Summary
A significant fraction of the eukaryotic genome is transcribed into RNAs that do not encode proteins, termed non-coding RNA (ncRNA). One class of ncRNA that is of particular interest is antisense RNAs, which are complementary to protein coding transcripts (mRNAs). In this article, we summarize recent studies using different yeasts that reveal a conserved pattern in which meiotically expressed genes have antisense transcripts in vegetative cells. These antisense transcripts repress the basal transcription of the mRNA during vegetative growth and are diminished as cells enter meiosis. While the mechanism(s) by which these antisense RNAs interfere with production of sense transcripts is not yet understood, the effects appear to be independent of the canonical RNAi machinery.
Early examples of antisense RNAs
Before the advent of tiling array and next-generation sequencing technology, relatively few ncRNAs had been identified in vegetatively growing yeast. One of the first antisense RNAs identified in S. pombe was one that overlaps the meiosis-specific gene, spo6+ [1]. In spo6 cells, meiotic progression is blocked after meiosis I, and no mature asci are formed [1]. The spo6-antisense RNA is polyadenylated and encompasses the entire spo6+ gene as well as its promoter region [1]. Though this study documented the presence of the antisense RNA, its possible regulatory role in spo6 sense expression was not explored.
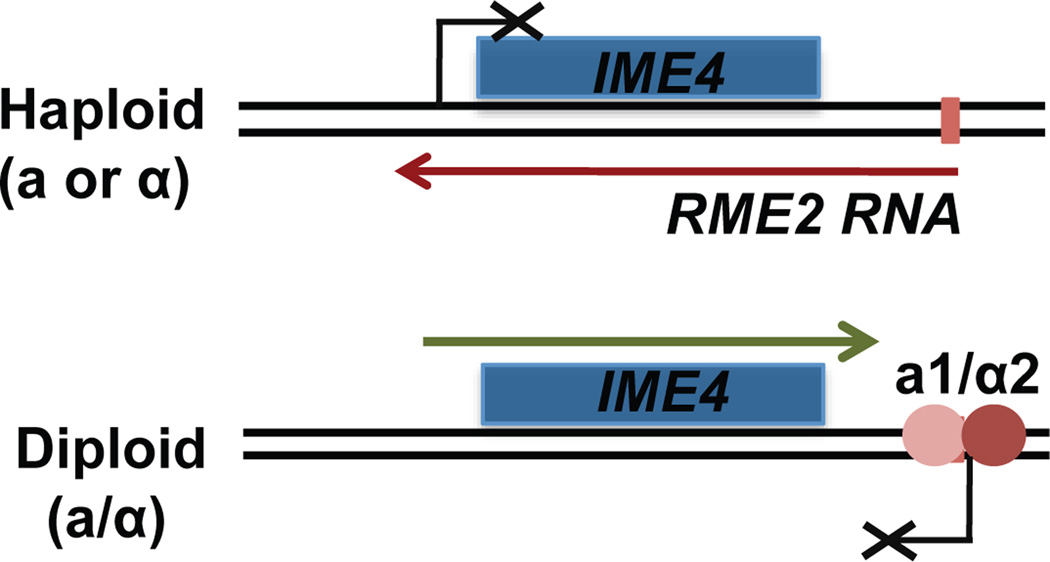
The first antisense RNA in yeast with a clear regulatory function was found in S. cerevisiae. This antisense transcript overlaps a meiotic regulator, IME4, that is required for the induction of genes early in meiosis [2,3]. The IME4 antisense transcript was subsequently named RME2 (Regulator of MEiosis) [4]. Similar to the spo6 antisense transcript, RME2 spans the coding region of IME4 and encompasses the IME4 promoter region (Figure 1). Transcription of IME4 is regulated by cell type and RME2 contributes directly to that regulation. In MATa or MATα cells, RME2 is actively transcribed and inhibits IME4 expression. In MATa/MATα cells, RME2 is repressed through an a1/α2 repressor binding site present in its promoter and IME4 is now transcribed ([2]; Figure 1). Deletion of the a1/α2 site allows expression of RME2 in MATa/MATα cells and results in loss of IME4 expression [2]. Thus, the RME2 antisense RNA regulates the cell type specificity of IME4.
Figure 1. The RME2 antisense RNA controls IME4 expression.
In cells of a or a mating type, RME2 is expressed and blocks expression of the IME4 coding gene. In a/a cells, the repressor a1/a2 inhibits RME2 transcription, allowing IME4 expression.
These two examples demonstrate that antisense RNAs exist in both S. pombe and S. cerevisiae and may have regulatory functions. More recently, genome-wide studies of transcription in both budding and fission yeasts indicate that, far from being isolated cases, antisense RNA is a common feature of the yeast transcriptome [5–9].
Transcriptome studies
While the generation of mRNAs used for translation is well understood, advances in genomic technologies have led to the uncovering of a surprisingly complex RNA transcriptome in eukaryotic cells [5,9]. The complementary strands of protein-coding genes as well as intergenic regions are frequently transcribed into ncRNAs. Estimates of how large a portion of the genome in yeast is transcribed vary somewhat with organism and experimental platform in yeast ranging from as high as 99.6% down to 74.5% [6,10]. Whatever the precise number, it is clear from these studies that there is extensive transcription of the genome outside well-defined coding regions.
A wide variety of ncRNAs have been identified including siRNAs and various small RNA species associated with promoter regions [11–14]. These different types of ncRNA vary in size, stability, and polyadenylation. Some ncRNAs have clear functions in regulating transcription or chromatin states (reviewed in [15,16]); however, for the majority of ncRNAs their function is unclear. For instance, transcripts from most intergenic regions or antisense strands are of very low abundance and may simply represent transcriptional background noise [6]. Of particular interest from a functional standpoint, is a class of longer, polyadenylated ncRNAs that are transcribed from the complementary strand of protein-coding genes, that is, antisense RNA. The prevalence of antisense RNAs and their overlap with coding regions suggests that they could play a regulatory role in the cell.
Expression of Meiotic Genes
In both fission and budding yeasts starvation can induce diploid cells to undergo meiosis and meiotic development requires the expression of specific genes [17,18]. Expression of these meiotic genes is repressed during vegetative growth and induced upon entry into the meiotic program. The mechanisms of induction are different in the two yeasts and include both increased transcription and mRNA stabilization [19–21]. However, studies described below suggest that antisense RNAs play a conserved role in maintaining low basal expression of these genes during vegetative growth.
Genome wide studies reveal a large number of antisense RNAs in yeast
The relatively small genome size of yeasts has allowed for detailed studies of the transcriptome under a variety of growth conditions, including synchronized cell cycle, meiosis, stress, different carbon sources, nutrient deprivation and individual mutant strains [9,14,22–25]. These studies have identified many antisense RNAs and revealed that expression of antisense RNAs, like regulated coding genes, is often restricted to certain conditions. Here we focus on those antisense RNAs that are relevant to meiotically-induced genes.
Fission yeast
Tiling array studies of S. pombe identified a large number (>2000) of ncRNAs in vegetative cells growing in rich medium [6,9]. A strong correlation was noted between coding and noncoding transcription suggesting that highly transcribed regions might be susceptible to "noise", producing antisense transcripts. However, when different growth conditions, for instance, heat shock [7] were examined, it was found that most sense/antisense pairs do not correlate in expression; i.e.., the sense and antisense transcripts are independently regulated indicating that the presence of the antisense transcript is not simply a product of an open chromatin state. Importantly, some sense/antisense pairs exhibit a strong anti-correlation that is, they are inversely transcribed, suggesting these particular pairs may regulate each other’s expression, as for IME4 and RME2 in S. cerevisiae.
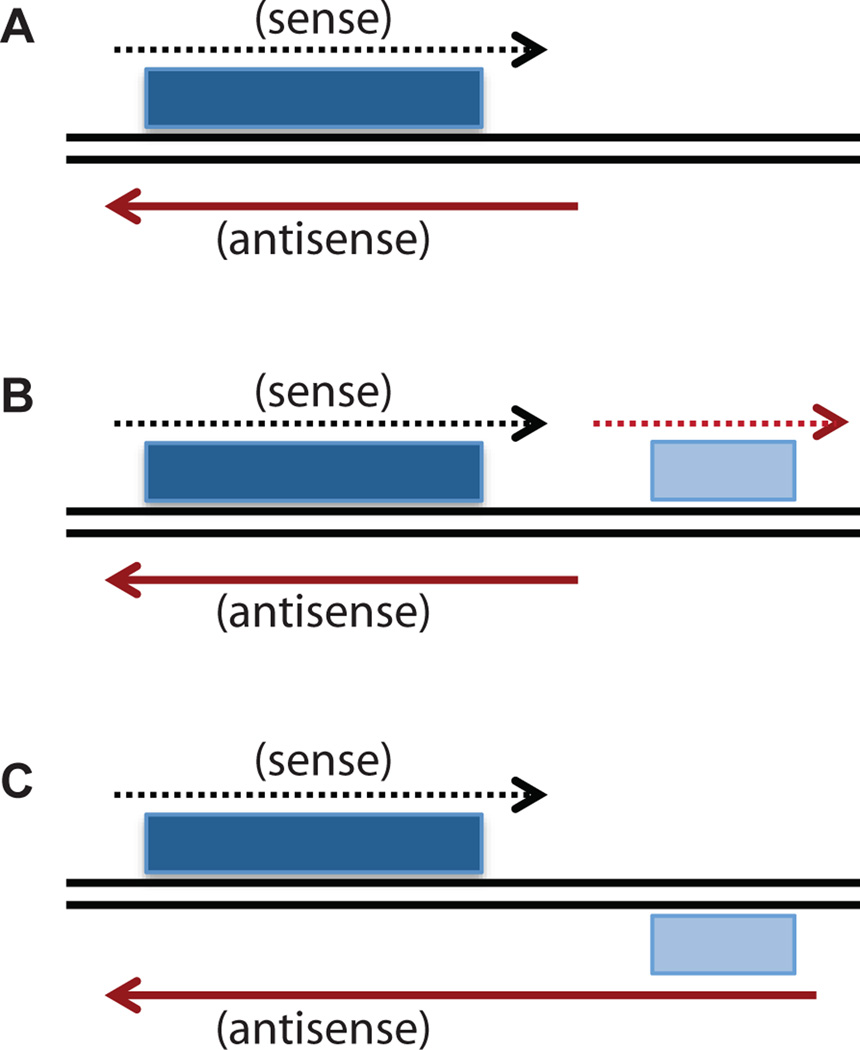
Two studies specifically looked at genes that have dominant antisense RNA, that is, higher levels of antisense than sense transcripts in vegetative cells [7,24]. Both studies found ~200 genes that fit this criterion. Analysis of the genes demonstrated that these antisense RNAs are highly enriched for genes whose sense expression is induced during meiosis. The transcriptional start sites of these antisense RNAs can be divided according to where their transcription initiates: (1) those that initiate at novel transcription start sites (Figure 2A); (2) those that initiate from a bidirectional promoter of a neighboring gene (Figure 2B); or (3) those that are created by an extended 3’ untranslated region (UTR) of a neighboring gene transcript. (Figure 2C). It is noteworthy that these extended 3’UTRs are unusual, given that the average length of a 3’UTR in S. pombe is 170 to 280 nucleotides [6,9], suggesting that their presence is not accidental. Interestingly, like the spo6 antisense RNA, most of these antisense RNAs cover the entire coding region of the sense gene and have their 3’ end positioned slightly beyond the transcription start site of the sense RNA, indicating that antisense transcription continues through the promoter region of the coding gene [24]. Moreover, these antisense RNAs generally show decreased expression and/or different transcript boundaries during meiosis when the sense genes are induced [24].
Figure 2. Sources of antisense RNA.
A) An antisense RNA with a unique promoter. The coding region of a gene that is associated with an antisense RNA is shown as a dark blue box. The dashed black line with an arrowhead represents the sense transcript. The solid red line with arrowhead indicates the antisense RNA. B) An antisense RNA from a bidirectional promoter of a neighboring gene. The coding region of a gene that is associated with an antisense RNA is shown as a dark blue box. The dashed black line with an arrowhead represents the sense transcript. The coding region of a neighboring gene is shown as a light blue box and the transcript of the neighboring gene is indicated by the dashed red line. The solid red line with arrowhead indicates the antisense RNA. C) An antisense RNA created by an extended 3'UTR of a neighboring gene. The coding region of a gene that is associated with an antisense RNA is shown as a dark blue box. The dashed black line with an arrowhead represents the sense transcript. The coding region of a neighboring gene is shown as a light blue box. The solid red line with arrowhead indicates the antisense RNA.
Importantly, these antisense RNAs are functionally significant. Disruption of the transcription of antisense RNAs associated with three different meiotic genes: spo4+, spo6+, and mug28+, led to a corresponding increase in the sense RNA in vegetative cells [24] though the level of expression was still low compared to the levels of the sense transcripts in meiotic cells. These results indicate that these antisense RNAs suppress the basal expression of meiotic genes in vegetative cells but that additional activation of sense transcription is required for full induction during meiosis.
A comparative transcriptome study of the fission yeast clade, comprising S. pombe, S. octosporus, S. cryophilus and S. japonicus, revealed that the set of genes with higher antisense than sense expression in vegetative cells is significantly enriched for meiotic genes across all four fission yeasts [8]. 60% of the specific sense/antisense pairs seen for S. pombe meiotic genes are conserved in at least one other species. For instance, the antisense RNA shown to suppress basal transcription of the spo4+ gene [24], is conserved in all four fission yeasts [8]. Taken together, these data suggest that antisense RNA may be a conserved mechanism for the repression of meiotic gene expression in vegetative cells.
Budding yeast
Tiling array studies of budding yeast reveal that the prevalence of ncRNA and the coincidence of antisense transcripts with meiotic genes is not limited to fission yeasts. Around 500 long, polyadenylated antisense RNAs have been identified in vegetatively growing budding yeast [5,22]. Similar to the situation in S. pombe, these antisense transcripts are frequently associated with meiotic genes [26]. The correspondence between meiotic genes and antisense transcripts is even evident in genome-wide studies of nucleosome distribution, as histone modifications indicative of promoter regions are frequently associated with the 3' ends of meiotic genes -- presumably representing the promoter region of the antisense transcript [27]. These studies make clear that the presence of antisense transcripts and their association with meiotic genes in vegetative cells is a feature of both fission and budding yeasts and, therefore, likely conserved throughout the ascomycetes.
Mechanisms of antisense RNA-mediated regulation
An important area that remains to be fully explored is the mechanism(s) by which antisense RNAs act to influence expression of sense genes. One obvious possibility would be that base pairing between the sense and antisense transcripts would create a double stranded RNA substrate to trigger the RNAi pathway. Indeed, S. pombe has a canonical RNAi pathway [28]. However, in S. pombe deletion of ago1, dcr1 or rdp1, encoding the three major components of the RNAi pathway, does not affect the ability of antisense RNA to inhibit sense expression [24], indicating that the RNAi pathway is not required for antisense regulation of meiotic genes. Moreover, S. cerevisiae lacks a functional RNAi system [29], ruling this out as the basis of antisense effects in budding yeast.
In the case of the IME4/RME2 sense/antisense pair, antisense transcription acts in cis to interfere with sense expression [2]. This suggests that it is the act of transcription, rather than the antisense RNA per se that is important for regulation. A common feature of antisense RNAs is that they extend through the promoter of the sense gene, suggesting that active transcription might occlude binding of transcriptional regulators to the sense promoter. Such a mechanism has been proposed for ncRNAs (though not antisense RNAs) at the SER3 and FLO11 loci in S. cerevisiae. In both cases, transcription of an ncRNA in the promoter region inhibits expression of the protein-coding gene [30,31]. However, studies of IME4/RME2 indicate that antisense transcription through the promoter region is not necessary for inhibition of sense expression [4]. Rather, antisense transcription through the coding region of the gene inhibits sense expression, suggesting that RME2 transcription blocks elongation rather than initiation of the IME4 transcript.
Analysis of tiling array data under different growth conditions in S. cerevisae reveals that, in addition to meiotic genes, antisense RNAs are associated with other genes that display large changes in expression level under some condition (eg. heat shock) [32]. Studies of one such gene, PHO84, normally induced by low phosphate, show that expression of the PHO84-antisense RNA leads to recruitment of a histone deacetylase complex. This is proposed to create a local chromatin environment that inhibits sense transcription [33]. Whether similar mechanisms function at meiotic genes remains to be tested.
Not all regulation by antisense RNAs is repressive. For the PHO5 gene, antisense transcription of the promoter region promotes rapid induction of the sense transcript, perhaps by maintaining a more open chromatin configuration at the promoter [34]. Thus, antisense transcription may fine tune sense expression in multiple ways, both inhibiting basal expression and priming the sense gene for rapid induction.
Conclusion
The mechanism(s) by which antisense RNAs influence sense transcription, and whether these mechanisms are conserved between yeasts, remain to be fully elucidated. Nonetheless, the explosion of yeast transcriptome data has made clear that antisense RNAs are widespread and, potentially, play important regulatory roles in the control of gene expression. Antisense RNA-mediated control of meiotic genes is a conserved regulatory scheme in ascomycetes. It will be of interest to learn if antisense-mediated regulation plays a similar role in other developmental transitions and in other fungi.
Highlights.
>Antisense RNAs are common in the yeast transcriptome. > Antisense RNAs are enriched at meiotically-induced genes during mitotic growth. > The antisense RNAs interfere with basal expression of these genes. > This is a conserved regulatory strategy for meiotic genes in budding and fission yeasts.
Acknowledgements
The authors wish to thank Angad Garg, Nancy Hollingsworth and Janet Leatherwood for helpful comments on the manuscript. Work in the Leatherwood and Neiman labs is supported by NIH Program Project Grant P01 GM088297.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nakamura T, Kishida M, Shimoda C. The Schizosaccharomyces pombe spo6+ gene encoding a nuclear protein with sequence similarity to budding yeast Dbf4 is required for meiotic second division and sporulation. Genes Cells. 2000;5:463–479. doi: 10.1046/j.1365-2443.2000.00343.x. [DOI] [PubMed] [Google Scholar]
- 2.Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 3.Shah JC, Clancy MJ. IME4, a gene that mediates MAT and nutritional control of meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1078–1086. doi: 10.1128/mcb.12.3.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gelfand B, Mead J, Bruning A, Apostolopoulos N, Tadigotla V, Nagaraj V, Sengupta AM, Vershon AK. Regulated antisense transcription controls expression of cell-type specific genes in yeast. Mol Cell Biol. 2011;31:1701–1709. doi: 10.1128/MCB.01071-10. Analysis of the sense/antisense pair IME4/RME2 indicates that RME2 interferes with IME4 expression by inhibiting elongation rather than inititation of the sense transcript.
- 5.David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci U S A. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dutrow N, Nix DA, Holt D, Milash B, Dalley B, Westbroek E, Parnell TJ, Cairns BR. Dynamic transcriptome of Schizosaccharomyces pombe shown by RNA-DNA hybrid mapping. Nat Genet. 2008;40:977–986. doi: 10.1038/ng.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ni T, Tu K, Wang Z, Song S, Wu H, Xie B, Scott KC, Grewal SI, Gao Y, Zhu J. The prevalence and regulation of antisense transcripts in Schizosaccharomyces pombe. PLoS One. 2010;5:e15271. doi: 10.1371/journal.pone.0015271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. Comparative functional genomics of the fission yeasts. Science. 2011;332:930–936. doi: 10.1126/science.1203357. The authors compare genome wide expression in four different fission yeast species. This study reveals the conserved association of antisense transcripts with meiotically-induced genes.
- 9.Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 10.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Djupedal I, Kos-Braun IC, Mosher RA, Soderholm N, Simmer F, Hardcastle TJ, Fender A, Heidrich N, Kagansky A, Bayne E, et al. Analysis of small RNA in fission yeast; centromeric siRNAs are potentially generated through a structured RNA. EMBO J. 2009;28:3832–3844. doi: 10.1038/emboj.2009.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buhler M, Spies N, Bartel DP, Moazed D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol. 2008;15:1015–1023. doi: 10.1038/nsmb.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 14.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev. 2010;20:134–141. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamamoto M. Regulation of meiosis in fission yeast. Cell Struct Funct. 1996;21:431–436. doi: 10.1247/csf.21.431. [DOI] [PubMed] [Google Scholar]
- 18.Kassir Y, Adir N, Boger-Nadjar E, Raviv NG, Rubin-Bejerano I, Sagee S, Shenhar G. Transcriptional regulation of meiosis in budding yeast. Int Rev Cytol. 2003;224:111–171. doi: 10.1016/s0074-7696(05)24004-4. [DOI] [PubMed] [Google Scholar]
- 19.Harigaya Y, Yamamoto M. Molecular mechanisms underlying the mitosis-meiosis decision. Chromosome Res. 2007;15:523–537. doi: 10.1007/s10577-007-1151-0. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell AP. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994;58:56–70. doi: 10.1128/mr.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherwood RK, Bennett RJ. Fungal meiosis and parasexual reproduction--lessons from pathogenic yeast. Curr Opin Microbiol. 2009;12:599–607. doi: 10.1016/j.mib.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Granovskaia MV, Jensen LJ, Ritchie ME, Toedling J, Ning Y, Bork P, Huber W, Steinmetz LM. High-resolution transcription atlas of the mitotic cell cycle in budding yeast. Genome Biol. 2010;11:R24. doi: 10.1186/gb-2010-11-3-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lardenois A, Liu Y, Walther T, Chalmel F, Evrard B, Granovskaia M, Chu A, Davis RW, Steinmetz LM, Primig M. Execution of the meiotic noncoding RNA expression program and the onset of gametogenesis in yeast require the conserved exosome subunit Rrp6. Proc Natl Acad Sci U S A. 2011;108:1058–1063. doi: 10.1073/pnas.1016459108. The paper describes a tiling array study of both vegetative and sporulating cells. The authors demonstrate an extensive, and changing, pattern of ncRNAs during meiotic development that is regulated by the exosome component Rrp6.
- 24. Chen HM, Rosebrock AP, Khan SR, Futcher B, Leatherwood JK. Repression of meiotic genes by antisense transcription and by the Fkh2 transcription factor in Schizosaccharomyces pombe. PLos One. 2011 doi: 10.1371/journal.pone.0029917. submitted. This study shows that the antisense RNAs associated with several meiotically induced genes are important for repression of basal expression of the genes in vegetative cells. It is also shown that the RNAi machinery is not necessary for this repression.
- 25.Nishizawa M, Komai T, Katou Y, Shirahige K, Ito T, Toh EA. Nutrient-regulated antisense and intragenic RNAs modulate a signal transduction pathway in yeast. PLoS Biol. 2008;6:2817–2830. doi: 10.1371/journal.pbio.0060326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yassour M, Pfiffner J, Levin JZ, Adiconis X, Gnirke A, Nusbaum C, Thompson DA, Friedman N, Regev A. Strand-specific RNA sequencing reveals extensive regulated long antisense transcripts that are conserved across yeast species. Genome Biol. 2010;11:R87. doi: 10.1186/gb-2010-11-8-r87. A deep sequencing approach demonstrates the association of antisense transcripts with meitoically-induced genes in vegetative S. cerevisiae cells and the conservation of this association in other budding yeasts.
- 27.Zhang L, Ma H, Pugh BF. Stable and dynamic nucleosome states during a meiotic developmental process. Genome Res. 2011 doi: 10.1101/gr.117465.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martienssen RA, Zaratiegui M, Goto DB. RNA interference and heterochromatin in the fission yeast Schizosaccharomyces pombe. Trends Genet. 2005;21:450–456. doi: 10.1016/j.tig.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 29.Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bumgarner SL, Dowell RD, Grisafi P, Gifford DK, Fink GR. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc Natl Acad Sci U S A. 2009;106:18321–18326. doi: 10.1073/pnas.0909641106. Two different ncRNAs in the promoter of the FLO11 gene collaborate to regulate expression of the FLO11 mRNA
- 31.Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 32. Xu Z, Wei W, Gagneur J, Clauder-Munster S, Smolik M, Huber W, Steinmetz LM. Antisense expression increases gene expression variability and locus interdependency. Mol Syst Biol. 2011;7:468. doi: 10.1038/msb.2011.1. The authors show that an associated antisense RNA can cause expression of the sense transcript to behave in a switch-like manner. A nice demonstration of how an antisense transcript can tune the behavior of the sense gene.
- 33.Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 34.Uhler JP, Hertel C, Svejstrup JQ. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc Natl Acad Sci U S A. 2007;104:8011–8016. doi: 10.1073/pnas.0702431104. [DOI] [PMC free article] [PubMed] [Google Scholar]