Abstract
Many machine learning and pattern classification methods have been applied to the diagnosis of Alzheimer’s disease (AD) and its prodromal stage, i.e., mild cognitive impairment (MCI). Recently, rather than predicting categorical variables as in classification, several pattern regression methods have also been used to estimate continuous clinical variables from brain images. However, most existing regression methods focus on estimating multiple clinical variables separately and thus cannot utilize the intrinsic useful correlation information among different clinical variables. On the other hand, in those regression methods, only a single modality of data (usually only the structural MRI) is often used, without considering the complementary information that can be provided by different modalities. In this paper, we propose a general methodology, namely Multi-Modal Multi-Task (M3T) learning, to jointly predict multiple variables from multi-modal data. Here, the variables include not only the clinical variables used for regression but also the categorical variable used for classification, with different tasks corresponding to prediction of different variables. Specifically, our method contains two key components, i.e., (1) a multi-task feature selection which selects the common subset of relevant features for multiple variables from each modality, and (2) a multi-modal support vector machine which fuses the above-selected features from all modalities to predict multiple (regression and classification) variables. To validate our method, we perform two sets of experiments on ADNI baseline MRI, FDG-PET, and cerebrospinal fluid (CSF) data from 45 AD patients, 91 MCI patients, and 50 healthy controls (HC). In the first set of experiments, we estimate two clinical variables such as Mini Mental State Examination (MMSE) and Alzheimer’s Disease Assessment Scale - Cognitive Subscale (ADAS-Cog), as well as one categorical variable (with value of ‘AD’, ‘MCI’ or ‘HC’), from the baseline MRI, FDG-PET, and CSF data. In the second set of experiments, we predict the 2-year changes of MMSE and ADAS-Cog scores and also the conversion of MCI to AD from the baseline MRI, FDG-PET, and CSF data. The results on both sets of experiments demonstrate that our proposed M3T learning scheme can achieve better performance on both regression and classification tasks than the conventional learning methods.
Keywords: Alzheimer’s disease (AD), multi-modal multi-task (M3T) learning, multi-task feature selection, multi-modality, MCI conversion, MMSE, ADAS-Cog
Introduction
Alzheimer’s disease (AD) is the most common form of dementia diagnosed in people over 65 years of age. It is reported that there are 26.6 million AD sufferers worldwide, and 1 in 85 people will be affected by 2050 (Ron et al., 2007). Thus, accurate diagnosis of AD and especially its early stage, i.e., mild cognitive impairment (MCI), is very important for timely therapy and possible delay of the disease. Over the past decade, many machine learning and pattern classification methods have been used for early diagnosis of AD and MCI based on different modalities of biomarkers, e.g., the structural brain atrophy measured by magnetic resonance imaging (MRI) (de Leon et al., 2007; Du et al., 2007; Fjell et al., 2010; McEvoy et al., 2009), metabolic alterations in the brain measured by fluorodeoxyglucose positron emission tomography (FDG-PET) (De Santi et al., 2001; Morris et al., 2001), and pathological amyloid depositions measured through cerebrospinal fluid (CSF) (Bouwman et al., 2007b; Fjell et al., 2010; Mattsson et al., 2009; Shaw et al., 2009), etc. In all these methods, classification models are learned from training subjects to predict categorical classification variable (i.e., class label) on test subjects.
Recently, rather than predicting categorical variables as in classification, several studies begin to estimate continuous clinical variables from brain images (Duchesne et al., 2009; Duchesne et al., 2005; Fan et al., 2010; Stonnington et al., 2010; Wang et al., 2010). This kind of research is important because it can help evaluate the stage of AD pathology and predict future progression. Different from classification that classifies a subject into binary or multiple categories, regression needs to estimate continuous values and are thus more challenging. In the literature, a number of regression methods have been used for estimating clinical variables based on neuroimaging data. For example, linear regression models were used to estimate the 1-year Mini Mental State Examination (MMSE) changes from structural MR brain images (Duchesne et al., 2009; Duchesne et al., 2005). High-dimensional kernel-based regression method, i.e., Relevance Vector Machine (RVM), were also used to estimate clinical variables, including MMSE and Alzheimer’s Disease Assessment Scale - Cognitive Subscale (ADAS-Cog), from structural MR brain images (Fan et al., 2010; Stonnington et al., 2010; Wang et al., 2010). Besides clinical variables, regression methods have also been used for estimating age of individual subject from MR brain images (Ashburner, 2007; Franke et al., 2010).
In the practical diagnosis of AD, multiple clinical variables are generally acquired, e.g., MMSE and ADAS-Cog, etc. Specifically, MMSE is used to examine the orientation to time and place, the immediate and delayed recall of three words, the attention and calculations, language, and visuoconstructional functions (Folstein et al., 1975), while ADAS-Cog is a global measure encompassing the core symptoms of AD (Rosen et al., 1984). It is known that there exist inherent correlations among multiple clinical variables of a subject, since the underlying pathology is the same (Fan et al., 2010; Stonnington et al., 2010). However, most existing regression methods model different clinical variables separately, without considering their inherent correlations that may be useful for robust and accurate estimation of clinical variables from brain images. Moreover, to our knowledge, none of the existing regression methods used for estimating clinical variables ever exploit the class labels which are often available in the training subjects and are helpful to aid the accurate estimation of regression variables, and vice versa.
On the other hand, although multi-modal data are often acquired for AD diagnosis, e.g., MRI, PET, and CSF biomarkers, nearly all existing regression methods developed for estimation of clinical variables were based only on one imaging modality, i.e., mostly on the structural MRI. Recent studies have indicated that the biomarkers from different modalities provide complementary information, which is very useful for AD diagnosis (Fjell et al., 2010; Landau et al., 2010; Walhovd et al., 2010b). More recently, a number of research works have used multi-modal data for AD or MCI classification and obtained the improved performance compared with the methods based only on single-modal data (Fan et al., 2008; Hinrichs et al., 2011; Vemuri et al., 2009; Walhovd et al., 2010a; Zhang et al., 2011). However, to the best of our knowledge, the same type of study in imaging-based regression, i.e., estimation of clinical variables from multi-modal data, was not investigated previously.
Inspired by the above problems, in this paper, we propose a general methodology, namely Multi-Modal Multi-Task (M3T) learning, to jointly predict multiple variables from multi-modal data. Here, the variables include not only the continuous clinical variables for regression (MMSE, ADAS-Cog) but also the categorical variable for classification (i.e., class label). We treat the estimation of different regression or classification variables as different tasks, and use a multi-task learning method (Argyriou et al., 2008; Obozinski et al., 2006) developed in the machine learning community for joint regression and classification learning. Specifically, at first, we assume that the related tasks share a common relevant feature subset but with a varying amount of influence on each task, and thus adopt a multi-task feature selection method to obtain a common feature subset for different tasks simultaneously. Then, we use a multi-modal support vector machine (SVM) method to fuse the above-selected features from each modality to estimate multiple regression and classification variables.
We validate our method on two sets of experiments. In the first set of experiments, we estimate two regression variables (MMSE and ADAS-Cog) and one classification variable (with value of ‘AD’, ‘MCI’ or ‘HC’) from the baseline MRI, PET, and CSF data. In the second of experiment, we predict the 2-year changes of MMSE and ADAS-Cog scores and also the conversion of MCI to AD from the baseline MRI, PET, and CSF data. We hypothesize that the joint estimation or prediction of multiple regression and classification variables would perform better than estimating or predicting each individual variable separately, and that the use of multi-modal data (MRI, PET and CSF) would perform better on joint regression and classification than the use of only single-modal data.
Method
The data used in the preparation of this paper were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (www.loni.ucla.edu/ADNI). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies, and non-profit organizations, as a $60 million, 5-year public-private partnership. The primary goal of ADNI has been to test whether the serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials.
ADNI is the result of efforts of many coinvestigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 adults, aged 55 to 90, to participate in the research – approximately 200 cognitively normal older individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years, and 200 people with early AD to be followed for 2 years (see www.adni-info.org for up-to-date information). The research protocol was approved by each local institutional review board and written informed consent is obtained from each participant.
Subjects
The ADNI general eligibility criteria are described at www.adni-info.org. Briefly, subjects are between 55–90 years of age, having a study partner able to provide an independent evaluation of functioning. Specific psychoactive medications will be excluded. General inclusion/exclusion criteria are as follows: 1) healthy subjects: MMSE scores between 24–30, a Clinical Dementia Rating (CDR) of 0, non-depressed, non MCI, and nondemented; 2) MCI subjects: MMSE scores between 24–30, a memory complaint, having objective memory loss measured by education adjusted scores on Wechsler Memory Scale Logical Memory II, a CDR of 0.5, absence of significant levels of impairment in other cognitive domains, essentially preserved activities of daily living, and an absence of dementia; and 3) Mild AD: MMSE scores between 20–26, CDR of 0.5 or 1.0, and meets the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s Disease and Related Disorders Association (NINCDS/ADRDA) criteria for probable AD.
In this paper, 186 ADNI subjects with all corresponding baseline MRI, PET, and CSF data are included. In particular, it contains 45 AD patients, 91 MCI patients (including 43 MCI converters (MCI-C) and 48 MCI non-converters (MCI-NC)), and 50 healthy controls. Table 1 lists the demographics of all these subjects. Subject IDs are also given in Supplemental Table 4.
Table 1.
Subject information.
| AD (n=45) |
HC (n=50) |
MCI-C (n=43) |
MCI-NC (n=48) |
|
|---|---|---|---|---|
| Female/Male | 16/29 | 18/32 | 15/28 | 16/32 |
| Age | 75.4 ± 7.1 | 75.3 ± 5.2 | 75.8 ± 6.8 | 74.7 ± 7.7 |
| Education | 14.9 ± 3.4 | 15.6 ± 3.2 | 16.1 ± 2.6 | 16.1 ± 3.0 |
| MMSE (baseline) | 23.8 ± 1.9 | 29.0 ± 1.2 | 26.6 ± 1.7 | 27.5 ± 1.6 |
| MMSE (2 years) | 19.3 ± 5.6 | 29.0 ± 1.3 | 23.8 ± 3.3 | 26.9 ± 2.6 |
| ADAS-Cog (baseline) | 18.3 ± 6.1 | 7.3 ± 3.3 | 12.9 ± 3.9 | 9.7 ± 4.0 |
| ADAS-Cog (2 years) | 27.3 ± 11.7 | 6.3 ± 3.5 | 16.1 ± 6.4 | 11.2 ± 5.7 |
AD = Alzheimer’s Disease, HC = Healthy Control, MCI = Mild Cognitive Impairment, MCI-C = MCI converter, MCI-NC = MCI non-converter, MMSE = Mini-Mental State Examination, ADAS-Cog = Alzheimer’s Disease Assessment Scale - Cognitive Subscale
MRI, PET, and CSF
A detailed description on acquiring MRI, PET and CSF data from ADNI as used in this paper can be found at (Zhang et al., 2011). Briefly, structural MR scans were acquired from 1.5T scanners. Raw Digital Imaging and Communications in Medicine (DICOM) MRI scans were downloaded from the public ADNI site (www.loni.ucla.edu/ADNI), reviewed for quality, and automatically corrected for spatial distortion caused by gradient nonlinearity and B1 field inhomogeneity. PET images were acquired 30–60 minutes post-injection, averaged, spatially aligned, interpolated to a standard voxel size, intensity normalized, and smoothed to a common resolution of 8mm full width at half maximum. CSF data were collected in the morning after an overnight fast using a 20- or 24-gauge spinal needle, frozen within 1 hour of collection, and transported on dry ice to the ADNI Biomarker Core laboratory at the University of Pennsylvania Medical Center. In this study, CSF Aβ42, CSF t-tau and CSF p-tau are used as features.
Image analysis
Image pre-processing is performed for all MR and PET images following the same procedures as in (Zhang et al., 2011). First, we do anterior commissure (AC) - posterior commissure (PC) correction on all images, and use the N3 algorithm (Sled et al., 1998) to correct the intensity inhomogeneity. Next, we do skull-stripping on structural MR images using both brain surface extractor (BSE) (Shattuck et al., 2001) and brain extraction tool (BET) (Smith, 2002), followed by manual edition and intensity inhomogeneity correction. After removal of cerebellum, FAST in the FSL package (Zhang et al., 2001) is used to segment structural MR images into three different tissues: grey matter (GM), white matter (WM), and cerebrospinal fluid (CSF). After registration using HAMMER (Shen and Davatzikos, 2002), we obtain the subject-labeled image based on a template with 93 manually labeled ROIs (Kabani et al., 1998). For each of the 93 ROI regions in the labeled MR image, we compute the GM tissue volume of that region as a feature. For PET image, we first align it to its respective MR image of the same subject using a rigid transformation, and then compute the average intensity of each ROI in the PET image as a feature. Therefore, for each subject, we totally obtain 93 features from MRI image, other 93 features from PET image, and 3 features from CSF biomarkers.
Multi-Modal Multi-Task (M3T) learning
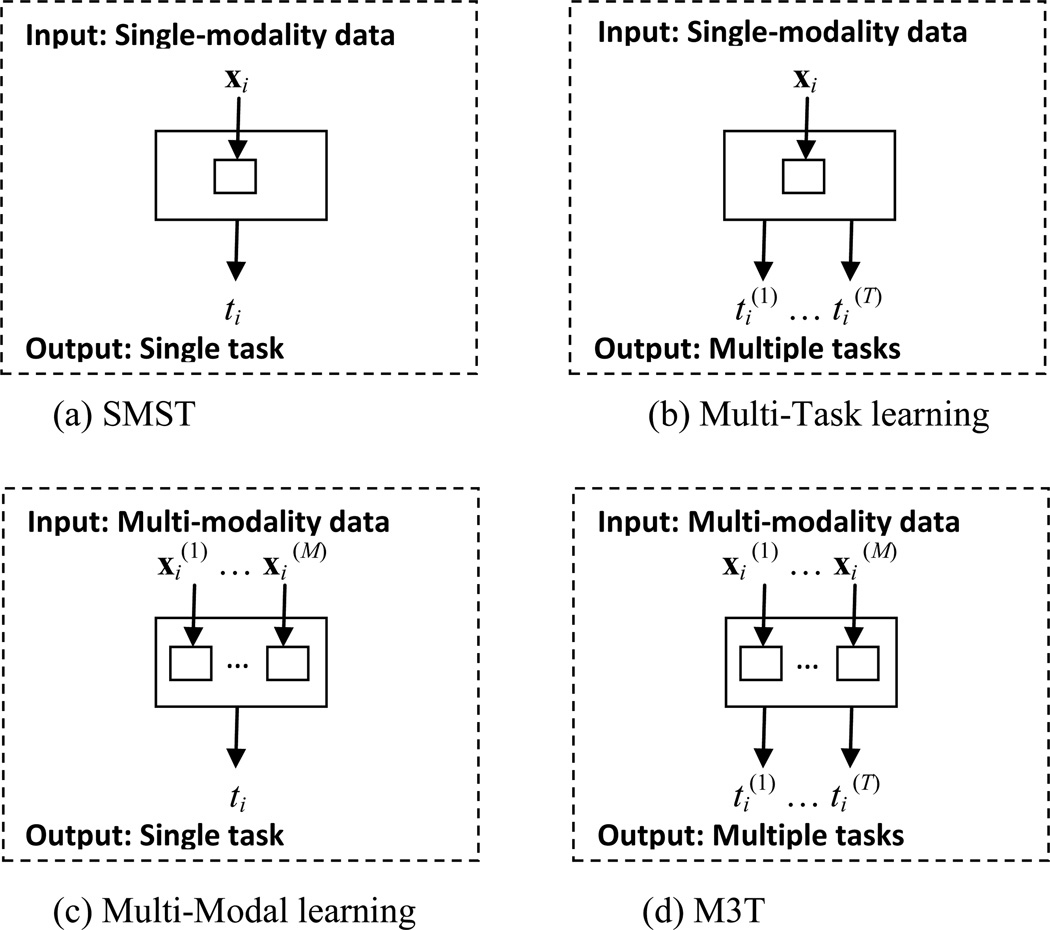
A new learning method, namely Multi-Modal Multi-Task (M3T) learning, is presented here to simultaneously learn multiple tasks from multi-modal data. Fig. 1 illustrates the new learning problem with comparison to the existing standard Single-Modal Single-Task (SMST) learning, Multi-Task learning, and Multi-Modal learning. As can be seen from Fig. 1, in SMST and Multi-Task learning (Fig. 1(a–b)), each subject has only one modality of data represented as xi, while, in M3T and Multi-Modal learning (Fig. 1(c–d)), each subject has multiple modalities of data represented as . On the other hand, Fig. 1 also shows that, in SMST and Multi-Modal learning (Fig. 1(a) and (c)), each subject corresponds to only one task denoted as ti, while, in M3T and Multi-Task learning (Fig. 1(b) and (d)), each subject corresponds to multiple tasks denoted as .
Fig. 1.
Illustration of the four different learning frameworks
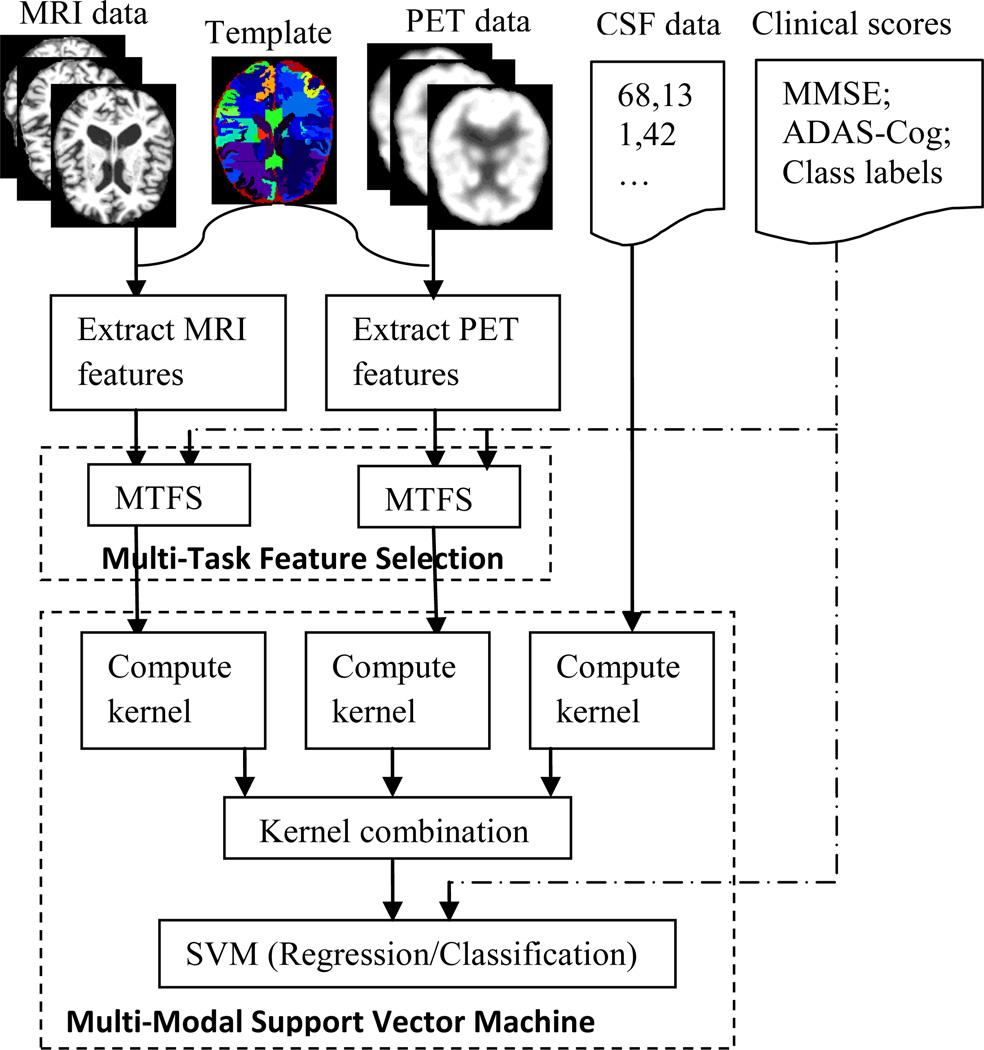
Now we can formally define the M3T learning as below. Given N training subjects and each having M modalities of data, represented as , our M3T method jointly learns a series of models corresponding to T different tasks denoted as . It is worth noting that M3T is a general learning framework, and here we implement it through two successive major steps, i.e., (1) multi-task feature selection (MTFS) and (2) multi-modal support vector machine (SVM) (for both regression and classification). Fig. 2 illustrates the flowchart of the proposed M3T method, where M = 3 modalities of data (e.g., MRI, PET, and CSF) are used for jointly learning models corresponding to different tasks. Note that, for CSF modality, the original 3 measures (i.e., Aβ42, t-tau, and p-tau) are directly used as features without any feature selection step.
Fig. 2.
Flowchart of the proposed M3T method
Multi-task feature selection (MTFS)
For imaging modalities such as MRI and PET, even after feature extraction, the number of features (extracted from brain regions) may be still large. Besides, not all features are relevant to the disease under study. So, feature selection is commonly used for dimensionality reduction, as well as for removal of irrelevant features. Different from the conventional single-task feature selection, the multi-task feature selection simultaneously selects a common feature subset relevant to all tasks. This point is especially important for diagnosis of neurological diseases, since multiple regression/classification variables are essentially determined by the same underlying pathology, i.e., the diseased brain regions. Also, simultaneously performing feature selection for multiple regression/classification variables is very helpful to suppress noises in the individual variables.
Denote as the training data matrix on the m-th modality from N training samples, and as the response vector on the j-th task from the same N training samples. Following the method proposed in (Obozinski et al., 2006), linear models are used to model the multi-task feature selection (MTFS) as below:
where is the weight vector for the j-th task on the m-th modality, and x(m) is the m-th modal data of a certain subject. The weight vectors for all T tasks form a weight matrix , which can be optimized by the following objective function:
where V(m)|d denotes the d-th row of V(m), D(m) is the dimension of the m-th modal data, and λ is the regularization coefficient controlling the relative contributions of the two terms. Note that λ also controls the ‘sparsity’ of the linear models, with the high value corresponding to more sparse models (i.e., more values in V(m) are zero). It is easy to know that the above equation reduces to the standard l1-norm regularized optimization problem in Lasso (Tibshirani, 1996) when there is only one task. In our case, this is a multi-task learning for the given m-th modal data.
The key point of the above objective function of MTFS is the use of l2-norm for V(m)|d, which forces the weights corresponding to the d-th feature (of the m-th modal data) across multiple tasks to be grouped together and tends to select features based on the strength of T tasks jointly. Note that, because of the characteristic of ‘group sparsity’, the solution of MTFS results in a weight matrix V(m) whose elements in some rows are all zeros. For feature selection, we just keep those features with non-zero weights. At present, there are many algorithms developed to solve MTFS; in this paper we adopt the SLEP toolbox (Liu et al., 2009), which has been shown very effective on many datasets.
Multi-modal support vector machine
In our previous work (Zhang et al., 2011), the multi-modal support vector classification (SVC) has been developed for multi-modal classification of AD and MCI. Following (Zhang et al., 2011), in this paper, we derive the corresponding multi-modal support vector regression (SVR) algorithm as below. Assume that we have N training subjects with the corresponding target output {zi ∈ ℝ, i = 1, …, N}, and each subject has M modalities of data with the features selected by the above proposed method and denoted as . Multi-modal SVR solves the following primal problem:
Where w(m), ϕ(m), and βm ≥ 0 denote the normal vector of hyperplane, the kernel-induced mapping function, and the combining weight on the m-th modality, respectively. Here, we constrain ∑m βm = 1. The parameter b is the offset. Note that ε-insensitive loss function is used in the above objective function, and ξ and ξ* are the two sets of slack variables.
Similar to the conventional SVR, the dual form of the multi-modal SVR can be represented as below:
Where is the kernel function for the two training samples on the m-th modality.
For a test sample with the selected features x = {x(1), …, x(m), …, x(M)}, we denote as the kernel between each training sample x′i and the test sample on the m-th modality. Then, the regression function takes the following form:
Similar to the multi-modal SVC (Zhang et al., 2011), the multi-modal SVR can also be solved with the conventional SVR, e.g., through the LIBSVM toolbox, if we define the mixed kernel between multi-modal training samples x′i and x′j, and between multimodal training sample x′i and test sample x. Here, βms are the nonnegative weight parameters used to balance the contributions of different modalities, and their values are optimized through a coarse-grid search by cross-validation on the training samples.
After obtaining the common feature subset for all different tasks by MTFS as described above, we use multi-modal SVM, including multi-modal SVC and multi-modal SVR, to train the final support vector classification and regression models, respectively. Here, we train a model for each corresponding variable (task). Specifically, we train support vector regression models corresponding to the regression variables, and support vector classification models corresponding to the classification variable, respectively. It is worth noting that, since we use the common subset of features (learned by MTFS during the feature selection stage) to train both regression and classification models, our models are actually performing the multi-modal multi-task learning.
Validation
To evaluate the performance of different methods, we perform two sets of experiments on 186 ADNI baseline MRI, PET, and CSF data, respectively, from 45 AD, 91 MCI (including 43 MCI-C and 48 MCI-NC), and 50 HC. In the first set of experiments (Experiment 1), we estimate two clinical variables (including MMSE and ADAS-Cog) and one categorical variable (with class label of ‘AD’, ‘MCI’ or ‘HC’) from the baseline brain data of all 186 subjects. It is worth noting that only the baseline data of MRI, PET, and CSF are used in our experiments, but, in order to alleviate the effect of noise in the measured clinical scores, we use the mean clinical score at both baseline and immediate follow-up time points as the ground truth for each subject. The same strategy has also been adopted in (Wang et al., 2010). In the second set of experiments (Experiment 2), we predict the 2-year changes of MMSE and ADAS-Cog scores and the conversion of MCI to AD from the baseline brain data of 167 subjects (since 19 subjects do not have the 2-year follow-up MMSE or ADAS-Cog scores and are thus discarded, as shown in Supplemental Table 4). Also, only the baseline data of MRI, PET, and CSF are used in the experiment, and the corresponding ground truths for the two regression tasks are the MMSE and ADAS-Cog changes from baseline to the 2-year follow-up. For classification task, we will discriminate between MCI-C and MCI-NC subjects, using the baseline MRI, PET, and CSF data.
We use 10-fold cross validation strategy by computing the Pearson’s correlation coefficient (for measuring the correlation between the predicted clinical scores and the actual clinical score in the regression tasks) and the classification accuracy (for measuring the proportion of subjects correctly classified in the classification task). Specifically, the whole set of subject samples are equally partitioned into 10 subsets, and each time the subject samples within one subset are selected as the testing samples and all remaining subject samples in the other 9 subsets are used for training the SVM models. This process is repeated for 10 times. It is worth noting that, in Experiment 1, two binary classifiers (i.e., AD vs. HC and MCI vs. HC, respectively) are built. Specifically, for AD vs. HC classification, we ignore the MCI subjects at each cross-validation trial and use only the AD and HC subjects. Similarly, for MCI vs. HC classification, we ignore the AD subjects at each cross-validation trial and use only the MCI and HC subjects. On the other hand, in Experiment 2, only one binary classifier (i.e., MCI-C vs. MCI-NC) is built involving only the MCI subjects. In both experiments, SVM is implemented using LIBSVM toolbox (Chang and Lin, 2001), and linear kernel is used after normalizing each feature vector with unit norm. For all respective methods, the values of the parameters (e.g., λ and βm) are determined by performing another round of cross-validation on the training data. Also, at preprocessing stage, we perform a common feature normalization step, i.e., subtracting the mean and then dividing the standard deviation (of all training subjects) for each feature value.
Results
Experiment 1: Estimating clinical stages (MMSE, ADAS-Cog, and class label)
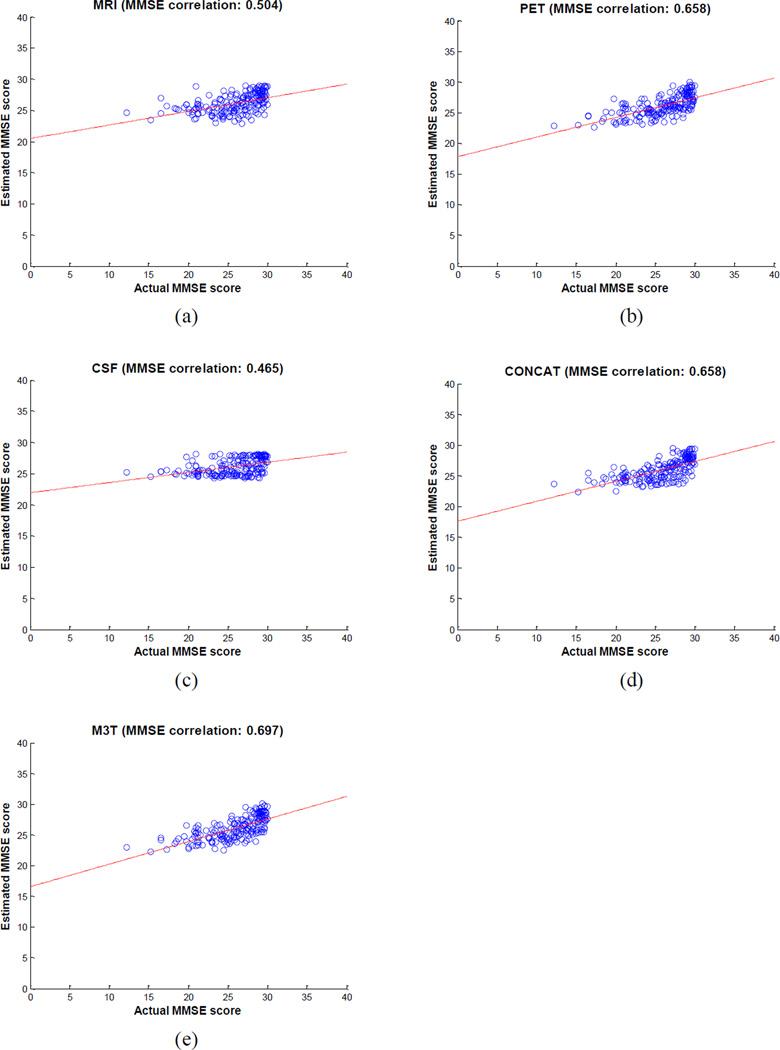
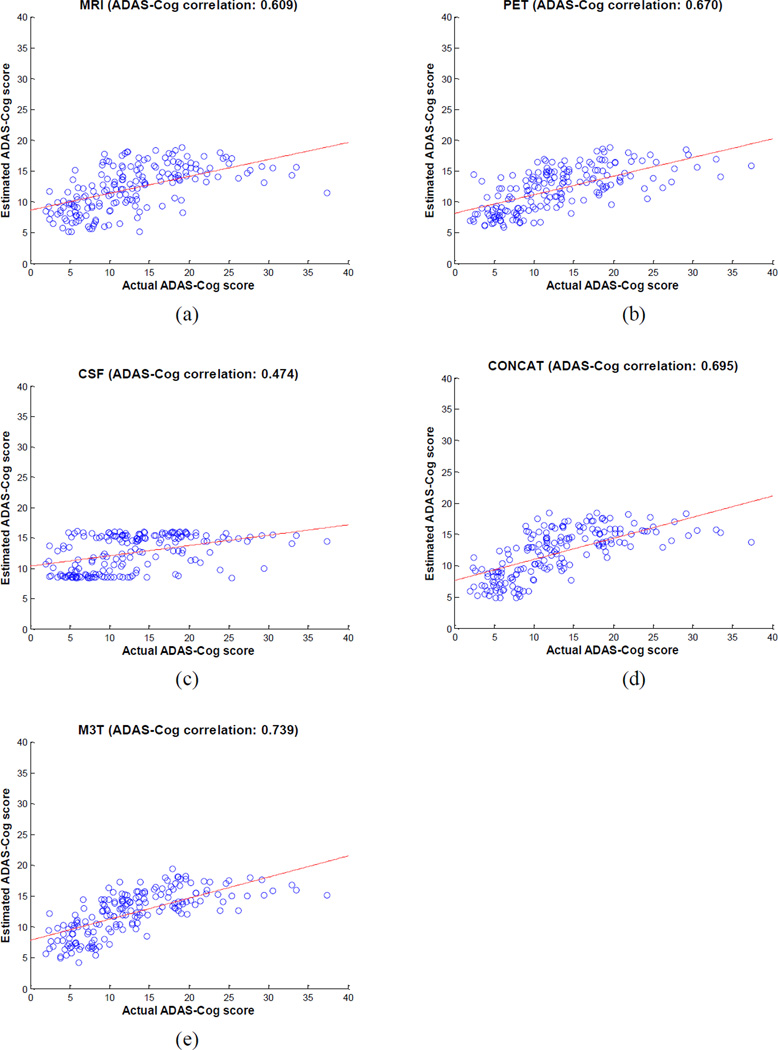
We first estimate the clinical stages, including two regression variables (MMSE and ADAS-Cog) and one classification variable (i.e., class label with a value of ‘AD’, ‘MCI’ or ‘HC’), from the baseline MRI, PET, and CSF data. It is worth noting that the original multi-class classification problem is formulated as two binary classification problems, i.e., AD vs. HC and MCI vs. HC, as mentioned above. Table 2 shows the performances of the proposed M3T method, compared with three methods each using individual modality, as well as the CONCAT method (as detailed below). Specifically, in Table 2, MRI-, PET-, and CSF-based methods denote the classification results using only the respective individual modality of data. For MRI-based and PET-based methods, similarly as our M3T method, they contain two successive steps, i.e., (1) the single-task feature selection method using Lasso (Tibshirani, 1996), and (2) the standard SVM for both regression and classification. For CSF-based method, it uses the original 3 features without any further feature selection, and performs the standard SVM for both regression and classification. Obviously, MRI-, PET- and CST-based methods all belong to the SMST learning as shown in Fig. 1. For comparison, we also implement a simple concatenation method (denoted as CONCAT) for using multi-modal data. In the CONCAT method, we first concatenate 93 features from MRI, 93 features from PET, and 3 features from CSF into a 189 dimensional vector, and then perform the same two steps (i.e., Lasso feature selection and SVM regression/classification) as in MRI-, PET- and CSF-based methods. It is worth noting that the same experimental settings are used in all five methods as compared in Table 2. Figs. 3–4 further show the scatter plots of the estimated scores vs. the actual scores by five different methods for MMSE and ADAS-Cog, respectively.
Table 2.
Comparison of performances of five different methods on Experiment 1. The reported values are the correlation coefficient (for MMSE and ADAS-Cog regression) and accuracy (for AD vs. HC and MCI vs. HC classification), averaged on 10-fold tests (with standard deviation also reported).
| Methods | Correlation coefficient | Classification accuracy | ||
|---|---|---|---|---|
| MMSE | ADAS-Cog | AD vs. HC | MCI vs. HC | |
| MRI-based | 0.504 ± 0.038 | 0.609 ± 0.014 | 0.848 ± 0.026 | 0.739 ± 0.028 |
| PET-based | 0.658 ± 0.027 | 0.670 ± 0.018 | 0.845 ± 0.035 | 0.797 ± 0.023 |
| CSF-based | 0.465 ± 0.019 | 0.474 ± 0.013 | 0.805 ± 0.022 | 0.536 ± 0.044 |
| CONCAT | 0.658 ± 0.023 | 0.695 ± 0.011 | 0.920 ± 0.033 | 0.800 ± 0.024 |
| Proposed M3T | 0.697 ± 0.022 | 0.739 ± 0.012 | 0.933 ± 0.022 | 0.832 ± 0.015 |
AD = Alzheimer’s Disease, HC = Healthy Control, MCI = Mild Cognitive Impairment, MMSE = Mini-Mental State Examination, ADAS-Cog = Alzheimer’s Disease Assessment Scale - Cognitive Subscale
Fig. 3.
Scatter plots of the estimated MMSE scores vs. the actual scores by five different methods
Fig. 4.
Scatter plots of the estimated ADAS-Cog scores vs. the actual scores by five different methods
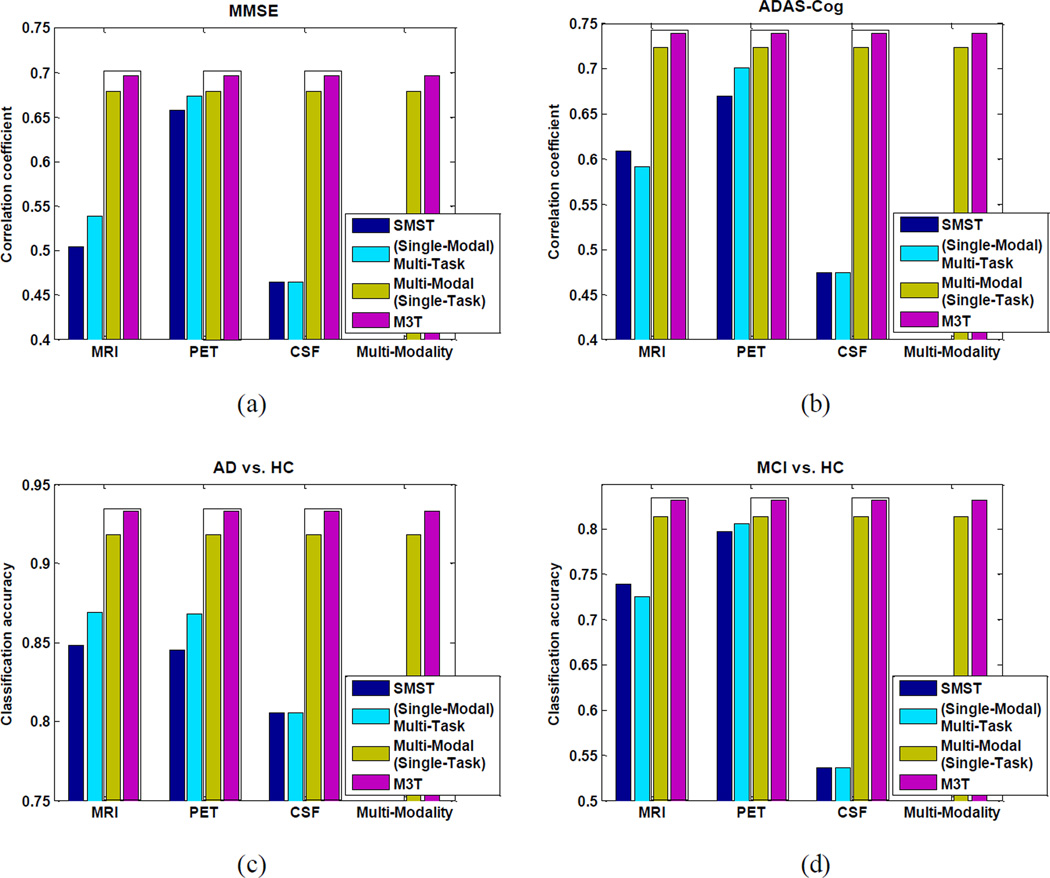
As can be seen from Table 2 and Figs. 3–4, our proposed M3T method consistently achieves better performance than other four methods. Specifically, for estimating MMSE and ADAS-Cog scores, our method achieves the correlation coefficients of 0.697 and 0.739, respectively, while the best performance using individual modality is only 0.658 and 0.670 (when using PET), respectively. On the other hand, for AD vs. HC and MCI vs. HC classification, our method achieves the accuracies of 0.933 and 0.832, respectively, while the best performance using individual modality is only 0.848 (when using MRI) and 0.797 (when using PET), respectively. Table 2 and Figs. 3–4 also indicate that our proposed M3T method consistently outperforms the CONCAT method on each performance measure, although the latter also achieves better performance than three MRI-, PET-, or CSF-based methods in most cases, because of using multimodal imaging data. However, CSF-based method always achieves the worst performances in all tasks, and is significantly inferior to MRI- and PET-based methods in this experiment. Finally, for each group (i.e., AD, MCI or HC), we compute its average estimated clinical scores using M3T, with respective values of 24.8 (AD), 25.5 (MCI) and 28.1 (HC) for MMSE, and 14.9 (AD), 13.3 (MCI) and 8.3 (HC) for ADAS-Cog. These results show certain consistency with the actual clinical scores as shown in Table 1.
We also compare M3T with its two variants, i.e., Multi-Modal (Single-Task) and (Single-Modal) Multi-Task methods as described in Fig. 1. Briefly, in Multi-Modal (Single-Task) method, the single-task feature selection method (Lasso) and the multi-modal SVM (for both regression and classification) are successively performed, while in Multi-Task method, the multi-task feature selection method (MTFS) and the standard SVM (for both regression and classification) are successively performed. In addition, we also contain the SMST methods (MRI-, PET-, or CSF-based) for comparison. Fig. 5 shows the comparison results of those methods on Experiment 1. It is worth noting that, using only CSF, (Single-Modal) Multi-Task method is equivalent to SMST since in this case the original CSF measures are directly used as features without any feature selection step. As can be seen from Fig. 5, our M3T method consistently outperforms the other three methods: SMST, Multi-Modal (Single-Task), and (Single-Modal) Multi-Task. Fig. 5 also indicates that, when performing MMSE regression and AD vs. HC classification using MRI-based or PET-based method, and when performing ADAS-Cog regression and MCI vs. HC classification using PET-based method, (Single-Modal) Multi-Task method, which jointly estimates multiple regression/classification variables, achieves better performance than SMST which estimates each variable separately. On the other hand, Fig. 5 also shows that Multi-Modal (Single-Task) method consistently outperforms SMST on both regression and classification, which validates and further complements the existing conclusion on the advantage of multi-modal classification using MRI, PET, and CSF data (Zhang et al., 2011). Finally, the t-test (at 95% significance level) results between M3T and the second best method, i.e., Multi-Modal (Single-Task) method, show that the former is significantly better than the latter on tasks of estimating ADAS-Cog score and classifying between MCI and HC.
Fig. 5.
Comparison of performances of four different methods on Experiment 1
Experiment 2: Predicting 2-year MMSE and ADAS-Cog changes and MCI conversion
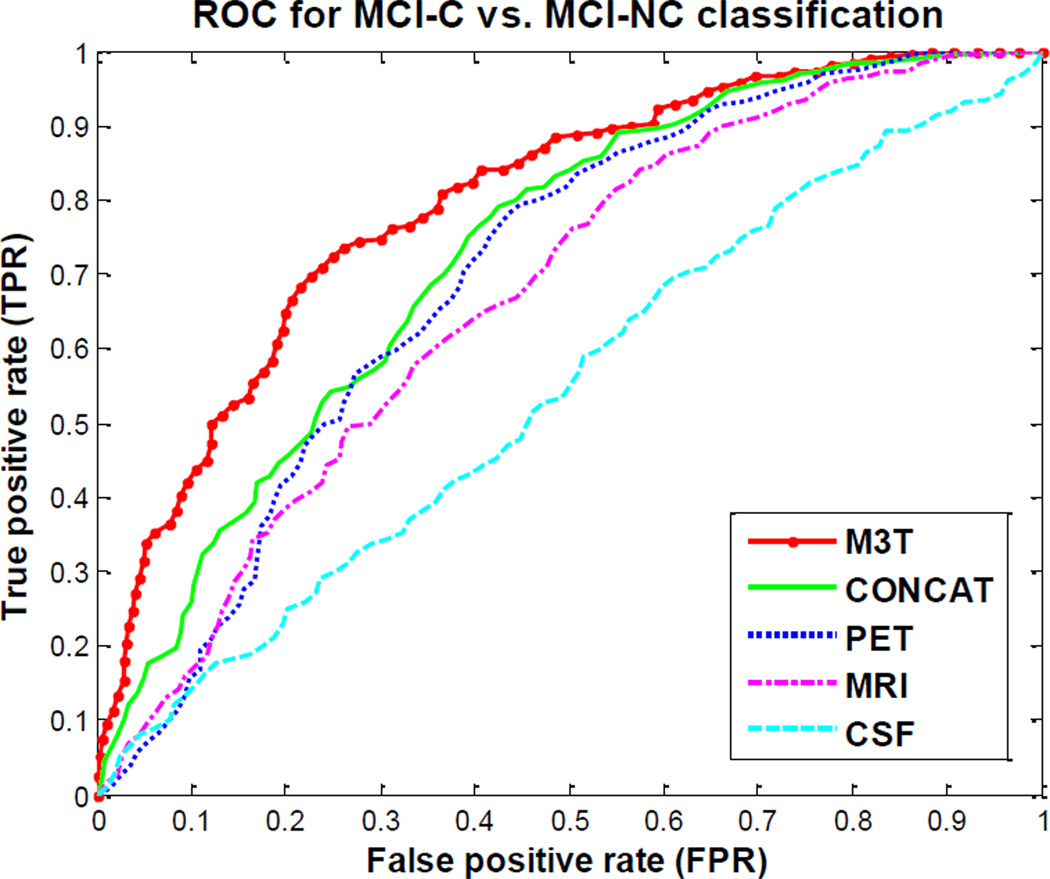
In this experiment, we predict the 2-year changes of MMSE and ADAS-Cog scores and the conversion of MCI to AD, from the baseline MRI, PET, and CSF data. Here, we have two regression tasks corresponding to the prediction of the regression variables of MMSE and ADAS-Cog changes from baseline to 2-year follow-up, respectively, and one classification task corresponding to prediction of the classification variable of MCI conversion to AD, i.e., MCI-C vs. MCI-NC. It is worth noting that as in Experiment 1, only the baseline MRI, PET, and CSF data are used for all prediction tasks. We use the same subjects as in Experiment 1, except for 19 subjects without 2-year MMSE or ADAS-Cog scores, thus reducing to totally 167 subjects with 40 AD, 80 MCI (38 MCI-C and 42 MCI-NC), and 47 HC that are finally used in Experiment 2. Table 3 shows the performance of the proposed M3T method compared with three individual-modality based methods and also the CONCAT method, which are the same methods as those used in Experiment 1. Here, for MCI-C vs. MCI-NC classification, besides reporting the classification accuracy, we also give other performance measures including sensitivity (i.e., the proportion of MCI-C subjects correctly classified) and the specificity (i.e., the proportion of MCI-NC subjects correctly classified). In addition, we also plot the ROC curves of five different methods for classification between MCI-C and MCI-NC, as shown in Fig. 6. Here, the individual-modality based methods (using MRI, CSF or PET) and the CONCAT method are defined in the same way as in Experiment 1.
Table 3.
Comparison of performances of five different methods on Experiment 2. The reported values are the correlation coefficient (for regressions of MMSE and ADAS-Cog change) and accuracy, sensitivity and specificity (for MCI-C vs. MCI-NC classification), averaged on 10-fold tests (with standard deviation also reported).
| Methods | Correlation coefficient | MCI-C vs. MCI-NC | |||
|---|---|---|---|---|---|
| MMSE change | ADAS-Cog change | Accuracy | Sensitivity | Specificity | |
| MRI-based | 0.419 ± 0.019 | 0.455 ± 0.037 | 0.620 ± 0.058 | 0.566 ± 0.069 | 0.602 ± 0.056 |
| PET-based | 0.434 ± 0.027 | 0.401 ± 0.046 | 0.639 ± 0.046 | 0.570 ± 0.067 | 0.623 ± 0.069 |
| CSF-based | 0.327 ± 0.018 | 0.425 ± 0.028 | 0.518 ± 0.086 | 0.454 ± 0.094 | 0.493 ± 0.089 |
| CONCAT | 0.484 ± 0.009 | 0.475 ± 0.045 | 0.654 ± 0.050 | 0.573 ± 0.062 | 0.651 ± 0.064 |
| Proposed M3T | 0.511 ± 0.021 | 0.531 ± 0.032 | 0.739 ± 0.038 | 0.686 ± 0.051 | 0.736 ± 0.045 |
MCI-C = MCI converter, MCI-NC = MCI non-converter, MMSE = Mini-Mental State Examination, ADAS-Cog = Alzheimer’s Disease Assessment Scale - Cognitive Subscale
Fig. 6.
ROC curves of five different methods: MRI-based, PET-based, CSF-based, CONCAT, and M3T methods, for classification between MCI-C and MCI-NC
Table 3 and Fig. 6 show that, as in Experiment 1, M3T also consistently outperform the individual-modality based methods and the CONCAT method, on both regression and classification tasks. Specifically, our method achieves the correlation coefficients of 0.511 and 0.531 and the accuracy of 0.739, for predicting the 2-year changes of MMSE and ADAS-Cog scores and the MCI conversion, respectively, while the best performance of individual-modality based methods are 0.434 (when using PET), 0.455 (when using MRI), and 0.639 (when using PET), respectively. In addition, the area under the ROC curve (AUC) is 0.797 for MCI-C vs. MCI-NC classification with our M3T method, while the best AUC using the individual-modality based method is 0.70 (when using PET) and the AUC of the CONCAT method is 0.729. On the other hand, if comparing Table 3 with Table 2, we can see that there is a significant decline in the corresponding performances. It implies that predicting future MMSE and ADAS-Cog changes and the MCI conversion is much more difficult and challenging than estimating the MMSE and ADAS scores and the class labels.
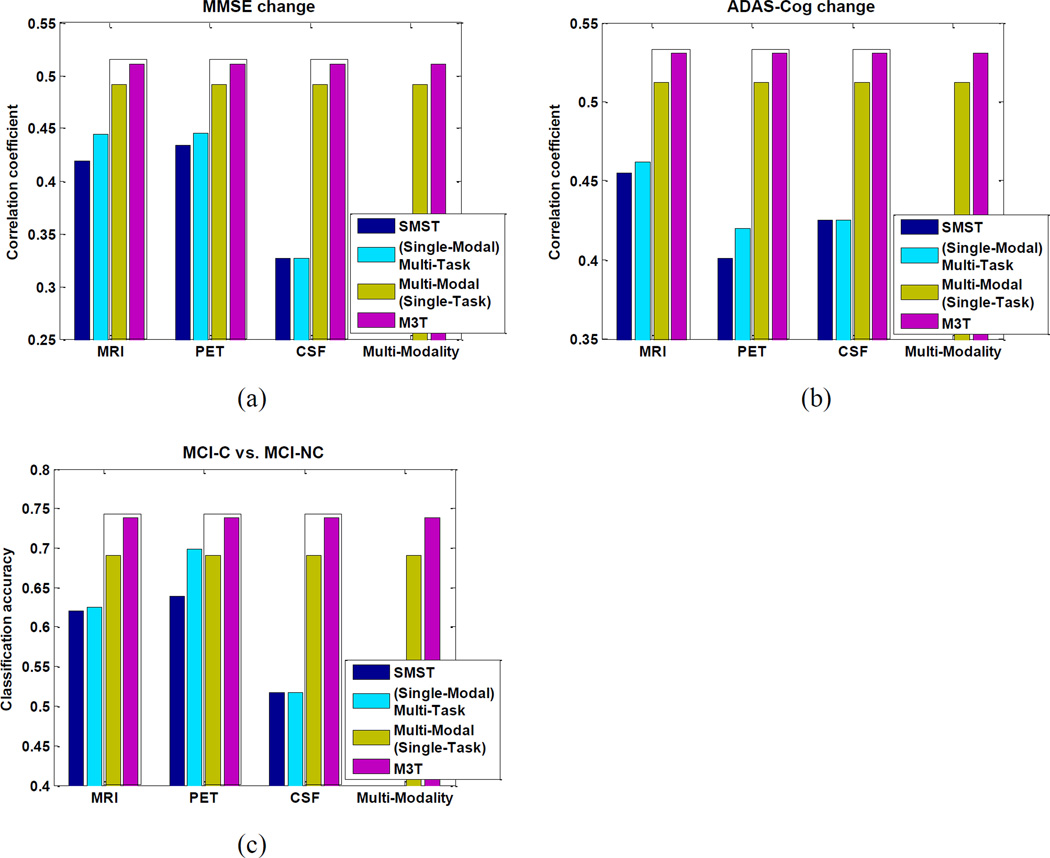
Finally, as in Experiment 1, we compare our M3T method with Multi-Modal (Single-Task), (Single-Modal) Multi-Task, and SMST (including MRI, CSF or PET) methods, with the results shown in Fig. 7. As can be seen from Fig. 7, M3T consistently achieves the best performances among all methods. Fig. 7 also shows that, in all cases except on CSF-based regression/classification (where no feature selection is performed), (Single-Modal) Multi-Task method achieves better performance than the corresponding SMST method. On the other hand, Fig. 7 also indicates that Multi-Modal (Single-Task) method consistently outperforms the corresponding SMST method in both regression and classification tasks. These results, respectively, validate the superiorities of Multi-Modal (Single-Task) and (Single-Modal) Multi-Task methods over SMST method, where both methods improve performance of SMST from different aspects. By fusing Multi-Modal (Single-Task) and (Single-Modal) Multi-Task methods together in a unified framework, M3T further improves the performance. In particular, the t-test (at 95% significance level) results between M3T and the second best method, i.e., Multi-Modal (Single-Task) method, show that the former is significantly better than the latter on tasks of predicting 2-year change of MMSE score and predicting the conversion of MCI to AD.
Fig. 7.
Comparison of performances of four different methods on Experiment 2
Group comparisons of multiple variables
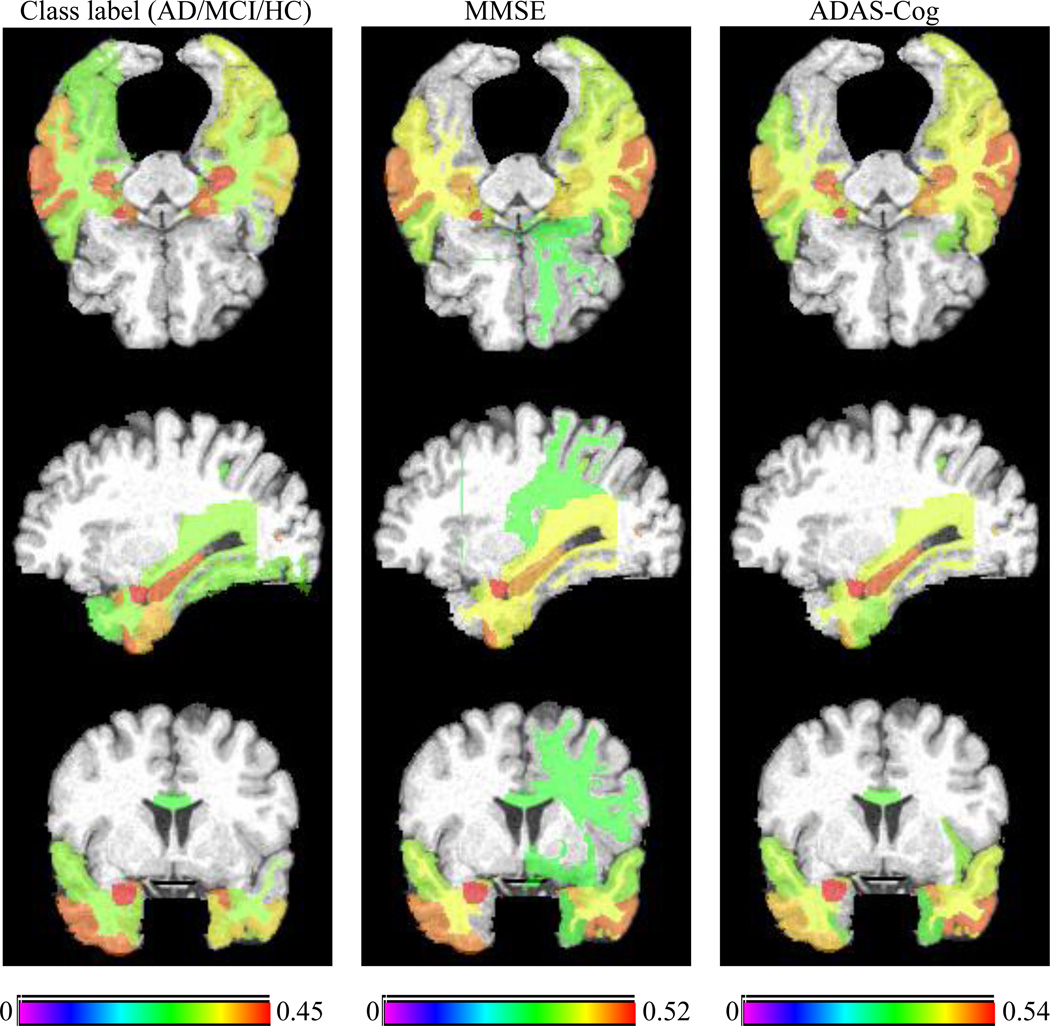
To investigate the relationship between multiple regression and classification variables in Experiment 1 (including MMSE score, ADAS-Cog score, and class label (AD/MCI/HC)) and Experiment 2 (including MMSE change, ADAS-Cog change, and class label (MCI-C/MCI-NC)), we perform group comparisons on them through the computation of the correlation between each brain region and each variable across all subjects. Fig. 8 shows the top 25% brain regions (with their names listed in Supplemental Table 5) that have the highest correlation with class label (AD/MCI/HC), MMSE and ADAS-Cog scores using MRI on Experiment 1, where different color represents correlation coefficient. For comparison, we also list the bottom 25% brain regions with the lowest correlation with class label, MMSE and ADAS-Cog scores in Supplemental Table 7. As can be seen from Fig. 8, the selected brain regions with the highest correlations are very consistent across multiple variables (i.e., class label, MMSE and ADAS-Cog scores). It implies that that there exist inherent correlations among multiple variables, since the underlying pathology is the same. A close observation on Fig. 8 indicates that most of the commonly selected top regions (from multiple variables), e.g., hippocampal, amygdale and uncus regions, are known to be related to the AD by many studies using group comparison methods (Chetelat et al., 2002; Convit et al., 2000; Fox and Schott, 2004; Jack et al., 1999; Misra et al., 2009).
Fig. 8.
Top 25% brain regions with the highest correlation with class label, MMSE, and ADAS-Cog on Experiment 1
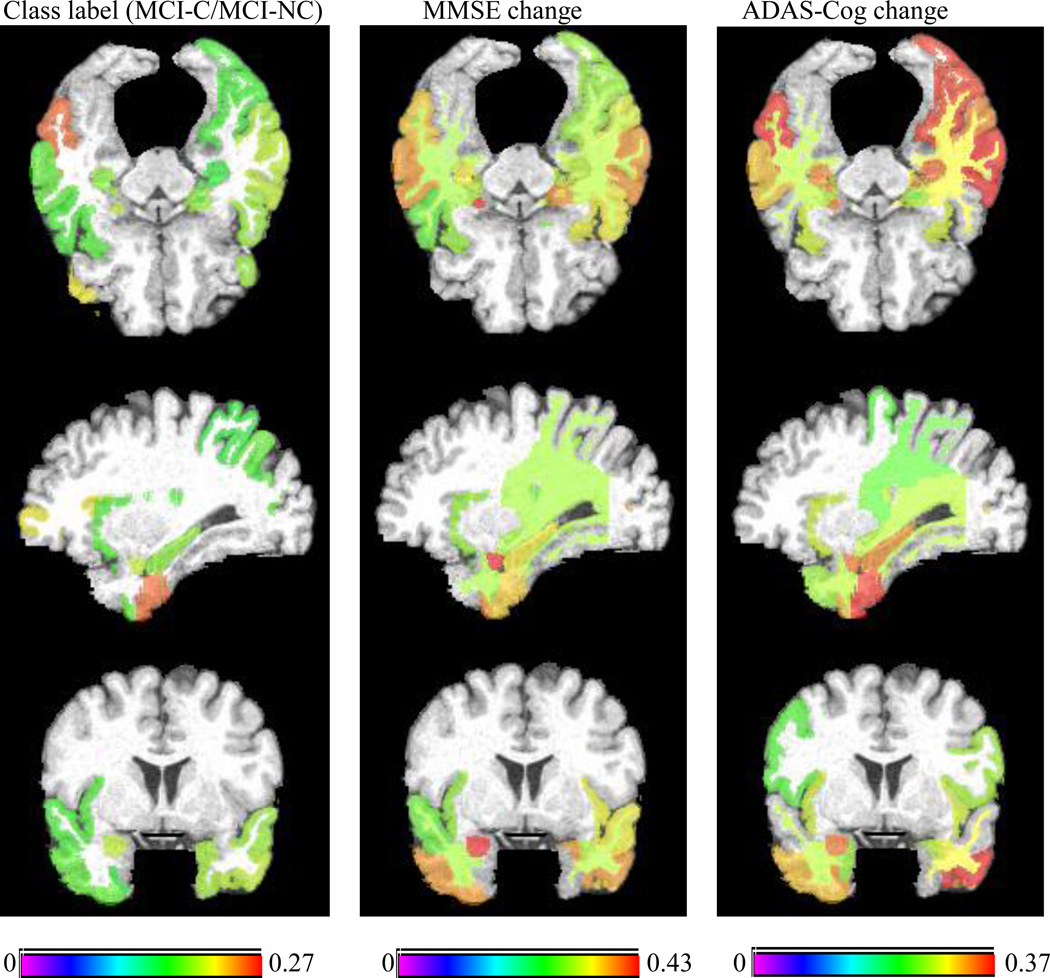
On the other hand, Fig. 9 shows the top 25% brain regions (with their names listed in Supplemental Table 6) that have the highest correlation with class label (MCI-C/MCI-NC), MMSE and ADAS-Cog changes using MRI on Experiment 2, where different color again represents correlation coefficient. For comparison, we also list the bottom 25% brain regions with the lowest correlation with class label, MMSE and ADAS-Cog changes in Supplemental Table 8. Fig. 9 indicates that there still exists consistency between the selected top regions across multiple variables (i.e., class label, MMSE and ADAS-Cog changes), but it is not as apparent as the regions obtained in Experiment 1. This partly explains the fact why the lower performance is achieved in predicting the future changes of MMSE and ADAS-Cog scores and the MCI conversion to AD, compared to estimating the MMSE and ADAS-Cog scores and the class labels, as shown in Tables 2–3. This is because the tasks of predicting the future changes of clinical variables and the conversion of MCI to AD are more challenging than the tasks of estimating clinical variables and class label.
Fig. 9.
Top 25% brain regions with the highest correlation with class label, MMSE change, and ADAS-Cog change on Experiment 2
Discussion
In this paper, we have proposed a new Multi-Modal Multi-Task (M3T) learning method with two successive steps, i.e., multi-task feature selection and multi-modal support vector machine, to jointly predict multiple regression and classification variables from multi-modal data. Our proposed method has been validated on 186 baseline subjects from ADNI through two different sets of experiments. In the first set of experiment, we tested its performance in jointly estimating the MMSE and ADAS-Cog scores and the class label (AD/MCI/HC) of subjects, from the baseline MRI, PET, and CSF data. In the second set of experiment, we tested its performance in jointly predicting the 2-year changes of MMSE and ADAS-Cog scores and the conversion of MCI to AD, also from the baseline MRI, PET, and CSF data.
Multi-task learning
Multi-task learning is a recent machine learning technique, which learns a set of related models for predicting multiple related tasks (Argyriou et al., 2008; Obozinski et al., 2006; Yang et al., 2009). Because multi-task learning uses the commonality among different tasks, it often leads to a better model than learning the individual tasks separately. In multi-task learning, one key issue is how to characterize and use the task relatedness among multiple tasks, with several strategies used in the existing multi-task learning methods: (1) sharing parameters or prior distributions of the hyperparameters of the models across multiple tasks (Bi et al., 2008), and 2) sharing a common underlying representation across multiple tasks (Argyriou et al., 2008; Obozinski et al., 2006; Yang et al., 2009). A few studies have used multi-tasking learning in medical imaging. For example, multi-task learning, which is based on sharing common prior distribution in the parameters of different models, is used to detect different types of clinically related abnormal structures in medical images (Bi et al., 2008).
In another work which is the most related to the current study, a joint Bayesian classifier by sharing the same hyperparameters for model parameters is used to estimate the MMSE and ADAS-Cog scores from the baseline MRI data (Fan et al., 2010). In contrast, the multi-task feature selection method used in this paper belongs to the second scenario, i.e., it assumes that multiple related tasks share a subset of relevant features and thus select them from a large common space of features based on group sparsity constraints. Moreover, besides the regression tasks on estimating clinical scores including MMSE and ADAS-Cog, our method also learns classification tasks in a unified multi-task learning framework. Our experimental results have shown the advantage of jointly estimating multiple regression and classification variables. Also, it is worth noting that, in (Fan et al., 2010), their method achieved the correlation coefficients of 0.569 and 0.522 in estimating MMSE and ADAS-Cog scores, respectively, from the ADNI baseline MRI data of 52 AD, 148 MCI, and 64 HC, which are inferior to our corresponding results in Table 2.
It is worth noting that feature selection (including both MTFS and Lasso) is performed on the training data only. Thus, the selected features at each cross-validation trial may be different. Accordingly, we checked the selected features by MTFS at each cross-validation trial in Experiment 1, and found that the selected features do vary across different cross-validation trials. But we also found that some important features such as hippocampal regions, which are highly relevant to the disease, are always selected in each cross-validation trial.
Multi-modal classification and regression
In recent studies on AD and MCI, it has been shown that biomarkers from different modalities contain complementary information for diagnosis of diseases (Apostolova et al., 2010; de Leon et al., 2007; Fjell et al., 2010; Foster et al., 2007; Landau et al., 2010; Walhovd et al., 2010b), and thus a lot of works on combining different modalities of biomarkers have been reported for multi-modal classification (Bouwman et al., 2007a; Chetelat et al., 2005; Fan et al., 2008; Fellgiebel et al., 2007; Geroldi et al., 2006; Vemuri et al., 2009; Visser et al., 2002; Walhovd et al., 2010a). Typically in those methods, features from all different modalities are concatenated into a longer feature vector for the purpose of multi-modal classification. More recently, multiple-kernel method is used for multi-modal data fusion and classification, and achieves better performance than the baseline feature concatenation method (Hinrichs et al., 2011; Zhang et al., 2011).
On the other hand, compared with the abundant works on multi-modal classification, to the best of our knowledge, there are no previous studies on using multi-modal data for estimating clinical variables, i.e., using multi-modal regression. Instead, nearly all existing works on estimating clinical variables use only the structural MRI. However, as shown in Tables 2–3, in some cases using PET data achieves better performance than using MRI data, and by further combining MRI, PET, and CSF data, the multi-modal regression methods always outperform the individual-modality based methods. Also, our experimental results suggest that, although using only CSF data alone achieves the worst performances in most cases, it can help build powerful multi-modal regression models when combined with MRI and PET data. Similar conclusions have also been drawn in the multi-modal classification (Zhang et al., 2011).
Another general scheme for fusing multi-modal data is the ensemble learning (Zhang et al., 2011) (denoted as ENSEMBLE in this paper), which trains multiple learners for each modality and then aggregates them by majority voting (for classification) or averaging (for regression) at the decision-making level. For comparison, we perform the ENSEMBLE method on Experiment 1, where the ENSEMBLE method achieves the correlation coefficients of 0.677 and 0.727 in estimating MMSE and ADAS-Cog scores, respectively, and the classification accuracies of 0.888 and 0.769 in classifying AD and MCI from HC, respectively. These results are inferior to the corresponding results of M3T in Table 2. Also, similar to regression, we found that the ENSEMBLE method cannot achieve satisfactory results on classification, which implies that the simple majority voting based on the only 3 individual classifiers (from MRI, PET and CSF modalities, respectively) may be not sufficient for achieving a better ensemble classification on this dataset.
Our current model adopts (multi-modal) SVM for both regression and classification. For SVM, a linear kernel is used after normalizing each feature vector with unit norm. The advantage of using linear kernel is that there is no free parameter to be adjusted. In fact, we also tried the Gaussian kernel under different values of the kernel width (i.e., sigma) (see Supplemental Fig. 10). We found that using the linear kernel plus our normalization step can achieve similar performance as using the Gaussian kernel with the best value of the kernel width. Moreover, besides SVM, there also exist other models for regression and classification, e.g., multiple regression, logistic regression, etc (Hastie et al., 2001). However, our experimental results indicate that these models achieve much poorer performance than SVM.
Finally, in this paper, for measuring the performance of different methods in regression tasks, we use the Pearson’s correlation coefficient throughout our experiments. In fact, besides correlation coefficient, there also exist other performance evaluation metrics, e.g., the PRESS statistic which is defined as the sum of squares of the prediction residuals computed under the Leave-One-Out (LOO) strategy. Here, we compare the performance of different regression methods on Experiment 1 using a variant of the PRESS metric, i.e., PRESS RMSE (root mean square prediction error). Specifically, for estimating MMSE score, the PRESS RMSE measures of MRI-based, PET-based, CSF-based, CONCAT and M3T methods are 3.073, 2.669, 3.112, 2.667 and 2.563, respectively. On the other hand, for estimating ADAS-Cog score, the PRESS RMSE measures of MRI-based, PET-based, CSF-based, CONCAT and M3T methods are 6.306, 5.966, 6.851, 5.834 and 5.652, respectively. The above results further show that under the PRESS RMSE metric, our M3T method still achieves the best performance on both regression tasks, followed by CONCAT and PET-based methods, which are consistent to our previous results in Table 2 which uses correlation coefficient as the performance measure.
Prediction of conversion and future decline of MCI
More and more of recent interests in early diagnosis of AD has been moved to identify the MCI subjects who will progress to clinical AD, i.e., MCI converters (MCI-C), from those who remain stable, i.e., MCI non-converters (MCI-NC) (Davatzikos et al., 2010; Leung et al., 2010; Misra et al., 2009). Although our method was not specifically aiming for prediction of MCI to AD, the achieved performances, i.e., an accuracy of 0.739 and an AUC of 0.797 on 38 MCI-C and 42 MCI-NC, are very comparable to the best results reported in several recent studies on ADNI. For example, in (Misra et al., 2009), the accuracy between 0.75 and 0.80 and an AUC of 0.77 were reported on 27 MCI-C and 76 MCI-NC using structural MRI data of ADNI. In (Davatzikos et al., 2010), the maximum accuracy of 0.617 and AUC of 0.734 were reported on 69 MCI-C and 170 MCI-NC using both MRI and CSF data. In (Leung et al., 2010), the maximum AUC of 0.67 was reported on 86 MCI-C and 128 MCI-NC using the hippocampal atrophy rates calculated by the boundary shift integral within ROIs.
On the other hand, a few recent studies also investigated the problem of predicting future cognitively decline of MCI subjects. For example, a study based on group comparison on 85 MCI from ADNI in (Landau et al., 2010) indicated that CSF and PET could predict longitudinal cognitive decline. In (Wang et al., 2010), a Bagging relevant vector machine (RVM) was adopted to predict the future decline of MMSE score from baseline MRI data and a correlation coefficient of 0.537 was achieved on 16 MCI-C, 5 MCI-NC, and 5 AD. In contrast, our method achieved the correlation coefficients of 0.511 and 0.531 on 38 MCI-C and 42 MCI-NC, as well as 40 AD and 47 HC, which are comparable to the result in (Wang et al., 2010). Finally, in (Duchesne et al., 2009), a principal component analysis (PCA) based model is used on MRI data of 49 MCI (including 20 MCI-C and 29 MCI-NC) to predict the one-year change in MMSE score, and a correlation coefficient of 0.31 was reported. This low correlation coefficient result indicated that it is more difficult to predict one-year changes than two-year changes, since the MCI converters (who convert to AD after two years) had not progress to AD completely after one year and thus the corresponding measured cognitive scores did not accurately reflect the underlying pathological changes in brain regions.
Limitations
The current study is limited by several factors as below. First, the proposed method is based on multi-modal data, i.e., MRI, PET, and CSF, and thus requires each subject to have the corresponding modality data, which limits the size of subjects that can be used for study. For example, there are approximately 800 subjects in ADNI database, while there are only around 200 subjects having all baseline MRI, PET, and CSF data. Second, besides MRI, PET, and CSF, there also exist other modalities of data, i.e., APOE. However, since not every subject has this data and the number of subjects with all modality data (including APOE) is too small for reasonable learning, the current study does not consider APOE for multimodal classification and regression. Finally, besides MMSE and ADAS-Cog scores, there exist other clinical scores in ADNI database. However, due to the similar reasons (i.e., not every subject has all clinical scores available), we did not investigate those clinical variables in the current study, although in principle including more related clinical variables is not difficult and would further improve the regression/classification performance.
Conclusion
In summary, our experimental results have showed that our proposed Multi-Modal Multi-Task (M3T) method can effectively perform multiple-tasks learning from multi-modal data. Specifically, it can effectively estimate the MMSE and ADAS-Cog scores and the classification label in both AD vs. HC and MCI vs. HC classifications, and can also predict the 2-year MMSE and ADAS-Cog changes and the classification label in MCI-C vs. MCI-NC classification. To the best of our knowledge, it made the first investigation on jointly predicting multiple regression and classification variables from the baseline multi-modal data. In the future work, we will investigate incomplete multi-modal multi-task learning with missing values in both modalities and tasks, to increase the number of ADNI subjects that can be used for training our method. Moreover, we will develop new models which can iteratively use multi-modal and multi-task information, i.e. using regression/classification results to guide feature selection, for further improving the final performance. This general wrapper-like framework can embrace a series of feature selection method, e.g., SVM-RFE (Guyon et al., 2002) which has been widely used in neuroimaging area. We will extend it for multi-modal multi-task learning and compare with our current model. Finally, it is interesting to investigate the integration of the existing domain knowledge in AD research into our current model, for not only achieving good prediction accuracy but also providing good interpretability in understanding the biology of AD.
Research Highlights.
-
➢
We jointly predict regression and classification variables from multi-modal data
-
➢
Two sets of experiments are performed on baseline MRI, PET, and CSF data from ADNI
-
➢
We first estimate MMSE and ADAS-Cog clinical scores and class label (AD/MCI/HC)
-
➢
We then predict 2-year change of MMSE and ADAS-Cog scores and MCI conversion to AD
-
➢
Our method achieves better performance on both experiments than conventional ones
Supplementary Material
Acknowledgment
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson and Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Novartis AG, Pfizer Inc, F. Hoffman-La Roche, Schering-Plough, Synarc, Inc., as well as non-profit partners the Alzheimer's Association and Alzheimer's Drug Discovery Foundation, with participation from the U.S. Food and Drug Administration. Private sector contributions to ADNI are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (www.loni.ucla.edu/ADNI). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: www.loni.ucla.edu\ADNI\Collaboration\ADNI_Authorship_list.pdf.
References
- Apostolova LG, Hwang KS, Andrawis JP, Green AE, Babakchanian S, Morra JH, Cummings JL, Toga AW, Trojanowski JQ, Shaw LM, Jack CR, Jr, Petersen RC, Aisen PS, Jagust WJ, Koeppe RA, Mathis CA, Weiner MW, Thompson PM. 3D PIB and CSF biomarker associations with hippocampal atrophy in ADNI subjects. Neurobiol Aging. 2010;31:1284–1303. doi: 10.1016/j.neurobiolaging.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyriou A, Evgeniou T, Pontil M. Convex multi-task feature learning. Machine Learning. 2008;73:243–272. [Google Scholar]
- Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Bi J, Xiong T, Yu S, Dundar M, Rao B. An improved multi-task learning approach with applications in medical diagnosis; Proceedings of the 2008 European Conference on Machine Learning and Knowledge Discovery in Databases; 2008. pp. 117–132. [Google Scholar]
- Bouwman FH, Schoonenboom SN, van der Flier WM, van Elk EJ, Kok A, Barkhof F, Blankenstein MA, Scheltens P. CSF biomarkers and medial temporal lobe atrophy predict dementia in mild cognitive impairment. Neurobiol Aging. 2007a;28:1070–1074. doi: 10.1016/j.neurobiolaging.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Bouwman FH, van der Flier WM, Schoonenboom NS, van Elk EJ, Kok A, Rijmen F, Blankenstein MA, Scheltens P. Longitudinal changes of CSF biomarkers in memory clinic patients. Neurology. 2007b;69:1006–1011. doi: 10.1212/01.wnl.0000271375.37131.04. [DOI] [PubMed] [Google Scholar]
- Chang CC, Lin CJ. LIBSVM: a library for support vector machines. 2001 [Google Scholar]
- Chetelat G, Desgranges B, de la Sayette V, Viader F, Eustache F, Baron J-C. Mapping gray matter loss with voxel-based morphometry in mild cognitive impairment. Neuroreport. 2002;13:1939–1943. doi: 10.1097/00001756-200210280-00022. [DOI] [PubMed] [Google Scholar]
- Chetelat G, Eustache F, Viader F, De La Sayette V, Pelerin A, Mezenge F, Hannequin D, Dupuy B, Baron JC, Desgranges B. FDG-PET measurement is more accurate than neuropsychological assessments to predict global cognitive deterioration in patients with mild cognitive impairment. Neurocase. 2005;11:14–25. doi: 10.1080/13554790490896938. [DOI] [PubMed] [Google Scholar]
- Convit A, de Asis J, de Leon MJ, Tarshish CY, De Santi S, Rusinek H. Atrophy of the medial occipitotemporal, inferior, and middle temporal gyri in non-demented elderly predict decline to Alzheimer's disease. Neurobiol Aging. 2000;21:19–26. doi: 10.1016/s0197-4580(99)00107-4. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, Bhatt P, Shaw LM, Batmanghelich KN, Trojanowski JQ. Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiol Aging. 2010 doi: 10.1016/j.neurobiolaging.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leon MJ, Mosconi L, Li J, De Santi S, Yao Y, Tsui WH, Pirraglia E, Rich K, Javier E, Brys M, Glodzik L, Switalski R, Saint Louis LA, Pratico D. Longitudinal CSF isoprostane and MRI atrophy in the progression to AD. J Neurol. 2007;254:1666–1675. doi: 10.1007/s00415-007-0610-z. [DOI] [PubMed] [Google Scholar]
- De Santi S, de Leon MJ, Rusinek H, Convit A, Tarshish CY, Roche A, Tsui WH, Kandil E, Boppana M, Daisley K, Wang GJ, Schlyer D, Fowler J. Hippocampal formation glucose metabolism and volume losses in MCI and AD. Neurobiol Aging. 2001;22:529–539. doi: 10.1016/s0197-4580(01)00230-5. [DOI] [PubMed] [Google Scholar]
- Du AT, Schuff N, Kramer JH, Rosen HJ, Gorno-Tempini ML, Rankin K, Miller BL, Weiner MW. Different regional patterns of cortical thinning in Alzheimer's disease and frontotemporal dementia. Brain. 2007;130:1159–1166. doi: 10.1093/brain/awm016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchesne S, Caroli A, Geroldi C, Collins DL, Frisoni GB. Relating one-year cognitive change in mild cognitive impairment to baseline MRI features. Neuroimage. 2009;47:1363–1370. doi: 10.1016/j.neuroimage.2009.04.023. [DOI] [PubMed] [Google Scholar]
- Duchesne S, Caroli A, Geroldi C, Frisoni G, Collins D. Predicting clinical variable from MRI features: application to MMSE in MCI. MICCAI. 2005:392–399. doi: 10.1007/11566465_49. [DOI] [PubMed] [Google Scholar]
- Fan Y, Kaufer D, Shen D. Joint estimation of multiple clinical variables of neurological diseases from imaging patterns; Proceedings of the 2010 IEEE international conference on Biomedical imaging: from nano to Macro; 2010. [Google Scholar]
- Fan Y, Resnick SM, Wu X, Davatzikos C. Structural and functional biomarkers of prodromal Alzheimer's disease: a high-dimensional pattern classification study. Neuroimage. 2008;41:277–285. doi: 10.1016/j.neuroimage.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellgiebel A, Scheurich A, Bartenstein P, Muller MJ. FDG-PET and CSF phospho-tau for prediction of cognitive decline in mild cognitive impairment. Psychiatry Res. 2007;155:167–171. doi: 10.1016/j.pscychresns.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Fjell AM, Walhovd KB, Fennema-Notestine C, McEvoy LK, Hagler DJ, Holland D, Brewer JB, Dale AM. CSF biomarkers in prediction of cerebral and clinical change in mild cognitive impairment and Alzheimer's disease. J Neurosci. 2010;30:2088–2101. doi: 10.1523/JNEUROSCI.3785-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. "Mini-mental state". A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Foster NL, Heidebrink JL, Clark CM, Jagust WJ, Arnold SE, Barbas NR, DeCarli CS, Turner RS, Koeppe RA, Higdon R, Minoshima S. FDG-PET improves accuracy in distinguishing frontotemporal dementia and Alzheimer's disease. Brain. 2007;130:2616–2635. doi: 10.1093/brain/awm177. [DOI] [PubMed] [Google Scholar]
- Fox N, Schott J. Imaging cerebral atrophy: normal ageing to Alzheimer's disease. Lancet. 2004;363:392–394. doi: 10.1016/S0140-6736(04)15441-X. [DOI] [PubMed] [Google Scholar]
- Franke K, Ziegler G, Kloppel S, Gaser C. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: exploring the influence of various parameters. Neuroimage. 2010;50:883–892. doi: 10.1016/j.neuroimage.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Geroldi C, Rossi R, Calvagna C, Testa C, Bresciani L, Binetti G, Zanetti O, Frisoni GB. Medial temporal atrophy but not memory deficit predicts progression to dementia in patients with mild cognitive impairment. Journal of Neurology Neurosurgery and Psychiatry. 2006;77:1219–1222. doi: 10.1136/jnnp.2005.082651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Machine Learning. 2002;46:389–422. [Google Scholar]
- Hastie T, Tibshirani R, Friedman J. The elements of statistical learning: Data mining, inference and prediction. New York: Springer-Verlag; 2001. [Google Scholar]
- Hinrichs C, Singh V, Xu G, Johnson SC. Predictive markers for AD in a multi-modality framework: an analysis of MCI progression in the ADNI population. Neuroimage. 2011;55:574–589. doi: 10.1016/j.neuroimage.2010.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Petersen RC, Y.C X, O'Brien PC, Smith GE, Ivnik RJ, Boeve BF, Waring SC, Tangalos E, Kokmen E. Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment. Neurology. 1999;52:1397–1403. doi: 10.1212/wnl.52.7.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani N, MacDonald D, Holmes CJ, Evans A. A 3D atlas of the human brain. Neuroimage. 1998;7:S717. [Google Scholar]
- Landau SM, Harvey D, Madison CM, Reiman EM, Foster NL, Aisen PS, Petersen RC, Shaw LM, Trojanowski JQ, Jack CR, Jr, Weiner MW, Jagust WJ. Comparing predictors of conversion and decline in mild cognitive impairment. Neurology. 2010;75:230–238. doi: 10.1212/WNL.0b013e3181e8e8b8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KK, Shen KK, Barnes J, Ridgway GR, Clarkson MJ, Fripp J, Salvado O, Meriaudeau F, Fox NC, Bourgeat P, Ourselin S. Increasing power to predict mild cognitive impairment conversion to Alzheimer's disease using hippocampal atrophy rate and statistical shape models. Med Image Comput Comput Assist Interv. 2010;13:125–132. doi: 10.1007/978-3-642-15745-5_16. [DOI] [PubMed] [Google Scholar]
- Liu J, Ji S, Ye J. SLEP: Sparse learning with efficient projections. Arizona State University; 2009. [Google Scholar]
- Mattsson N, Zetterberg H, Hansson O, Andreasen N, Parnetti L, Jonsson M, Herukka SK, van der Flier WM, Blankenstein MA, Ewers M, Rich K, Kaiser E, Verbeek M, Tsolaki M, Mulugeta E, Rosen E, Aarsland D, Visser PJ, Schroder J, Marcusson J, de Leon M, Hampel H, Scheltens P, Pirttila T, Wallin A, Jonhagen ME, Minthon L, Winblad B, Blennow K. CSF biomarkers and incipient Alzheimer disease in patients with mild cognitive impairment. Jama. 2009;302:385–393. doi: 10.1001/jama.2009.1064. [DOI] [PubMed] [Google Scholar]
- McEvoy LK, Fennema-Notestine C, Roddey JC, Hagler DJ, Jr, Holland D, Karow DS, Pung CJ, Brewer JB, Dale AM. Alzheimer disease: quantitative structural neuroimaging for detection and prediction of clinical and structural changes in mild cognitive impairment. Radiology. 2009;251:195–205. doi: 10.1148/radiol.2511080924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra C, Fan Y, Davatzikos C. Baseline and longitudinal patterns of brain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: results from ADNI. Neuroimage. 2009;44:1415–1422. doi: 10.1016/j.neuroimage.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC, Storandt M, Miller JP, McKeel DW, Price JL, Rubin EH, Berg L. Mild Cognitive Impairment Represents Early-Stage Alzheimer Disease. Archives of Neurology. 2001;58:397–405. doi: 10.1001/archneur.58.3.397. [DOI] [PubMed] [Google Scholar]
- Obozinski G, Taskar B, Jordan MI. Technical report. UC Berkeley: Statistics Department; 2006. Multi-task feature selection. [Google Scholar]
- Ron B, Elizabeth J, Kathryn Z-G, Arrighi HM. Forecasting the global burden of Alzheimer's disease. Alzheimer's & dementia : the journal of the Alzheimer's Association. 2007;3:186–191. doi: 10.1016/j.jalz.2007.04.381. [DOI] [PubMed] [Google Scholar]
- Rosen WG, Mohs RC, Davis KL. A new rating scale for Alzheimer's disease. Am J Psychiatry. 1984;141:1356–1364. doi: 10.1176/ajp.141.11.1356. [DOI] [PubMed] [Google Scholar]
- Shattuck DW, Sandor-Leahy SR, Schaper KA, Rottenberg DA, Leahy RM. Magnetic resonance image tissue classification using a partial volume model. Neuroimage. 2001;13:856–876. doi: 10.1006/nimg.2000.0730. [DOI] [PubMed] [Google Scholar]
- Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, Blennow K, Soares H, Simon A, Lewczuk P, Dean R, Siemers E, Potter W, Lee VM, Trojanowski JQ. Cerebrospinal fluid biomarker signature in Alzheimer's disease neuroimaging initiative subjects. Ann Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D, Davatzikos C. HAMMER: Hierarchical attribute matching mechanism for elastic registration. IEEE Trans Med Imaging. 2002;21:1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonnington CM, Chu C, Kloppel S, Jack CR, Jr, Ashburner J, Frackowiak RS. Predicting clinical scores from magnetic resonance scans in Alzheimer's disease. Neuroimage. 2010;51:1405–1413. doi: 10.1016/j.neuroimage.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society Series B. 1996;58:267–288. [Google Scholar]
- Vemuri P, Wiste HJ, Weigand SD, Shaw LM, Trojanowski JQ, Weiner MW, Knopman DS, Petersen RC, Jack CR., Jr MRI and CSF biomarkers in normal, MCI, and AD subjects: predicting future clinical change. Neurology. 2009;73:294–301. doi: 10.1212/WNL.0b013e3181af79fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser PJ, Verhey FRJ, Hofman PA, Scheltens P, Jolles J. Medial temporal lobe atrophy predicts Alzheimer's disease in patients with minor cognitive impairment. Journal of Neurology, Neurosurgery, and Psychiatry. 2002;72:491–497. doi: 10.1136/jnnp.72.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhovd KB, Fjell AM, Brewer J, McEvoy LK, Fennema-Notestine C, Hagler DJ, Jr, Jennings RG, Karow D, Dale AM. Combining MR imaging, positron-emission tomography, and CSF biomarkers in the diagnosis and prognosis of Alzheimer disease. AJNR Am J Neuroradiol. 2010a;31:347–354. doi: 10.3174/ajnr.A1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhovd KB, Fjell AM, Dale AM, McEvoy LK, Brewer J, Karow DS, Salmon DP, Fennema-Notestine C. Multi-modal imaging predicts memory performance in normal aging and cognitive decline. Neurobiol Aging. 2010b;31:1107–1121. doi: 10.1016/j.neurobiolaging.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Fan Y, Bhatt P, Davatzikos C. High-dimensional pattern regression using machine learning: from medical images to continuous clinical variables. Neuroimage. 2010;50:1519–1535. doi: 10.1016/j.neuroimage.2009.12.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Kim S, Xing EP. Heterogeneous multitask learning with joint sparsity constraints. Advances in Neural Information Processing Systems. 2009 [Google Scholar]
- Zhang D, Wang Y, Zhou L, Yuan H, Shen D. Multimodal classification of Alzheimer's disease and mild cognitive impairment. Neuroimage. 2011;55:856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation maximization algorithm. IEEE Trans Med Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.