Abstract
Cholangiocarcinoma (CCA) cells paradoxically express the death ligand TRAIL, and, therefore, are dependent upon potent survival signals to circumvent TRAIL cytotoxicity. CCAs are also highly desmoplastic cancers with a tumor microenvironment rich in myofibroblasts (MFBs). Herein, we examine a role for MFB-derived CCA survival signals. We employed human KMCH-1, KMBC, HuCCT-1, TFK-1, and Mz-ChA-1 CCA cells as well as human primary hepatic stellate and myofibroblastic LX-2 cells for these studies. In vivo experiments were conducted using a syngeneic rat orthotopic CCA model. Co-culturing CCA cells with myofibroblastic human primary HSCs or LX-2 cells significantly decreased TRAIL-induced apoptosis in CCA cells, a cytoprotective effect abrogated by neutralizing PDGF-BB-antiserum. Cytoprotection by PDGF-BB was dependent upon Hedgehog (Hh) signaling as it was abolished by the smoothened (the transducer of Hh signaling) inhibitor cyclopamine. PDGF-BB induced PKA-dependent trafficking of smoothened to the plasma membrane resulting in GLI2 nuclear translocation and activation of a consensus GLI reporter gene-based luciferase assay. A genome-wide mRNA expression analysis identified 67 target genes to be commonly up- (50 genes) or downregulated (17 genes) by both SHH and PDGF-BB in a cyclopamine-dependent manner in CCA cells. Finally, in a rodent CCA in vivo-model, cyclopamine administration increased apoptosis in CCA cells resulting in tumor suppression.
Conclusions
Myofibroblast-derived PDGF-BB protects CCA cells from TRAIL cytotoxicity by a Hh signaling-dependent process. These results have therapeutical implications for the treatment of human cholangiocarcinoma.
Keywords: Desert hedgehog, Hepatic stellate cells, Indian hedgehog, Platelet-derived growth factor receptor beta, Sonic hedgehog
Cholangiocarcinoma (CCA) is a highly lethal malignancy with limited treatment options.1-3 It is the most common biliary cancer and epidemiologic studies suggest that its incidence is increasing in several Western Countries.4 Human CCA in vivo paradoxically expresses the death ligand tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and its cognate death receptors 5 suggesting that these cancers are reliant on potent survival signals for tumor maintenance and progression. However, the mechanisms by which CCA evades apoptosis by TRAIL and other pro-apoptotic stimuli is incompletely understood.
CCAs are highly desmoplastic cancers suggesting cancer-associated fibroblasts within the tumor microenvironment contribute to their development and progression as has been proposed for other cancers (e.g. breast cancer, prostate cancer, etc.).6, 7 Cancer-associated fibroblasts are perpetually “activated” and express α-smooth muscle actin (α-SMA); cells exhibiting this activated phenotype are often referred to as myofibroblasts (MFBs).8 In the liver, MFBs are derived from periportal fibroblasts, hepatic stellate cells (HSCs), and perhaps an epithelial-to-mesenchymal transition of cholangiocytes, hepatocytes, and/or the tumor itself.9, 10 A role for MFBs in carcinogenesis and tumor biology has only recently received attention.8, 11-13 Cross-talk between the cancer and MFBs appears to be exploited by cancers as a tumor promoting mechanism. For example, in CCA the number of MFBs correlates with patient survival. 14 MFBs also appear capable of providing survival signals as they reduce apoptosis of non-malignant cholangiocytes in co-culture experiments.15 However, information regarding the nature of the cross-talk, and in particular the identity of the potential survival signals, remains obscure.
Platelet-derived growth factor (PDGF) paracrine signaling between MFBs and cholangiocytes occurs in rodent models of biliary tract inflammation and fibrogenesis.15, 16 Five different ligands of PDGF exist including PDGF-AA, -BB, -AB, -C and –D. However, PDGF-BB appears to be the predominant isoform secreted by liver MFBs.17 Of the two cognate receptors, platelet-derived growth factor receptor (PDGFR)-α and –β, PDGFR-β is the cognate receptor for PDGF-BB. PDGFR-β is a receptor tyrosine kinase that is also known to alter plasma membrane dynamics associated with cell migration by a cyclic adenosine monophosphate (cAMP)-dependent kinase (PKA)-dependent process; 18 thus, PDGF-BB effects on intracellular signaling cascades are pleiotropic. Given an emerging role for PDGF-BB in MFB-to-cholangiocyte cross-talk, a role for PDGF-BB as a survival factor for CCA warrants further investigation.
The Hedgehog (Hh) signaling pathway has been strongly implicated in gastrointestinal tumor biology including CCA.19, 20 Hh signaling is initiated by any of the three ligands Sonic (SHH), Indian (IHH), and Desert (DHH) hedgehog. These ligands bind to the Hh receptor Patched1 (PTCH1) resulting in activation of Smoothened (SMO) and subsequently the transcription factors glioma-associated oncogene (GLI) 1, 2, and 3.21 How PTCH1 modulates SMO was enigmatic until quite recently, as the two proteins do not physically associate. SMO trafficking from an intracellular compartment to the plasma membrane apparently results in its activation.22 Hh ligand binding to PTCH1 increases the concentration of intracellular messengers (lipid phosphates), which in turn promote SMO trafficking to the plasma membrane.23, 24 PKA affects SMO trafficking and activation, raising the unexplored possibility that cues from other ligand:receptor systems such as PDGF-BB may also augment SMO activation by facilitating its trafficking to the plasma membrane.22 Interestingly, SHH mRNA expression is increased by PDGF-BB in immature cholangiocytes 16 providing an additional link between Hh signaling and PDGF. Hh signaling may also be a master switch mediating resistance of CCA cells to TRAIL cytotoxicity. 25, 26 Taken together, these observations suggest MFB-derived PDGF-BB may modulate Hh survival signaling in CCA cells.
The objective of this study was to examine the role for MFB-to-CCA cell paracrine signaling in mediating CCA resistance to TRAIL cytotoxicity. The results suggest that PDGF-BB secreted by MFBs protects CCA cells from TRAIL-induced apoptosis. PDGF-BB appears to exert its cytoprotective effects by a Hh signaling-dependent manner. These observations have implications for the treatment of human CCA.
MATERIALS AND METHODS
Cell lines/culture and human samples
The human CCA cell lines KMCH-1, KMBC, HuCCT-1, TFK-1, and Mz-ChA-1 and as well as the erythroblastic leukemia viral oncogene homolog (ErbB-2)/neu transformed malignant rat cholangiocyte cell line BDEneu (in vivo experiment) and the LX-2 cells, an immortalized myofibroblast cell line derived from human HSCs, were cultured as previously described.5, 27-30 The human primary myofibroblastic HSCs were kindly provided by V.H. Shah (Division of Gastroenterology and Hepatology, Mayo Clinic, Rochester, MN) and cultured in Dulbecco’s modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum, penicillin G (100 U/mL), and streptomycin (100 μg/mL) under standard conditions. Human samples were collected in accordance with the Declaration of Helsinki.
Generation of enhanced green fluorescent protein–tagged SMO (GFP-SMO)
A pRK7 plasmid containing the human SMO sequence (GenBank accession no.: NM_005631) was a generous gift from M. Fernandez-Zapico (Division of Oncology Research, Mayo Clinic, Rochester, MN). The pRK7-SMO plasmid was modified to accept the GFP tag first by inserting recognition sites for EcoRI and NotI at the C-terminus of SMO, replacing the stop codon. For this, a PCR-generated EcoRI/NotI modified SMO C-terminal coding sequence was inserted into pRK7-SMO. Next, GFP from the pEGFP-N1 protein fusion vector (Clontech Laboratories, Inc., Mountain View, CA; Catalog no.: 6085-1; GenBank accession no.: U55762) was digested and inserted into the modified pRK7-SMO plasmid to generate a SMO construct fused to GFP at the C-terminal cytoplasmic domain. The plasmid was sequenced to confirm that the construct was in frame and no polymerase chain reaction artifacts were introduced.
SMO trafficking
HuCCT-1 cells were cultured on coverslips, treated as indicated, and fixed with PBS containing 4% paraformaldehyde for 20 min at 37°C. After being washed with PBS, cells were incubated with 0.5% Triton X-100 in PBS for 15 min at RT and then blocked with PBS containing 5% BSA for 60 min at 37°C. Cells were subsequently incubated with anti-SMO antiserum (1:250; Santa Cruz, Santa Cruz, CA; H-300) at 4°C overnight. After being washed, coverslips were incubated with Texas Red®-X goat anti-rabbit IgG (1:1000; Invitrogen, Camarillo, CA; T6391) for 1 hr in the dark. Cells were then washed three times in PBS, one time in water and mounted using Prolong Antifade (Invitrogen). The slides were analyzed by fluorescent confocal microscopy (LSM 510; Zeiss, Jena, Germany). In additional experiments, SMO trafficking was examined by total internal reflection microscopy (TIRF).31 KMCH-1 cells cultured on coverslips were transfected with GFP-SMO plasmid 48 hours prior to study. Cells were treated as indicated, and fixed with ddH2O containing 2.5% formaldehyde, 0.1 M PIPES, 1.0mM EGTA, and 3.0 mM MgSO4 for 20 min at 37°C. Cells were then washed three times in PBS, one time in water and mounted using Prolong Antifade (Invitrogen). The slides were analyzed with a TIRF microscop (Zeiss AxioObserver.Z1, Munich, Germany). GFP-SMO localized to the plasma membrane was quantified using image analysis software (Carl Zeiss AxioVision 4.8.2.0, Munich, Germany). Data were expressed as the average fluorescence intensity in the cell multiplied by the number of pixels above the background.
GLI reporter construct and promoter-reporter assay
To determine GLI activity, a reporter containing eight directly repeated copies of a consensus GLI-binding site (8x-GLI) downstream of the luciferase gene was employed (pδ51LucII plasmid; δ-crystalline promoter). 32 The 8x-GLI reporter was kindly provided by M. Fernandez-Zapico (Division of Oncology Research, Mayo Clinic, Rochester, MN). The plasmid was transfected into normal, stable scrambled, or shSMO KMCH-1 cells (0.5 μg/well) using FuGene HD (Roche Diagnosis, Basel, Switzerland). Cells were co-transfected with 50 ng of a plasmid expressing Renilla luciferase (pRL-CMV; Promega, Madison, WI). 24 hours after transfection, cells were treated as indicated, cell lysates prepared, and both firefly and Renilla luciferase activities quantified using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Firefly luciferase activity was normalized to Renilla luciferase activity to control for transfection efficiency and cell numbers. Data (firefly/Renilla luciferase activity) are expressed as fold increase over vehicle-treated cells transfected with the 8x-GLI/pRL-CMV reporter constructs.
Animal experiments
All animal studies were performed in accordance with and approved by the Institutional Animal Care and Use Committee. In vivo intrahepatic cell implantation (syngeneic rat orthotopic CCA model) was carried out in male adult Fischer 344 rats (Harlan, Indianapolis, IN) with initial body weights between 190 and 220 g as previously described.28-30 Cyclopamine (2.5 mg/kg BW; 0.5 mL) complexed with 2-hydroxypropyl-β-cyclodextrin (Tocris, Ellisville, MO) as previously described 33, 34 or vehicle was given intraperitoneally every day for one week (1st injection: 7th post-operative day; 7th injection: 13th post-operative day). Twenty-four hours after receiving the last injection, the rats were euthanized and the livers removed for further analysis including histopathology and mRNA extraction. To assess the numbers of metastases-free and metastases-bearing rats, the abdominal cavities, the retroperitoneal spaces and the thoracic cavities were thoroughly examined as previously described.29
Supplemental methods
Materials, generation of shSMO KMCH-1 cells, quantitation of PDGF-BB and cAMP, co-culture experiments, quantitation of apoptosis, immunoblot analysis, immunohistochemistry for α-SMA, PDGFR-β, PDGF-BB, and cytokeratin 7 as well as RT-PCR, the genome-wide mRNA expression assay, and statistical analysis are described in the online supplementary material.
RESULTS
Expression of PDGF-BB by MFBs and PDGFR-β by CCA cells
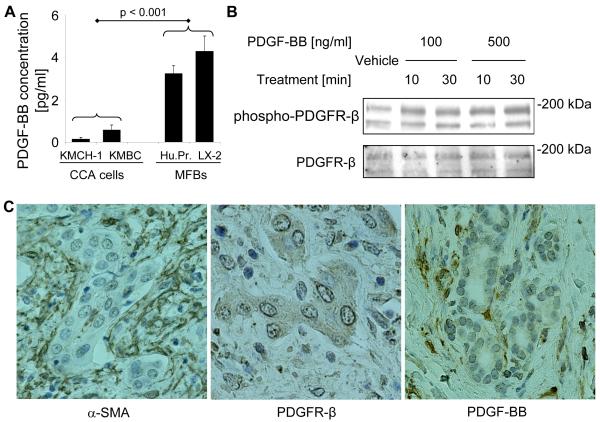
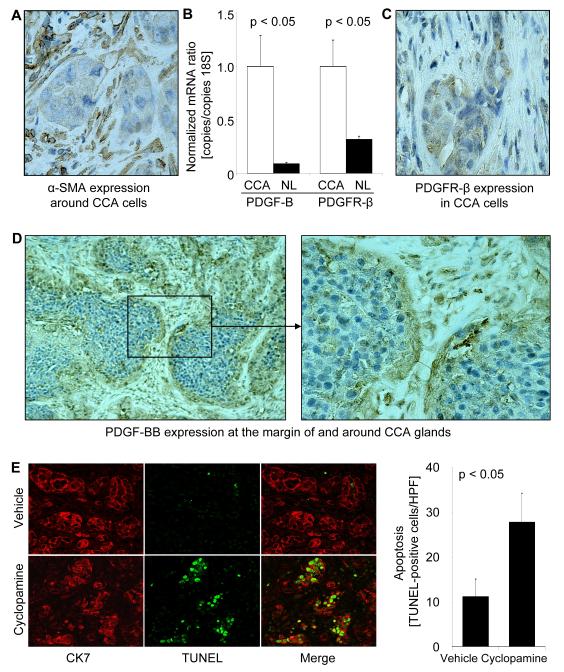
Initially, we assessed basal PDGF-BB secretion by two human CCA cell lines, KMCH-1 and KMBC, primary HSC cells, and the human MFB cell line LX-2 by ELISA (monoculture conditiones, Fig. 1A). The MFB cells secreted significantly higher levels of PDGF-BB than the CCA cell lines. Because many cancer cells do not express PDGF receptors, 35 we next examined KMCH-1 cells for the presence of PDGFR-β and its activating phosphorylation by PDGF-BB (Fig. 1B). Immunoblot analysis confirmed protein expression of PDGFR-β in KMCH-1 cells (Fig. 1B, lower), while PDGFR-α was not detectable (data not shown). PDGFR-β also displayed receptor phosphorylation (Tyr857) upon PDGF-BB treatment (Fig. 1B, upper). In addition, we confirmed mRNA expression of PDGFR-β in KMCH-1 cells and 4 other human CCA cell lines (KMBC, HuCCT-1, TFK-1, and MzChA-1) as well as in the ErbB-2/neu transformed malignant rat cholangiocyte cell line BDEneu (employed in the in vivo CCA model; Suppl. Fig. 1). To characterize the expression of α-SMA, PDGFR-β, and PDGF-BB in vivo, we performed immunohistochemistry for these proteins in human CCA specimens (Fig. 1C). Numerous α-SMA-positive MFBs were present in the stromal tumor microenvironment in all human CCA samples examined (Fig. 1C, left). Moreover, PDGFR-β immunoreactivity was confirmed in CCA cell glands in approximately half of the samples (Fig. 1C, middle), whereas PDGF-BB was expressed in MFBs in two-thirds of the samples (Fig. 1C, right). Thus, PDGF-BB was shown to be secreted by MFBs and its receptor expressed by CCA cells.
Figure 1.
Cellular expression of PDGF-BB, PDGFR-β, and α-SMA in CCA. (A, B) PDGF-BB is secreted by MFBs and phosphorylates PDGFR-β in CCA cells in vitro (A) Basal PDGF-BB secretion (24 hours; corrected for total protein expression) by two human CCA cell lines, KMCH-1 and KMBC, a human myofibroblast (MFB) cell line, LX-2, and myofibroblastic human primary HSC cells (Hu.Pr.) was assessed in the serum-free supernatant by ELISA (monoculture conditiones). Mean ± s.e.m. (n=3). (B) KMCH-1 cells (serum-starved for 2 days) were treated with vehicle or PDGF-BB at the concentrations and time intervals indicated. Cell treatment was followed by immunoblot analysis for PDGFR-β and phospho-PDGFR-β (Tyr857).Upper bands show the N-linked glycosylated and lower bands the unglycosylated forms of PDGFR-β. (C) α-SMA (MFB marker; left), PDGFR-β (middle), and PDGF-BB (right) protein expression (brown) was examined by immunohistochemistry (counterstaining with Mayers’ Hematoxylin) in human CCA specimens.
MFB-derived PDGF-BB promotes resistance to TRAIL cytotoxicity
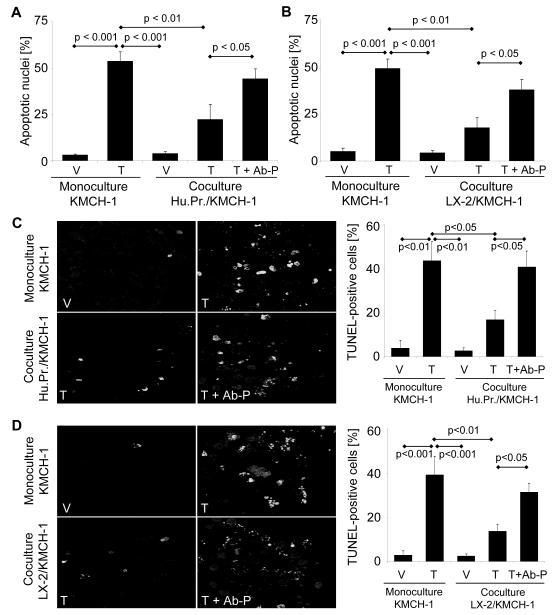
Next, we examined the effect of co-culturing KMCH-1 cells with PDGF-BB-secreting myofibroblastic human primary HSCs (Fig. 2A and C) or LX-2 cells (Fig. 2B and D) on TRAIL-induced CCA cell apoptosis. As assessed by either nuclear morphology (Fig. 2 A and B) or the TUNEL assay (Fig. 2C and D), KMCH-1 cells were more resistant to TRAIL-induced apoptosis when co-cultured with human primary HSCs or LX-2 cells as compared to monoculture conditions. Interestingly, the KMCH-1 cells were resensitized to TRAIL (10 ng/mL) when co-cultured in the presence of neutralizing anti-human PDGF-BB antiserum (Fig. 2A-D). Similar results were obtained co-culturing KMBC cells with LX-2 cells (Supplemental Fig. 1A). These findings were somewhat selective for TRAIL-induced apoptosis, as less cytotoxic potentiation by the neutralizing anti-human PDGF-BB antiserum was observed in etoposide-treated cells (Supplemental Fig. 1B). Thus, PDGF-BB secreted by co-cultured MFB cells, reduces the susceptibility of CCA cells to TRAIL-induced apoptosis.
Figure 2.
PDGF-BB promotes resistance to TRAIL cytotoxicity. KMCH-1 cells were plated alone (monoculture) or together with PDGF-BB-secreting human primary HSCs (Hu.Pr.; A and C) or LX-2 (B and D) cells in a transwell insert co-culture system (KMCH-1 cells in the bottom and human primary HSCs or LX-2 cells in the top wells; 1:1 ratio) for 6 days. Cells were treated as indicated with vehicle (V), rhTRAIL (T; 10ng/mL for 6 hrs on day 6) or rhTRAIL plus anti-human PDGF-BB antiserum (T + AB-P; rhTRAIL: 10ng/mL for 6 hrs on day 6; anti-human PDGF-BB antiserum: 10μg/mL for 24 hrs on day 5). After rhTRAIL treatment for 6 hrs, KMCH-1 cells were analyzed for apoptotic nuclear morphology by DAPI-staining (A and B) and for DNA fragmentation by TUNEL-staining (C and D) with quantitation of apoptotic nuclei by fluorescence microscopy. Mean ± s.e.m. (n=5). (C and D left) Representative fluorescent photomicrographs of TUNEL-positive KMCH-1 cells are depicted.
PDGF-BB cytoprotection is dependent on Hh signaling
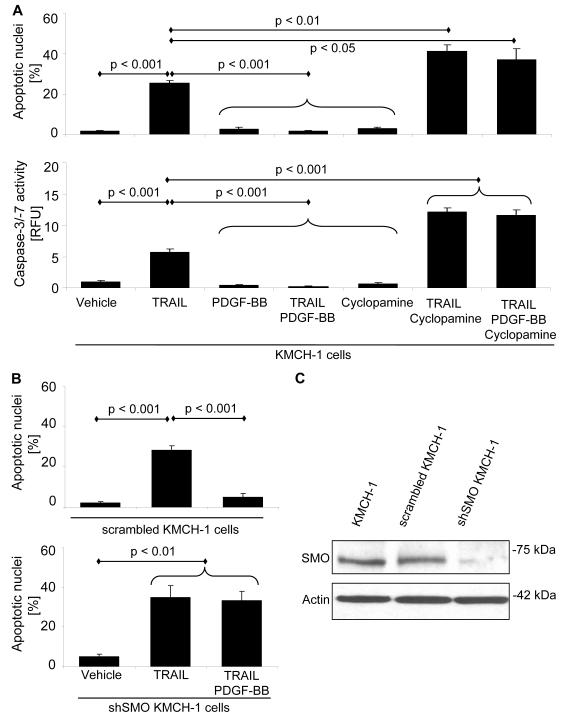
Given that PDGF-BB modulates antiapoptotic Hh signaling in immature cholangiocytes 16 and Hh signaling appears to be a potent survival signal for CCA cells, 25, 26 we explored the effect of Hh signaling inhibition on PDGF-BB-mediated cytoprotection against TRAIL cytotoxicity. Apoptosis was assessed morphologically following DAPI-staining (Fig. 3A upper and B) and biochemically by a caspase-3/-7 activity assay (Fig. 3A lower). Exogenous PDGF-BB protected KMCH-1 cells from TRAIL-induced apoptosis (Fig. 3A). In contrast, cyclopamine, an inhibitor of SMO (the transducer of Hh signaling) 36 sensitized KMCH-1 cells to TRAIL-induced apoptosis (Fig. 3A). Moreover, cyclopamine completely abrogated PDGF-BB inhibition of TRAIL-induced apoptosis (Fig. 3A). Likewise, shSMO KMCH-1 cells also underwent TRAIL-mediated apoptosis despite exogenous PDGF-BB treatment (Fig. 3B lower; compare with stable scrambled KMCH-1 cells Fig. 3B upper). Taken together, these observations suggest that PDGF-BB-mediated protection from TRAIL-induced apoptosis is dependent upon an intact Hh signaling pathway.
Figure 3.
Cytoprotection by PDGF-BB is dependent on Hh signaling. (A) First, KMCH-1 cells (serum-starved for 2 days) were treated with vehicle or PDGF-BB (200 ng/mL) for 8 hrs. PDGF-BB was retained in the culture media, and the cells were then treated with rhTRAIL (5 ng/mL), cyclopamine (a Hh [SMO] inhibitor; 5 μM) or TRAIL plus cyclopamine (5 ng/mL; 5 μM) for additional 6 hrs. (B) Similar experiments (same PDGF-BB and TRAIL treatment) were performed with shSMO KMCH-1 instead of cyclopamine-treated KMCH-1 cells (lower) and stable scrambled KMCH-1 instead of normal KMCH-1 cells (controls, upper). (C) Stable knockdown of SMO only in shSMO KMCH-1 cells was confirmed by immunoblot analysis. Apoptosis was measured by DAPI-staining with quantitation of apoptotic nuclei by fluorescence microscopy (A upper and B; mean ± s.e.m.; n=3) or fluorescent analysis of caspase-3/-7 activity (A lower; mean ± s.e.m.; n=6; RFU, relative fluorescence unit).
PDGF-BB induces translocation of SMO to the plasma membrane
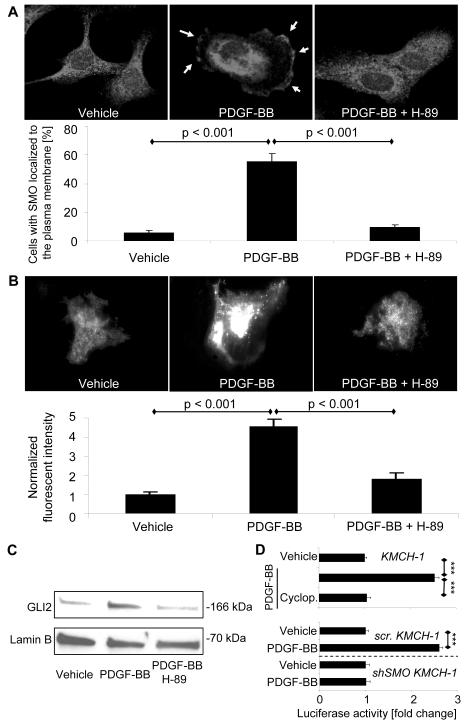
We next sought to explore how PDGF-BB stimulates Hh signaling in order to promote CCA cell survival. Initially, we analyzed the direct effect of PDGF-BB on mRNA expression of the Hh signaling ligands SHH, IHH and DHH as well as PTCH1, SMO, and GLI1-3 by quantitative RT-PCR (Supplemental Fig. 3A). PDGF-BB did not significantly alter mRNA expression of the three Hh ligands nor that of PTCH1, SMO, or GLI1-3 in KMCH-1 and HuCCT-1 cells. To investigate if PDGF-BB promotes Hh signaling by downregulation of known negative regulators of this pathway, we also measured the mRNA levels of hedgehog-interacting protein (HIP) and suppressor of fused (SUFU). PDGF-BB had no effect on these negative regulators in KMCH-1 cells (Supplemental Fig. 3B). Because translocation of SMO from intracellular vesicles to the plasma membrane results in its activation during Hh signaling, 22 we next examined the cellular localization of SMO upon PDGF-BB treatment by immunocytochemistry (Fig. 4A). PDGF-BB significantly induced translocation of SMO from intracellular compartments to the plasma membrane (Arrows; Fig 4A, middle). This process appears to be PKA-dependent as it was effectively attenuated by the PKA inhibitor H-89. Similar results were obtained when we employed KMCH-1 cells transiently transfected with a plasmid expressing GFP-tagged human SMO and analyzed GFP-SMO localized at the plasma membrane by TIRF microscopy 26 (Fig. 4B). Moreover, PDGF-BB derived from co-cultured LX-2 cells also induced SMO trafficking as assessed by TIRF microscopy (Suppl. Fig. 4A). As a further indicator for Hh signaling activation, we examined the effect of PDGF-BB on GLI2 nuclear translocation in KMCH-1 cells by immunoblot analysis (Fig 4C). PDGF-BB treatment increased GLI2 abundance in nuclear protein extracts, an effect that again was attenuated by the PKA inhibitor H-89. Consistent with these results, KMCH-1 cells transiently transfected with a GLI reporter construct displayed GLI activation upon PDGF-BB treatment. The SMO inhibitor cyclopamine effectively blocked PDGF-BB-mediated GLI activation (Fig. 4D upper). Likewise, stable scrambled KMCH cells also displayed PDGF-BB-induced GLI activation, whereas no PDGF-BB effect was observed in shSMO KMCH-1 cells (Fig. 4D lower).
Figure 4.
PDGF-BB induces SMO trafficking to the plasma membrane resulting in GLI2 nuclear translocation and transcriptional activation of a GLI reporter gene. (A) HuCCT-1 cells were treated with vehicle, PDGF-BB (200 ng/mL) or PDGF-BB plus the PKA inhibitor H-89 (5 μM) for 2 hrs. SMO cellular localization was analyzed by immunocytochemistry. Note that PDGF-BB induces translocation of SMO from intracellular compartments (left) to the plasma membrane (yellow arrows, middle), an effect that is inhibited by the PKA inhibitor H-89 (right). Cells with membranes positive for SMO immunocytochemistry were counted in each of these conditions. Mean ± s.e.m. (n=7). (B) KMCH-1 cells were transiently transfected with a plasmid expressing GFP-tagged human SMO (GFP-SMO; 48h) and then treated with vehicle or PDGF-BB (200 ng/mL) with and without H-89 (5 μM) for 2 hrs. GFP-SMO localized at the plasma membrane was analyzed by TIRF microscopy and the fluorescent intensity quantified using image analysis software. Mean ± s.e.m. (n=20). (C) GLI2 nuclear translocation in KMCH-1 cells was assessed by immunoblot analysis after treating the cells with vehicle or PDGF-BB (200 ng/mL) with and without H-89 (5 μM) for 8 hrs. Lamin B was used as loading control for the nuclear protein extracts. (D) KMCH-1 cells (normal, stable scrambled [scr.], or shSMO) were transiently transfected (24 hrs) with a reporter construct containing eight consecutive consensus GLI-binding sites (8x-GLI) and co transfected with pRL-CMV. Cells were then treated as indicated with vehicle or PDGF BB (200 ng/mL) with and without cyclopamine (5 μM) for 24 hrs. Both firefly and Renilla luciferase activities were quantified and data (firefly/Renilla luciferase activity) are expressed as fold increase over vehicle-treated cells transfected with the 8x-GLI/pRL CMV reporter constructs. Mean ± s.e.m. (n=3; ***p<0.001).
Since PKA function is cAMP-dependent,37 we measured the effect of PDGF-BB on intracellular cAMP levels (Suppl. Fig. 4B). Indeed, PDGF-BB-treated cells rapidly displayed significant increases of cAMP levels as compared to controls, an effect that was blocked by the PDGFR(-β) inhibitor imatinib. Thus, PDGF-BB appears to promote Hh signaling-dependent cytoprotection by inducing cAMP/PKA-mediated SMO trafficking to the plasma membrane. To further confirm the PDGF-BB-stimulated SMO-dependent gene regulation, we identified 67 target genes to be commonly up- (50 genes) or downregulated (17 genes) by both SHH and PDGF-BB in a cyclopamine-dependent manner in KMCH-1 cells via an Affymetrix U133 Plus 2.0 GeneChip analysis (Table 1).
Table 1.
Gene targets regulated by both SHH and PDGF-BB in a cyclopamine-dependent manner in KMCH-1 cells (alphabetical order).
| Gene symbol |
Gene name | GenBank accession no. |
|---|---|---|
|
| ||
| upregulated * | ||
|
| ||
| C7orf40 | chromosome 7 open reading frame 40 | AI937446 |
| CARD10 | caspase recruitment domain family, member 10 | AY028896 |
| CCL2 | chemokine (C-C motif) ligand 2 | S69738 |
| CYR61 | cysteine-rich, angiogenic inducer, 61 | AF003114 |
| DKK1 | dickkopf homolog 1 (Xenopus laevis) | NM_012242 |
| ENC1 | ectodermal-neural cortex (with BTB-like domain) | NM_003633 |
| EPHA2 | EPH receptor A2 | NM_004431 |
| F3 | coagulation factor III (thromboplastin, tissue factor) | NM_001993 |
| GPATCH4 | G patch domain containing 4 | BE794289 |
| HAS3 | hyaluronan synthase 3 | AF232772 |
| HBEGF | heparin-binding EGF-like growth factor | NM_001945 |
| HEATR1 | HEAT repeat containing 1 | NM_018072 |
| ISG20L2 | interferon stimulated exonuclease gene 20kDa-like 2 | AW294587 |
| JAG1 | jagged 1 (Alagille syndrome) | U77914 |
| LAMB3 | laminin, beta 3 | L25541 |
| LUC7L | LUC7-like (S. cerevisiae) | BE049621 |
| LYAR | Ly1 antibody reactive homolog (mouse) | AW958593 |
| LYAR | Ly1 antibody reactive homolog (mouse) | AL136750 |
| MAT2A | methionine adenosyltransferase II, alpha | BC001686 |
| METAP2 | methionyl aminopeptidase 2 | AW003997 |
| MKI67IP | MKI67 (FHA domain) interacting nucleolar phosphoprotein | AL577809 |
| MORC4 | MORC family CW-type zinc finger 4 | NM_024657 |
| MRPS2 | mitochondrial ribosomal protein S2 | NM_016034 |
| NOB1 | NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) | BC000050 |
| NOC3L | nucleolar complex associated 3 homolog (S. cerevisiae) | NM_022451 |
| NSUN2 | NOL1/NOP2/Sun domain family, member 2 | BC001041 |
| NUP35 | nucleoporin 35kDa | AL529634 |
| PFKFB3 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | NM_004566 |
| PLK2 | polo-like kinase 2 (Drosophila) | NM_006622 |
| PNN | pinin, desmosome associated protein | BF508848 |
| PNPT1 | polyribonucleotide nucleotidyltransferase 1 | AI967971 |
| POLR1E | polymerase (RNA) I polypeptide E, 53kDa | NM_022490 |
| POP1 | processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) | D31765 |
| PRKCH | protein kinase C, eta | NM_024064 |
| PUS3 | pseudouridylate synthase 3 | NM_031307 |
| PWP2 | PWP2 periodic tryptophan protein homolog (yeast) | U56085 |
| RBM34 | RNA binding motif protein 34 | AA887480 |
| RND3 | Rho family GTPase 3 | BG054844 |
| RRAD | Ras-related associated with diabetes | NM_004165 |
| SERPINE1 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
AL574210 |
| SLC25A32 | solute carrier family 25, member 32 | NM_030780 |
| SLC41A1 | solute carrier family 41, member 1 | AW439816 |
| SNHG7 | small nucleolar RNA host gene 7 (non-protein coding) | AL533103 |
| SYTL2 | synaptotagmin-like 2 | AB046817 |
| TGFB2 | transforming growth factor, beta 2 | M19154 |
| UTP14A | UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) | BC001149 |
| VEGFC | vascular endothelial growth factor C | U58111 |
| WDR73 | WD repeat domain 73 | AF161382 |
| ZNF239 | zinc finger protein 239 | NM_005674 |
| ZYX | zyxin | BC002323 |
|
| ||
| downregulated * | ||
|
| ||
| BNIP3L | BCL2/adenovirus E1B 19kDa interacting protein 3-like | AL132665 |
| CCNG1 | cyclin G1 | BC000196 |
| CCNG2 | cyclin G2 | AW134535 |
| ELMOD2 | ELMO/CED-12 domain containing 2 | BG477315 |
| IFIT2 | interferon-induced protein with tetratricopeptide repeats 2 | AA131041 |
| INSIG1 | insulin induced gene 1 | BG292233 |
| LMO4 | LIM domain only 4 | BC003600 |
| LQK1 | hypothetical LOC642946 | N51468 |
| MEGF9 | multiple EGF-like-domains 9 | W68084 |
| NBR1 | neighbor of BRCA1 gene 1 | NM_005899 |
| NHSL1 | NHS-like 1 | AA503387 |
| PCMTD1 | protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 | AA453163 |
| SCD | stearoyl-CoA desaturase (delta-9-desaturase) | AB032261 |
| SCD | stearoyl-CoA desaturase (delta-9-desaturase) | AF132203 |
| TCEAL8 | transcription elongation factor A (SII)-like 8 | AI743979 |
| TMEM135 | transmembrane protein 135 | AK000684 |
| TP53INP1 | tumor protein p53 inducible nuclear protein 1 | AW341649 |
Affymetrix U133 Plus 2.0 GeneChip analysis was performed. Genes were considered to be upregulated when they a) displayed significant upregulation (p<0.05, compared with the control group) upon SHH (single treatment) as well as PDGF-BB (single treatment) stimulation, and b) displayed significant downregulation (p<0.05, compared with the SHH and PDGF-BB group, respectively) upon addition of Hh inhibitor cyclopamine to the SHH as well as to the PDGF-BB treatment. Downregulated genes were regulated vice versa to a) and b).
Hh signaling inhibition is tumor suppressive in vivo
To determine if the proapoptotic in vitro-effect of Hh signaling inhibition by cyclopamine observed in co-cultures is translatable to an in vivo model, we employed a syngeneic rat orthotopic CCA model (BDEneu malignant cells injected into the liver of male Fischer 344 rats). Like human CCA, the BDEneu cells also express TRAIL in vivo.28-30 We confirmed that BDEneu cells express mRNA of members of the Hh signaling pathway, i.e. SHH, IHH, DHH, PTCH1, SMO, and GLI 1-3 (Supplemental Fig. 5). This rodent model of CCA also duplicates the desmoplasia characteristic of the human disease with numerous α-SMA-positive MFBs present in the stromal tumor microenvironment (Fig. 5A). We confirmed that the tumor samples (including MFBs and CCA cells) in this in vivo model also richly expresses mRNA for PDGF-BB and its cognate receptor PDGFR-β as compared non-tumor liver tissue (Fig. 5B). Moreover, PDGFR-β immunoreactivity was identified in CCA cells (Fig. 5C), whereas PDGF-BB expression was apparent in the MFBs and at the margin of CCA glands (Fig. 5D). Thus, this preclinical, rodent model of CCA mimics the characteristic features observed in human CCA tissue and cell lines.
Figure 5.
Hh signaling inhibition promotes apoptosis in PDGFR-β-expressing CCA cells in vivo. A syngeneic rat orthotopic model of CCA (BDEneu cells; Fischer 344 rats) was employed for this examination. (A-D) The rodent CCA model recapitulates cellular expression patterns of α-SMA, PDGF-BB, and PDGFR-β observed in human CCA. (A) α-SMA protein expression (brown) in tumors of untreated tumor-bearing rats was examined by immunohistochemistry (counterstaining with Mayers’ Hematoxylin). (B) CCA and normal liver specimens of untreated tumor-bearing rats (14 days after tumor cell implantation into the left lateral liver lobe) were analyzed for mRNA expression of PDGF-B and PDGFR-β by quantitative RT-PCR. Mean ± s.e.m. (n=3). (C) PDGFR-β protein expression (brown) in tumors of untreated tumor-bearing rats was examined by immunohistochemistry. PDGFR-β-positive CCA cell nests are surrounded by a paucicellular stromal matrix. (D) PDGF-BB immunoreactivity (brown) was similarly analyzed and the most intense signal was observed in stromal cells and the tumor-stromal interface. (E) Apoptosis of CCA cells was assessed in tumor tissues by TUNEL staining (green) and the identity of cells determined by co-staining via immunohistochemistry for CK7 (a CCA marker; red) Animals were treated with vehicle (upper) or cyclopamine (lower; 2.5 mg/kg BW intraperitoneally daily for one week; 1st injection: 7th post-operative day, 7th injection: 13th post-operative day). Quantitation of TUNEL- and CK7-positive cells (expressed as number per HPF) 14 days after CCA cell implantation demonstrated that in cyclopamine-treated animals CCA cell apoptosis was increased as compared to controls (bar graph). Mean ± s.e.m. (n=10). HPF, high power field.
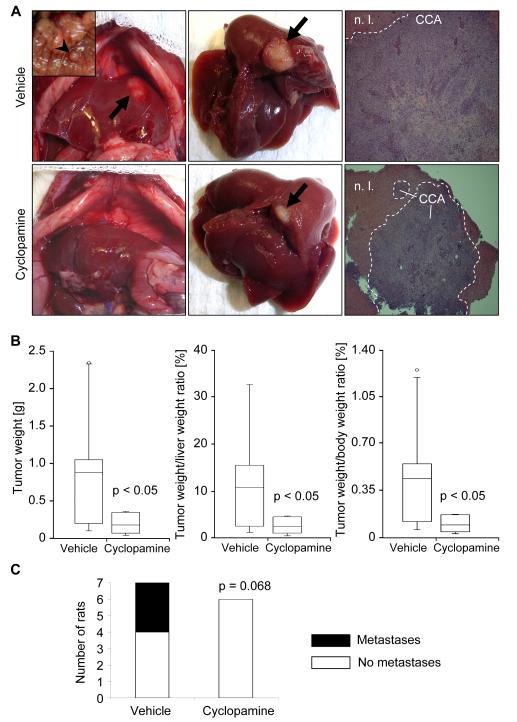
Next, we examined the potential therapeutic effects of the hedgehog signaling inhibitor cyclopamine in this in vivo model of CCA. In cyclopamine-treated animals, CCA cell apoptosis was increased as compared to controls. Apoptosis of CCA cells was confirmed by demonstrating colocalization of TUNEL-positive cells with cells displaying CK7 (a biliary epithelial cell marker expressed by CCA cells; Fig. 5E). Consistent with the proapoptotic effects of cyclopamine in this model, cyclopamine also had an effect on tumor size. Indeed, tumor weight, and tumor/liver as well as tumor/body weight ratios were significantly decreased in cyclopamine-treated rats (Fig. 6A and B). In addition, animals treated with cyclopamine displayed no extrahepatic metastases whereas 43% of vehicle-treated animals had extrahepatic metastases, predominantly occurring in the greater omentum and peritoneum (Fig. 6C, inset Fig. 6A left upper). In aggregate, these data suggest that cyclopamine promotes CCA cell apoptosis and decreases tumor growth as well as metastasis in an in vivo rodent model of CCA.
Figure 6.
Hh signaling inhibition reduces tumor growth and metastasis in vivo. A syngeneic rat orthotopic model of CCA (BDEneu cells; Fischer 344 rats) was employed for this examination. In cyclopamine- (2.5 mg/kg BW intraperitoneally daily for one week; 1st injection: 7th post-operative day, 7th injection: 13th post-operative day) or vehicle-treated tumor-bearing rats (n=7 rats/group) tumor/liver/body weight and extrahepatic metastasis were assessed 14 days after tumor cell implantation into the left lateral liver lobe. (A) Depicted are representative abdominal cavities (left), explanted livers (middle), and hematoxylin/eosin sections (right) of vehicle- (upper) and cyclopamine-treated (lower) rats. Arrows indicate the liver tumors, while the arrowhead (insert in the upper left photomicrograph) displays a representative example of extrahepatic metastasis (peritoneum). N.l., normal liver; CCA, cholangiocarcinoma. (B) Changes in tumor weight as well as tumor/liver and tumor/body weight ratios are depicted as box-and-whisker plots showing lowest value, 25th percentile, median, 75th percentile, highest value, and in some cases outliers. (C) The stacked column plot indicates the numbers of animals with and without metastases for vehicle- and cyclopamine-treated groups (p = 0.068 by χ2 test).
DISCUSSION
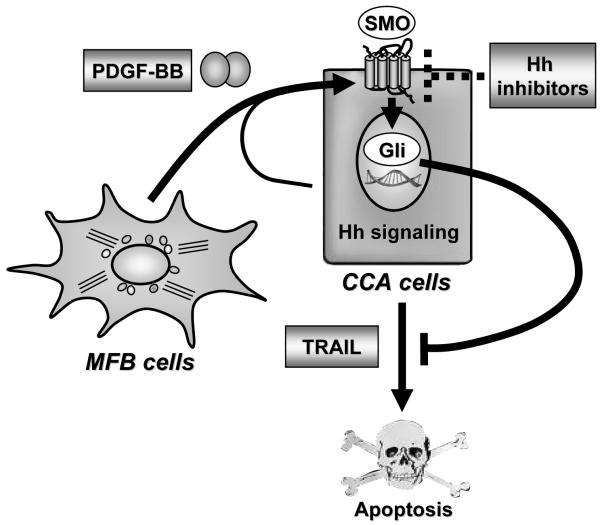
The results of this study provide new mechanistic insights regarding cytoprotective MFB-to-tumor cell paracrine signaling in CCA. These data indicate that MFB-derived PDGF-BB: (1) protects CCA cells from TRAIL-induced cell death in vitro; (2) exerts this cytoprotection in a Hh signaling-dependent manner by inducing cAMP/PKA-mediated SMO trafficking to the plasma membrane resulting in GLI2 nuclear translocation and GLI transcriptional activity; (3) and appears to act similarly in a rodent in vivo-model of CCA, where Hh signaling inhibition by cyclopamine promotes CCA cell apoptosis and is tumor suppressive. These findings are illustrated in Fig. 7 and discussed in greater detail below.
Figure 7.
Schematic diagram illustrating the role of platelet-derived growth factor (PDGF)-BB in promoting apoptosis resistance in cholangiocarcinoma (CCA) cells. PDGF-BB, mainly secreted by myofibroblasts (MBFs), promotes hedgehog (Hh) signaling by inducing trafficking of Hh signaling mediator smoothened (SMO) to the plasma membrane and subsequent glioma-associated oncogene (GLI) activation resulting in apoptosis resistance. Hh signaling inhibitors such as cyclopamine block Hh survival signaling and, thus, promote apoptosis in CCA cells. TRAIL; tumor necrosis factor-related apoptosis-inducing ligand.
In this study, we explored a role for PDGF-BB as a MFB-derived survival factor for CCA cells. Indeed, in coculture experiments, MFB cytoprotection against TRAIL-induced apoptosis was abrogated by neutralizing antisera to PDGF-BB suggesting MFB-derived PDGF-BB is a potent anti-TRAIL survival factor for CCA cells. Although many cancer cells may not express PDGF receptors,35 our data indicate CCA cells express PDGFR-β and respond to PDGF-BB by activating (phosphorylation) this receptor. These observations suggest the existence of a distinctive paracrine survival signaling pathway between MFB and CCA cells.
Coactivation networks are being increasingly recognized in cancer biology.38 We had previously implicated a major role for Hh-signaling directed survival signals against TRAIL cytotoxicity of CCA cells in vitro. 25, 26 Also, others have suggested PDGF-BB increases Hh ligand generation in immature bile ductular cells.16 Given this information, we posited that a PDGF-BB and Hh signaling co-activation network could contribute to survival signaling in CCA cells. Somewhat surprisingly, we found that PDGF-BB does not induce Hh ligand expression. 15, 16 Instead, PDGF-BB appears to increase Hh signaling by promoting SMO trafficking to the plasma membrane (an event known to increase SMO activation 22). Moreover, these processes were blocked by H89 (an inhibitor of the cAMP-regulated kinase PKA), suggesting that PDGF-BB-induced SMO trafficking is PKA-mediated. We note that the role of PKA in the Hh pathway is complex and likely depends upon the cell type and cellular context. For example, although PKA has been reported to promote Hh signaling at the level of SMO, it may act as a negative regulator by promoting the cleavage of GLI proteins into their repressor forms. 22 However, in CCA cells treated with PDGF-BB, PKA does not repress PDGF-BB-mediated GLI transcriptional activity as we observed activation of a GLI reporter gene assay, and common gene expression between SHH and PDGF-BB stimulation in a cyclopamine-inhibitable manner.
Consistent with a requirement for PKA during PDGF-BB stimulation of SMO trafficking, we also were able to demonstrate an increase of intracellular cAMP by PDGF-BB. Because receptor tyrosine kinases - as opposed to G-protein coupled receptors - do not directly stimulate adenylate cyclase (the enzyme generating cAMP), the mechanism by which PDGFR-β signaling enhances PKA activity in CCA cells will require further elucidation. A plausible mechanism would be the PDGF-BB/mitogen-activated protein kinase (MAPK)/prostaglandin E2/cAMP axis described in arterial smooth muscle cells. 39
The SMO inhibitor, cyclopamine, significantly increased apoptosis in CCA cells and achieved suppression of CCA tumor growth and metastasis in a preclinical rodent model of CCA. The orthotopic rodent model of CCA employed in these studies reflects a similar molecular signature and TRAIL expression as human CCA, 29, 30 exhibits a tumor microenvironment rich in activated α-SMA-secreting MFBs, and also recapitulates the cellular expression patterns of PDGF-BB and PDGFR-β found in many human CCA samples. Berman et al. also reported cyclopamine suppresses digestive tract tumors including CCA in vivo (in a xenograft tumor model).19 Herein, we expand this observation and provide evidence of functional interactions between tumor microenvironment and CCA cells. Moreover, we demonstrate that Hh signaling inhibition increases apoptosis of CCA cells in vivo. The mechanism by which cyclopamine induces apoptosis in vivo likely involves TRAIL expression in the tumor tissue, because cyclopamine does not increase apoptosis of monocultured CCA cells in the absence of TRAIL. Hh signaling has also been implicated in altering the tumor microenvironment.40 For example, Hh inhibitors increase the efficiency of cytotoxic chemotherapy in rodent models of pancreatic cancer by modulating the microenvironment of the cancer.13 In contrast to those studies, we did not observe a decrease in α-SMA positive MFBs in cyclopamine treated tumors (data not shown). Moreover, the importance of Hh signaling in cancer cells as opposed to stromal cells has recently been emphasized. 41 Our observations are most consistent with a direct effect of cyclopamine on the tumor cells in vivo, although we cannot exclude a non-cytotoxic effect of cyclopamine on MFB function.
In conclusion, MFB-derived PDGF-BB protects CCA cells from TRAIL-induced apoptosis. This cytoprotection is exerted through a coactivation network involving Hh signaling. These observations support the examination of selective Hh inhibitors (currently in clinical development 42, 43) in human CCA.
Supplementary Material
ACKNOWLEDGEMENT
The Affymetrix U133 Plus 2.0 GeneChip analysis was performed in collaboration with the Genomics Technology Center Core and Dr. Y. Li from the Division of Biomedical Statistics & Informatics (both Mayo Clinic, Rochester, MN). The assistance of Dr. U. Yaqoob with the immunoblot for (phospho-)PDGFR-β is also gratefully acknowledged as well as the superb secretarial service of C. Riddle.
This work was supported by grants NIH DK59427 (GJG), NIH R01 CA 83650, R01 CA 39225 (AES), the optical microscopy, clinical, and genetics core of NIH DK84567, and the Mayo Foundation. C.D. Fingas is a German Research Foundation fellow (Deutsche Forschungsgemeinschaft; grant FI 1630/1-1).
Abbreviations
- α-SMA
α-smooth muscle actin
- cAMP
cyclic adenosine monophosphate
- CCA
cholangiocarcinoma
- CK7
cytokeratin 7
- DHH
Desert hedgehog
- ErbB-2
erythroblastic leukemia viral oncogene homolog
- GLI
glioma-associated oncogene
- Hh
hedgehog
- HIP
hedgehog-interacting protein
- HSC
hepatic stellate cells
- IHH
Indian hedgehog
- MAPK
mitogen-activated protein kinase
- MBF
myofibroblast
- PDGF
platelet-derived growth factor
- PDGFR
platelet-derived growth factor receptor
- PKA
cAMP-dependent protein kinase
- PTCH1
patched1
- SHH
Sonic hedgehog
- SMO
smoothened
- SUFU
suppressor of fused
- TIRF
total internal reflection fluorescence
- TRAIL
tumor necrosis factor-related apoptosis-inducing ligand
REFERENCES
- 1.de Groen PC, Gores GJ, LaRusso NF, Gunderson LL, Nagorney DM. Biliary tract cancers. N Engl J Med. 1999;341:1368–1378. doi: 10.1056/NEJM199910283411807. [DOI] [PubMed] [Google Scholar]
- 2.Fingas CD, Katsounas A, Kahraman A, Siffert W, Jochum C, Gerken G, et al. Prognostic assessment of three single-nucleotide polymorphisms (GNB3 825C>T, BCL2-938C>A, MCL1-386C>G) in extrahepatic cholangiocarcinoma. Cancer Invest. 2010;28:472–478. doi: 10.3109/07357900903095714. [DOI] [PubMed] [Google Scholar]
- 3.Roberts SK, Ludwig J, Larusso NF. The pathobiology of biliary epithelia. Gastroenterology. 1997;112:269–279. doi: 10.1016/s0016-5085(97)70244-0. [DOI] [PubMed] [Google Scholar]
- 4.Blechacz B, Gores GJ. Cholangiocarcinoma: advances in pathogenesis, diagnosis, and treatment. Hepatology. 2008;48:308–321. doi: 10.1002/hep.22310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishimura N, Isomoto H, Bronk SF, Gores GJ. Trail induces cell migration and invasion in apoptosis-resistant cholangiocarcinoma cells. Am J Physiol Gastrointest Liver Physiol. 2006;290:G129–136. doi: 10.1152/ajpgi.00242.2005. [DOI] [PubMed] [Google Scholar]
- 6.Kuperwasser C, Chavarria T, Wu M, Magrane G, Gray JW, Carey L, et al. Reconstruction of functionally normal and malignant human breast tissues in mice. Proc Natl Acad Sci U S A. 2004;101:4966–4971. doi: 10.1073/pnas.0401064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, Cunha GR. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999;59:5002–5011. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasanen K, Vaheri A. Activation of fibroblasts in cancer stroma. Exp Cell Res. 2010 doi: 10.1016/j.yexcr.2010.04.032. [DOI] [PubMed] [Google Scholar]
- 9.Dranoff JA, Wells RG. Portal fibroblasts: Underappreciated mediators of biliary fibrosis. Hepatology. 2010;51:1438–1444. doi: 10.1002/hep.23405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wells RG. The epithelial-to-mesenchymal transition in liver fibrosis: here today, gone tomorrow? Hepatology. 2010;51:737–740. doi: 10.1002/hep.23529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erez N, Truitt M, Olson P, Arron ST, Hanahan D. Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-kappaB-Dependent Manner. Cancer Cell. 2010;17:135–147. doi: 10.1016/j.ccr.2009.12.041. [DOI] [PubMed] [Google Scholar]
- 12.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 13.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chuaysri C, Thuwajit P, Paupairoj A, Chau-In S, Suthiphongchai T, Thuwajit C. Alpha-smooth muscle actin-positive fibroblasts promote biliary cell proliferation and correlate with poor survival in cholangiocarcinoma. Oncol Rep. 2009;21:957–969. doi: 10.3892/or_00000309. [DOI] [PubMed] [Google Scholar]
- 15.Omenetti A, Yang L, Li YX, McCall SJ, Jung Y, Sicklick JK, et al. Hedgehog-mediated mesenchymal-epithelial interactions modulate hepatic response to bile duct ligation. Lab Invest. 2007;87:499–514. doi: 10.1038/labinvest.3700537. [DOI] [PubMed] [Google Scholar]
- 16.Omenetti A, Popov Y, Jung Y, Choi SS, Witek RP, Yang L, et al. The hedgehog pathway regulates remodelling responses to biliary obstruction in rats. Gut. 2008;57:1275–1282. doi: 10.1136/gut.2008.148619. [DOI] [PubMed] [Google Scholar]
- 17.Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deming PB, Campbell SL, Baldor LC, Howe AK. Protein kinase A regulates 3-phosphatidylinositide dynamics during platelet-derived growth factor-induced membrane ruffling and chemotaxis. J Biol Chem. 2008;283:35199–35211. doi: 10.1074/jbc.M804448200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berman DM, Karhadkar SS, Maitra A, Montes De Oca R, Gerstenblith MR, Briggs K, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 20.Saqui-Salces M, Merchant JL. Hedgehog signaling and gastrointestinal cancer. Biochim Biophys Acta. 2010;1803:786–795. doi: 10.1016/j.bbamcr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walterhouse DO, Yoon JW, Iannaccone PM. Developmental pathways: Sonic hedgehog-Patched-GLI. Environ Health Perspect. 1999;107:167–171. doi: 10.1289/ehp.99107167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milenkovic L, Scott MP. Not lost in space: trafficking in the hedgehog signaling pathway. Sci Signal. 2010;3:pe14. doi: 10.1126/scisignal.3117pe14. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, Lin X. Hedgehog signaling uses lipid metabolism to tune smoothened activation. Dev Cell. 2010;19:3–4. doi: 10.1016/j.devcel.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 24.Yavari A, Nagaraj R, Owusu-Ansah E, Folick A, Ngo K, Hillman T, et al. Role of lipid metabolism in smoothened derepression in hedgehog signaling. Dev Cell. 2010;19:54–65. doi: 10.1016/j.devcel.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurita S, Mott JL, Almada LL, Bronk SF, Werneburg NW, Sun SY, et al. GLI3-dependent repression of DR4 mediates hedgehog antagonism of TRAIL-induced apoptosis. Oncogene. 2010;29:4848–4858. doi: 10.1038/onc.2010.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirasaki S, Koide N, Ujike K, Shinji T, Tsuji T. Expression of Nov, CYR61 and CTGF genes in human hepatocellular carcinoma. Hepatology Research. 2001;19:294–305. doi: 10.1016/s1386-6346(00)00101-7. [DOI] [PubMed] [Google Scholar]
- 27.Anan A, Baskin-Bey ES, Bronk SF, Werneburg NW, Shah VH, Gores GJ. Proteasome inhibition induces hepatic stellate cell apoptosis. Hepatology. 2006;43:335–344. doi: 10.1002/hep.21036. [DOI] [PubMed] [Google Scholar]
- 28.Blechacz BR, Smoot RL, Bronk SF, Werneburg NW, Sirica AE, Gores GJ. Sorafenib inhibits signal transducer and activator of transcription-3 signaling in cholangiocarcinoma cells by activating the phosphatase shatterproof 2. Hepatology. 2009;50:1861–1870. doi: 10.1002/hep.23214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fingas CD, Blechacz BR, Smoot RL, Guicciardi ME, Mott J, Bronk SF, et al. A smac mimetic reduces TNF related apoptosis inducing ligand (TRAIL)-induced invasion and metastasis of cholangiocarcinoma cells. Hepatology. 2010;52:550–561. doi: 10.1002/hep.23729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sirica AE, Zhang Z, Lai GH, Asano T, Shen XN, Ward DJ, et al. A novel “patient-like” model of cholangiocarcinoma progression based on bile duct inoculation of tumorigenic rat cholangiocyte cell lines. Hepatology. 2008;47:1178–1190. doi: 10.1002/hep.22088. [DOI] [PubMed] [Google Scholar]
- 31.Steyer JA, Almers W. A real-time view of life within 100 nm of the plasma membrane. Nat Rev Mol Cell Biol. 2001;2:268–275. doi: 10.1038/35067069. [DOI] [PubMed] [Google Scholar]
- 32.Sasaki H, Hui C, Nakafuku M, Kondoh H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- 33.van den Brink GR, Bleuming SA, Hardwick JC, Schepman BL, Offerhaus GJ, Keller JJ, et al. Indian Hedgehog is an antagonist of Wnt signaling in colonic epithelial cell differentiation. Nat Genet. 2004;36:277–282. doi: 10.1038/ng1304. [DOI] [PubMed] [Google Scholar]
- 34.van den Brink GR, Hardwick JC, Tytgat GN, Brink MA, Ten Kate FJ, Van Deventer SJ, et al. Sonic hedgehog regulates gastric gland morphogenesis in man and mouse. Gastroenterology. 2001;121:317–328. doi: 10.1053/gast.2001.26261. [DOI] [PubMed] [Google Scholar]
- 35.Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22:1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, et al. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 37.Wojtal KA, Hoekstra D, van Ijzendoorn SC. cAMP-dependent protein kinase A and the dynamics of epithelial cell surface domains: moving membranes to keep in shape. Bioessays. 2008;30:146–155. doi: 10.1002/bies.20705. [DOI] [PubMed] [Google Scholar]
- 38.Xu AM, Huang PH. Receptor tyrosine kinase coactivation networks in cancer. Cancer Res. 2010;70:3857–3860. doi: 10.1158/0008-5472.CAN-10-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graves LM, Bornfeldt KE, Sidhu JS, Argast GM, Raines EW, Ross R, et al. Platelet-derived growth factor stimulates protein kinase A through a mitogen-activated protein kinase-dependent pathway in human arterial smooth muscle cells. J Biol Chem. 1996;271:505–511. doi: 10.1074/jbc.271.1.505. [DOI] [PubMed] [Google Scholar]
- 40.Yauch RL, Gould SE, Scales SJ, Tang T, Tian H, Ahn CP, et al. A paracrine requirement for hedgehog signalling in cancer. Nature. 2008;455:406–410. doi: 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- 41.Singh S, Wang Z, Liang Fei D, Black KE, Goetz JA, Tokhunts R, et al. Hedgehog-Producing Cancer Cells Respond to and Require Autocrine Hedgehog Activity. Cancer Res. 2011;71:4454–4463. doi: 10.1158/0008-5472.CAN-10-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hyman JM, Firestone AJ, Heine VM, Zhao Y, Ocasio CA, Han K, et al. Small-molecule inhibitors reveal multiple strategies for Hedgehog pathway blockade. Proc Natl Acad Sci U S A. 2009;106:14132–14137. doi: 10.1073/pnas.0907134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng JM, Curran T. The Hedgehog’s tale: developing strategies for targeting cancer. Nat Rev Cancer. 2011;11:493–501. doi: 10.1038/nrc3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.