerning the RNA-protein interactions in the vicinity of the active site and their dynamics during processive extension of telomeric DNA.
Supplementary Material
Acknowledgments
This work was partially supported by a Jane Coffin Childs Memorial Fund for Medical Research postdoctoral fellowship to Andrea J. Berman, National Institutes of Health grant GM095850 to Michael D. Stone, and National Institutes of Health training grant T32 GM8646 to Benjamin M. Akiyama.
Footnotes
Author contributions
AJB performed all biochemical experiments; AJB and TRC designed and analyzed biochemical experiments. BMA performed smFRET experiments; BMA and MDS designed and analyzed smFRET experiments. All authors participated in writing the manuscript.
References
- 1.Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–98. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 2.Lingner J, et al. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–7. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 3.Romero DP, Blackburn EH. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–53. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- 4.Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–14. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 5.ten Dam E, van Belkum A, Pleij K. A conserved pseudoknot in telomerase RNA. Nucleic acids research. 1991;19:6951. doi: 10.1093/nar/19.24.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen JL, Greider CW. An emerging consensus for telomerase RNA structure. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:14683–4. doi: 10.1073/pnas.0406204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tzfati Y, Fulton TB, Roy J, Blackburn EH. Template boundary in a yeast telomerase specified by RNA structure. Science. 2000;288:863–7. doi: 10.1126/science.288.5467.863. [DOI] [PubMed] [Google Scholar]
- 8.Autexier C, Greider CW. Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes & development. 1995;9:2227–39. doi: 10.1101/gad.9.18.2227. [DOI] [PubMed] [Google Scholar]
- 9.Gilley D, Blackburn EH. Specific RNA residue interactions required for enzymatic functions of Tetrahymena telomerase. Molecular and cellular biology. 1996;16:66–75. doi: 10.1128/mcb.16.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Licht JD, Collins K. Telomerase RNA function in recombinant Tetrahymena telomerase. Genes & development. 1999;13:1116–25. doi: 10.1101/gad.13.9.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shefer K, et al. A triple helix within a pseudoknot is a conserved and essential element of telomerase RNA. Molecular and cellular biology. 2007;27:2130–43. doi: 10.1128/MCB.01826-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Molecular cell. 2005;17:671–82. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 13.Tzfati Y, Knight Z, Roy J, Blackburn EH. A novel pseudoknot element is essential for the action of a yeast telomerase. Genes & development. 2003;17:1779–88. doi: 10.1101/gad.1099403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mason DX, Goneska E, Greider CW. Stem-loop IV of tetrahymena telomerase RNA stimulates processivity in trans. Molecular and cellular biology. 2003;23:5606–13. doi: 10.1128/MCB.23.16.5606-5613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller MC, Collins K. Telomerase recognizes its template by using an adjacent RNA motif. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6585–90. doi: 10.1073/pnas.102024699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berman AJ, Gooding AR, Cech TR. Tetrahymena telomerase protein p65 induces conformational changes throughout telomerase RNA (TER) and rescues telomerase reverse transcriptase and TER assembly mutants. Molecular and cellular biology. 2010;30:4965–76. doi: 10.1128/MCB.00827-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai CK, Miller MC, Collins K. Template boundary definition in Tetrahymena telomerase. Genes & development. 2002;16:415–20. doi: 10.1101/gad.962602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen JL, Greider CW. Template boundary definition in mammalian telomerase. Genes & development. 2003;17:2747–52. doi: 10.1101/gad.1140303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stone MD, et al. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007;446:458–61. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Witkin KL, Collins K. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes & development. 2004;18:1107–18. doi: 10.1101/gad.1201704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Connor CM, Lai CK, Collins K. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. The Journal of biological chemistry. 2005;280:17533–9. doi: 10.1074/jbc.M501211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu JY, Stone MD, Zhuang X. A single-molecule assay for telomerase structure-function analysis. Nucleic acids research. 2010;38:e16. doi: 10.1093/nar/gkp1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collins K, Gandhi L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:8485–90. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaushik N, et al. Biochemical analysis of catalytically crucial aspartate mutants of human immunodeficiency virus type 1 reverse transcriptase. Biochemistry. 1996;35:11536–46. doi: 10.1021/bi960364x. [DOI] [PubMed] [Google Scholar]
- 25.Patel PH, et al. Insights into DNA polymerization mechanisms from structure and function analysis of HIV-1 reverse transcriptase. Biochemistry. 1995;34:5351–63. doi: 10.1021/bi00016a006. [DOI] [PubMed] [Google Scholar]
- 26.Cheetham GM, Jeruzalmi D, Steitz TA. Transcription regulation, initiation, and “DNA scrunching” by T7 RNA polymerase. Cold Spring Harbor symposia on quantitative biology. 1998;63:263–7. doi: 10.1101/sqb.1998.63.263. [DOI] [PubMed] [Google Scholar]
- 27.Kapanidis AN, et al. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314:1144–7. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brieba LG, Sousa R. T7 promoter release mediated by DNA scrunching. The EMBO journal. 2001;20:6826–35. doi: 10.1093/emboj/20.23.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaug AJ, Podell ER, Cech TR. Mutation in TERT separates processivity from anchor-site function. Nature structural & molecular biology. 2008;15:870–2. doi: 10.1038/nsmb.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forstemann K, Lingner J. Telomerase limits the extent of base pairing between template RNA and telomeric DNA. EMBO reports. 2005;6:361–6. doi: 10.1038/sj.embor.7400374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammond PW, Cech TR. Euplotes telomerase: evidence for limited base-pairing during primer elongation and dGTP as an effector of translocation. Biochemistry. 1998;37:5162–72. doi: 10.1021/bi972988o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.