Abstract
The current study tests a hypothesis that nuclear receptor signaling is altered in chronic hepatitis C patients and that the altered pattern is alcohol drinking history specific. The expression of a panel of more than 100 genes encoding nuclear receptors, co-regulators, and their direct/indirect targets was studied in human livers. (1) Gene expression pattern was compared between 15 normal donor livers and 23 HCV genotype 1 positive livers from patients without a drinking history (age, gender, and BMI matched). HCV infection increased the expression of nuclear receptors small heterodimer partner and constitutive androstane receptor (CAR) as well as genes involved in fatty acid trafficking, bile acid synthesis and uptake, and inflammatory response. However, the expression of retinoid x receptor (RXR) α, peroxisomal proliferator activated receptor (PPAR) α and β as well as SREBP-1c was decreased in HCV-infected livers. (2) Gene expression pattern was compared in chronic hepatitis C patients with (21) and without (13) a drinking history. Alcohol drinking increased the expression of genes involved in fatty acid uptake, trafficking, and oxidation, but decreased the expression of genes responsible for gluconeogenesis. These changes were consistent with reduced fasting plasma glucose levels and altered expression of upstream regulators that include RXRα, PPARα, and CAR. (3) The mRNA levels of fibroblast growth factor 21, IL-10, and fatty acid synthase, which are all regulated by nuclear receptors, showed independent correlation with hepatic HCV RNA levels. Our findings suggest that those genes and pathways that showed altered expression could potentially be therapeutic targets for HCV infection and/or alcohol drinking-induced liver injury.
Keywords: Hepatitis C virus, lipid, alcohol, peroxisome proliferator activated receptor, small heterodimer partner
Introduction
Hepatitis C is the principal cause of death from liver disease and the leading indication for liver transplantation in the U.S. (1). Advances have been made in anti-viral treatment with the combination of pegylated interferon and ribavirin, but less than half of the patients infected with genotype 1 achieve sustained virological response (SVR) (2-4). With the recent development of HCV protease inhibitors (telaprevir and boceprevir), only about 70 percent of the treatment naïve patients and half of the patients who failed standard treatment achieve SVR (5-8). There is an urgent need to understand the virus-host interaction in order to develop novel intervention strategies.
An intriguing feature of HCV infection is its relationship with lipids: (1) HCV virions circulate in serum bound to lipoproteins, called lipoviroparticles (9). (2) Steatosis is prevalent in HCV infected patients (10, 11). (3) Lipids are essential for the HCV life cycle and the virus was named a “metabolovirus” (12, 13). Nuclear receptors, which are transcriptional factors, play pivotal roles in lipid homeostasis. In addition, nuclear receptors also play important roles in regulating inflammatory response and fibrogenesis (13-15). HCV infection is associated with changes in nuclear receptor-mediated signaling. However, because various in vitro and animal models were used for most of the studies, inconsistent findings were obtained (13, 15, 16). The goal of the current study is to use human livers to test a hypothesis that HCV infection is associated with alteration of hepatic nuclear receptor-mediated pathways, which may in turn contribute to viral replication and the pathological process.
At least moderate alcohol consumption is found in two-thirds of patients with chronic hepatitis C and only half of them stop alcohol drinking upon counseling and initiation of hepatitis C treatment (EASL Clinical Practice Guidelines, 2011). Heavy alcohol intake is associated with an accelerated fibrosis progression, a higher incidence of cirrhosis and hepatocellular carcinoma (HCC) as well as a lower rate of SVR (17, 18). Nuclear receptor-mediated pathways not only play a role in alcohol detoxification, but also contribute to alcohol-induced liver pathogenesis in animal models (19, 20). Thus, another goal of this study is to identify biomarkers for alcohol drinking in HCV-infected patients.
In current study, differential expression genes encoding hepatic nuclear receptors, co-regulators, and their downstream targets were identified in hepatitis C patients and normal liver donors. The differential gene expressions were also examined in chronic hepatitis C patients with and without a history of alcohol drinking. Our data showed that the expression of many studied genes correlated with hepatic HCV RNA level. Our findings suggest that nuclear receptor-mediated pathways play an important role in HCV replication and pathogenesis and thus can be potential therapeutic targets to control the disease process.
Patients and Methods
Studied Populations
Forty-four liver specimens were obtained from the KU Liver Center Tissue Bank (http://www.kumc.edu/livercenter/liver_tissue_bank.html). All the specimens deposited received proper patient consent with an IRB approved protocol. These specimens were from patients with genotype 1 HCV infection. Inclusion criteria were as follows: patients older than 18 years, positive for both anti-HCV antibody (Abbott ARCHITECT anti-HCV test) and serum HCV RNA (Roche Cobas Ampliprep). The following patients were excluded: positive for hepatitis B virus surface antigen; primary biliary cirrhosis, autoimmune hepatitis, Wilson's disease, hemochromatosis, co-infection with human immunodeficiency virus; treatment with antiviral or immunosuppressive agents within 6 months of when liver tissues were obtained. Among the 44 HCV-infected patients, 23 patients were not current or former alcohol users (group A). Thirteen people in group A were male (group A1). Twenty-one out of 44 HCV-infected patients were either current or former alcohol drinkers and they were all male (group B). Group A1 and B are gender-, age-, and BMI-matched (Table 1). Fifteen liver specimens obtained from the donors at the Liver Transplant Program at the University of Kansas Hospital were used as normal controls (group C). Groups A and C were age-, gender-, and BMI-matched.
Table 1. Patient characteristics.
| Group | C (Normal livers from donors) | A (HCV patients without a reported history of drinking) | A1 (male) (HCV patients without a reported history of drinking) | B (male) (HCV patients with a reported history of drinking) |
|---|---|---|---|---|
| N | 15 | 23 | 13 | 21 |
| Age | 47.6 (31-78) | 52.3 (33-69) | 52.3 (41-69) | 50.2 (32-68) |
| Gender (M/F) | 7/8 | 13/10 | 13/0 | 21/0 |
| BMI | 26.9 ± 6.38 | 28.5 ± 5.0 | 27.2 ± 4.2 | 26.8 ± 4.1 |
| Ethnicity (Caucasian/Hispanic | 11/1/3 | 13/2/8 | 8/2/3 | 16/2/3 |
| HCV RNA level (log10) | 6.3 ± 0.86 | 6.3 ± 0.99 | 6.2 ± 0.55 | |
| Platelet count (×109/L) | 136.3 ± 68.1 | 128.2 ± 59.0 | 172.5 ± 77.8 | |
| AST | 196.2 ± 363.9 | 285.9 ± 468.2 | 124.2 ± 196.3 | |
| ALT | 147.3 ± 238.3 | 218.6 ± 299.8 | 120.2 ± 135.5 | |
| ALP | 90.0 ± 66.3 | 71.1 ± 38.3 | 75.3 ± 31.2 | |
| TBILI | 2.30 ± 3.20 | 1.48 ± 1.38 | 3.70 ± 9.40 | |
| GLU | 131.3 ± 49.7 | 125.6 ± 36.9# | 94.4 ± 35.1# | |
| TRIG | 117.8 ± 66.8 (N=13)* | 129.0 ± 80.5 (n=5)* | 106.0 ± 73.9 (N=11)* | |
| CHOL | 150.2 ± 28.2 (N=13)* | 134.0 ± 19.6 (n=5)* | 143.6 ± 54.1 (N=11)* | |
| Histological activity (≤4/>5) | 10/6** | 7/2** | 8/9** | |
| Fibrosis (≤2/3-5/6) | 10/4/9 | 6/3/4 | 10/6/5 | |
| Steatosis (0-4%/5-33%/>33%) | 15/8/0 | 7/5/1 | 10/9/2 |
the data of other cases are not available.
cirrhotic patients are not required to report histological activity.
p <0.05, comparison between group A1 and group B.
Laboratory Tests
Serum aspartate aminotransferase (AST), alanine aminotransferase (ALT), total bilirubin (TBILI), alkaline phosphatase (ALP), total cholesterol (CHOL), triglyceride (TRIG), and fasting plasma glucose were obtained from patients' charts and all the tests were performed within 3 months of liver biopsy. The HCV genotype was determined by sequencing using the TRUGENE HCV 5'NC Genotyping Kit.
Pathological Examination
Hematoxylin and eosin as well as Masson's trichrome stained liver sections were used for diagnosis by the pathologists. The degrees of inflammation and fibrosis were evaluated according to the criteria proposed by Ishak K et al. (21). Steatosis was graded based on percentage of hepatocytes involved: none (<5%), mild (5-33%), moderate (>33-66%), or severe (>66%).
Hepatic mRNA quantification
Hepatic RNA was extracted for studying of gene expression by real-time PCR. The studied genes are listed in Table S1. Data were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA level.
Statistical Analysis
The student's t-test was used for gene expression comparisons between two groups. For the correlation analysis, Δ Ct values were used. Pearson correlation analysis was used to study the correlation between gene expression and hepatic HCV RNA (according to the Kolmogorov Smirnov Z test, the Δ Ct data are within normal distribution). Multivariate linear regression analysis was used to identify the independent correlations for genes which had significant correlation as identified by bivariate correlation analysis. P<0.05 was considered statistically significant.
Results
Characteristics of Studied Patients
Demographical information and clinical data of the 44 studied patients and 15 liver donors are summarized in Table 1. Except for fasting plasma glucose level, which was reduced in patients with a drinking history, other parameters were not different between Group A1 and B or between Group C and A. Most patients in Group B had a heavy drinking history and were binge drinkers. Only 28.6% patients reported that they were current drinkers (Table 2).
Table 2. Alcohol consumption profile in group B (n = 21 males with HCV infection).
| Years of alcohol consumption | Range: 3-46 years 28.00 ±11.64 |
|---|---|
| Drinking onset age | 17.57 ± 3.57 |
| Current/noncurrent drinker | 6/15 (28.6% current) |
| Average drinks per day (light/moderate/heavy drinking#) | 3/10/8 |
| History of heavy drinking# | 21 (90.47%) |
| Binge drinking$ history | 14 (66.70%) |
Heavy drinking: 3+ drinks on most days or every day in a week; Moderate drinking: 2 drinks on most days or every day in a week; Light drinking: No more than 1 drink on most days or every day in a week.
consumption of 5 or more drinks during a single occasion for men or 4 or more drinks during a single occasion for women.
Gene signatures for HCV Infection
Among studied 17 nuclear receptors and 5 co-regulators (Table S1), nuclear receptors retinoid x receptor, RXRα; peroxisome proliferator activated receptor α and β, PPARα and β; small heterodimer partner, SHP; and constitutive androstane receptor, CAR as well as co-regulator (nuclear receptor coactivator 3, NCOA3) showed difference at their mRNA levels (Fig. 1A). SHP, CAR, and NCOA3 mRNA levels were increased in HCV-infected patients in comparison with normal controls. Hepatic RXRα, PPARα, and PPARβ mRNA levels were decreased in HCV-infected patients compared with normal controls. There was no change in the expression levels of other nuclear receptors including FXR (Fig. S1A).
Figure 1.
The expressions of nuclear receptors and their downstream target genes were studied in 23 hepatitis C patients' livers and 15 donor livers. (A) Nuclear receptor and co-regulator genes. (B) Genes that regulate lipid homeostasis. (C) Genes that regulate bile acid synthesis and uptake. (D) Gene expression in inflammatory pathways. Mean ± SEM are shown. *: p<0.05 in comparisons between two groups. NL: normal livers; HC: hepatitis C.
The expressions of genes (Table S1) that play pivotal roles in regulating lipid homeostasis were studied. The mRNA level of SREBP-1c was decreased in HCV-infected livers (Fig. 1B). The expression levels of SREBP-1c target genes (fatty acid synthase, FAS; acyl-coA carboxylase, ACC; Fatty acyl-CoA elongase, FAE) were modestly reduced in HCV-infected livers, but did not reach statistical significance (Fig. S1B). Genes responsible for fatty acid uptake (fatty acid transport protein 2 and 5, FATP2 and FATP5) and glucose uptake (facilitated glucose transporter 2, GLUT2) were up-regulated in HCV-infected livers (Fig. 1B). The expression of low-density lipoprotein receptor (LDLR), which plays a key role for viral entry to the hepatocyte (22), was decreased by 4-fold in HCV-infected patients (Fig. 1B). In addition, the mRNA levels of microsomal triglyceride transfer protein (MTP), which is essential for very low-density lipoprotein (VLDL) and HCV secretion from the infected cells (23), was increased in HCV-infected livers (Fig. 1B).
Cytochrome P450, family 7, subfamily A, polypeptide 1 (CYP7A1) catalyzes a rate-limiting step in cholesterol catabolism and bile acid biosynthesis. Na+/taurocholate cotransporter (NTCP) controls the uptake of bile acid. The expression levels of both genes were increased in HCV-infected livers (Fig. 1C). The mRNA level of CYP7A1 increased more than thirteen fold. CYP7A1 and NTCP are negatively regulated by SHP. However, HCV-infected patients have increased CYP7A1 and NTCP as well as SHP mRNA levels.
Inflammatory pathway gene expression was altered in HCV-infected livers (Fig. 1D). HCV-infected patients had increased expression of tumor necrosis factor α (TNF, 3 fold) and nitric oxide synthase 2 (NOS2, 7 fold). The expression of genes in the fatty acid oxidation pathway and antioxidant system showed no significant difference (Fig. S1C, D).
Biomarkers for Alcohol Drinking in HCV-infected Men
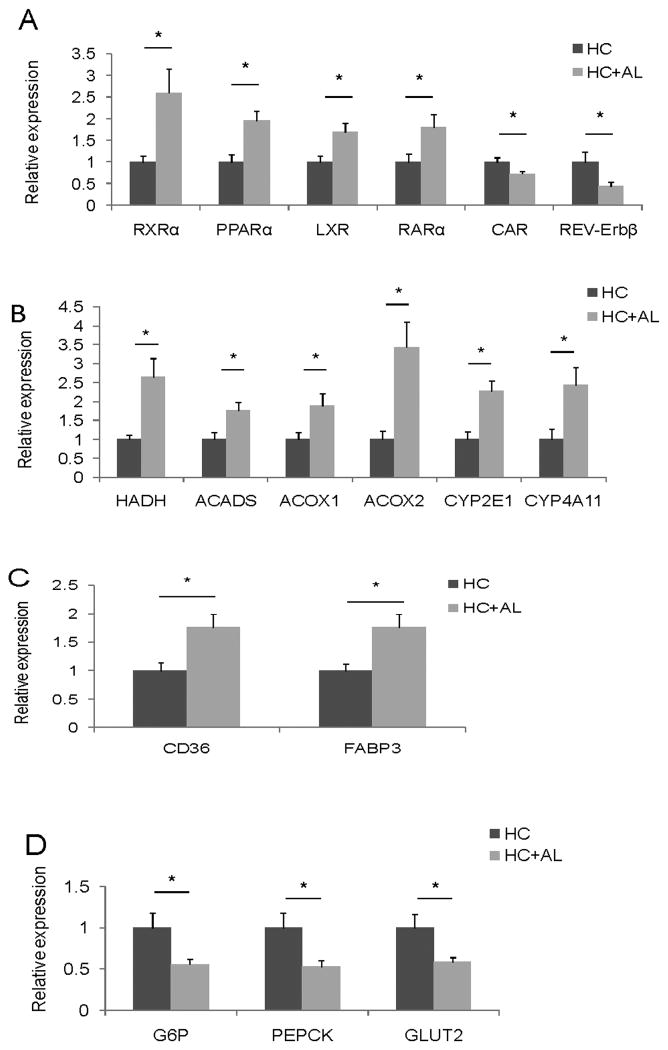
There is a gender difference in alcohol intake and alcoholic liver disease; female gender is a protective factor for drinking and yet females are more susceptible than males to develop alcoholic liver disease (19). It is essential to use gender-matched populations to study biomarkers for alcohol use. Thus, we compared gene expression patterns between HCV-infected men with and without an alcohol drinking history. In patients who had a history of drinking, hepatic RXRα and PPARα mRNA levels were increased whereas CAR mRNA level was decreased in comparison with those who did not have such a history (Fig. 2A). RXRα mRNA levels increased more than 2.5 fold implying the importance of retinoid signaling as a response to alcohol drinking. In addition, liver X receptor (LXR), retinoic acid receptor (RARα), and nuclear receptor subfamily 1, group D, member 2 (Rev-Erbβ) mRNA levels were different between these two cohorts (Fig. 2A). LXR plays a key role in fatty acid synthesis and regulates the expression of SREBP-1c (24, 25). Rev-Erbβ negatively regulates the expression of CD36, fatty acid binding protein 3 and 4 (FABP3 and FABP4), uncoupling protein 3, SREBP-1c, and stearyl-coA dehydrogenase (SCD-1) (26). The decreased Rev-Erbβ is consistent with the up-regulation of CD36 and FABP3 (Fig. 2C). Nuclear receptor corepressor 2 (NCOR2), and NCOA3 mRNA levels were significantly different between the two groups. Patients who had a drinking history had decreased NCOR2 and NCOA3 mRNA levels (Fig. S2A).
Figure 2.
The expression level of nuclear receptors and their downstream target genes were studied in the livers of male hepatitis C patients with (n=21) and without (n=13) an alcohol drinking history. (A) Nuclear receptor genes. (B) Genes in fatty acid oxidation pathway. (C) Genes in fatty acid uptake and intracellular trafficking pathways. (D) Genes in glucose uptake and gluconeogenesis pathways. HC: chronic hepatitis C patients without a history of drinking. HC+AL: chronic hepatitis C patients with a drinking history. Mean ± SEM are shown.*: p<0.05 in comparisons between HC and HC+AL.
Consistent with the changes in RXRα and PPARα, the expression levels of genes related to fatty acid oxidation were increased in alcoholic patients (Fig. 2 B). These up-regulated genes are involved in the mitochondrial β oxidation pathway (hydroxyacyl-CoA dehydrogenase, HADHα; acyl-CoA dehydrogenase, ACADS), peroxisomal oxidation pathway (acyl-CoA oxidase 1 and 2, ACO× and 2), and microsomal oxidation pathway (CYP2E1 and CYP4A11). Intriguingly, gene expression in the anti-oxidant and inflammatory systems did not change significantly (Fig. S2B).
In the fatty acid uptake and intracellular trafficking pathway, CD36 and FABP3 mRNA levels were increased in patients who had a history of drinking (Fig. 2C). There was no change in the expression of genes that are involved in the fatty acid synthesis or VLDL secretion pathways (Fig. S2C, D, E). In the hepatic gluconeogenesis pathway, both glucose-6-phosphatase (G6P) and phosphoenolpyruvate carboxykinase (PEPCK) mRNA levels were reduced in alcoholic patients (Fig. 2 D). These changes along with the reduction of GLUT2 mRNA level are consistent with reduced plasma glucose level found in alcoholic patients (Fig. S3).
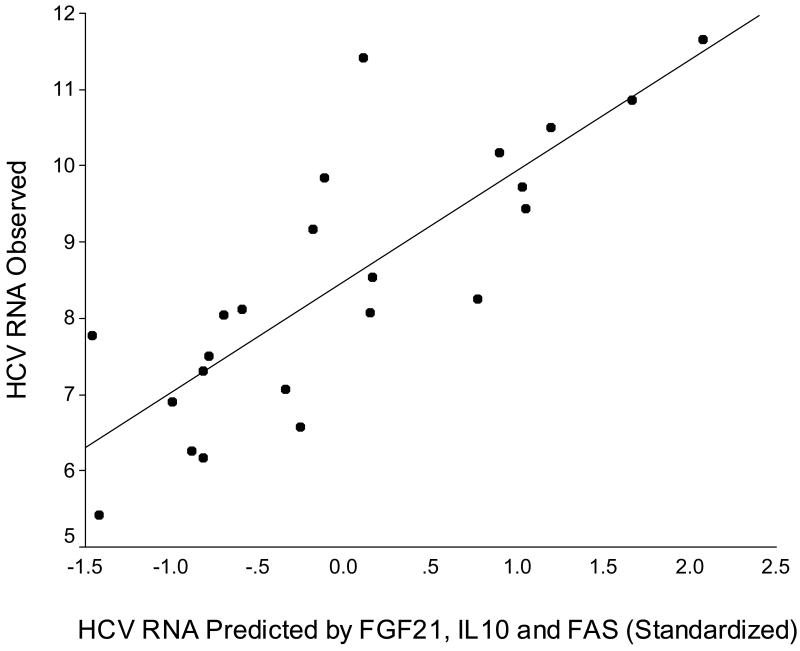
Correlations between Hepatic HCV RNA level and the Expression of Hepatic Genes Using bivariate correlation analysis, the mRNA levels of PPARγ, RARβ, RARγ, liver receptor homolog-1 (LRH-1), farnesoid x receptor (FXR), SCD1, FAS, fibroblast growth factor 21 (FGF21), G6P, IL-10, and retinoid-inducible gene 1 protein (RIG1) were correlated with hepatic HCV RNA levels. All the correlation coefficients were higher than 0.4, and RARγ had the best correlation coefficient (0.57) (Table 3). Stepwise multivariate linear regression analysis showed that FGF21, IL-10, and FAS mRNA levels were independently correlated with hepatic HCV RNA (Table 4). The adjusted R2 of this model was 0.63. Fig. 3 shows the predictability.
Table 3. Bivariate correlation analysis for HCV RNA and genes expression.
| Genes functions | Genes | Correlation coefficients | P value (2 tails) |
|---|---|---|---|
| Nuclear Receptor | PPARγ | 0.42 | 0.045 |
| RARβ | 0.48 | 0.021 | |
| RARγ | 0.57 | 0.004 | |
| LRH-1 | 0.45 | 0.031 | |
| FXR | 0.46 | 0.028 | |
| Lipid and glucose metabolism pathway | SCD | 0.47 | 0.020 |
| FASN | 0.46 | 0.027 | |
| FGF21 | -0.48 | 0.019 | |
| G6P | 0.43 | 0.039 | |
| Immune response and inflammatory pathway | IL10 | 0.48 | 0.021 |
| RIG1 | 0.42 | 0.048 |
Table 4. Multivariate linear regression analysis for HCV RNA.
| B* | Std. Error | Beta** | t | Sig | |
|---|---|---|---|---|---|
| Constant | 2.45 | 2.28 | 1.07 | 0.29 | |
| FGF21 | -0.22 | 0.1 | -0.31 | -2.16 | 0.04 |
| IL10 | 0.44 | 0.11 | 0.56 | 3.89 | <0.01 |
| FASN | 0.85 | 0.23 | 0.54 | 3.67 | <0.01 |
unstandardized coefficients.
standardized coefficients.
Figure 3.
Observed hepatic HCV RNA values versus predicted HCV RNA values based on hepatic FGF21, IL10, and FAS mRNA levels (standardized to have mean 0 and standard deviation 1). The adjusted R2 of this model was 0.63.
Discussion
The molecular mechanisms involved in HCV disease progression are not well understood. Our data indicate that viral infection can alter nuclear receptor-mediated signaling, which may lead to HCV infection-associated pathological conditions. Nuclear receptors are popular drug targets, and drugs that modulate nuclear receptor activity are among the most prescribed pharmaceuticals on the market (27). Nuclear receptors may have the potential to be therapeutic targets to control HCV-associated disease process (15, 16).
Among the studied nuclear receptor genes, RXRα, PPARα and β, SHP, and CAR showed significant changes in their expression levels in HCV-infected livers. These nuclear receptors play important roles in lipid metabolism or related pathways (Fig. 4). Our published data show that serum cholesterol and triglyceride levels are increased due to hepatocyte RXRα deficiency (28, 29). PPARα modulates genes encoding lipid metabolism enzymes, lipid transporters, and apolipoproteins, and PPARα mRNA level is reduced in HCV-infected livers. An interesting finding is the concomitant down-regulation of RXRα and PPARβ in HCV-infected patients. It has shown that the expression of RXRα is not altered in HCV-associated cirrhotic livers (30). The difference may be due to lack of advanced cirrhosis in most patients included in the current study.
Figure 4.
Nuclear receptor-mediated pathways in chronic hepatitis C patients. (1) Fat influx and efflux. FATP and CD36 are involved in fatty acid uptake and MTP is important to VLDL secretion. The expression levels of FATP5, FATP2, and MTP are up-regulated in livers with chronic hepatitis C infection compared with controls. (2) Fatty acid synthesis and oxidation. SREBP-1c and PPARα are the key regulators of fatty acid synthesis and oxidation. Their expression is down-regulated in livers with chronic hepatitis C infection compared with controls. (3) Glucose uptake. GLUT2 mediates facilitated glucose uptake. Its expression was up-regulated in livers with chronic hepatitis C infection compared with controls, but de novo fatty acid synthesis was not up-regulated. (4) Bile acid synthesis and uptake. CYP7A1 catalyzes the rate-limiting step in cholesterol catabolism and bile acid biosynthesis and NTCP is involved in hepatic sodium/bile acid uptake. Expression of both genes was up-regulated in livers with chronic hepatitis C infection compared with controls. Bile acids enhance genotype 1 HCV replication in an HCV-replication cell model (38, 39). (5) SHP plays a key role in the regulation of hepatic lipid metabolism (37, 55). SHP was up-regulated in livers with chronic hepatitis C infection compared with controls, but it was accompanied by up-regulated expression of CYP7A1, NTCP, and MTP. Gene names in yellow indicate expression increase in hepatitis C patients compared with controls. Gene names in white indicate expression decrease in hepatitis C patients compared with controls. Small arrows indicate the direction of the regulatory pathways. Big arrows suggest pathways are stimulated in hepatitis C patients compared with controls. X: The inhibitory function is compromised in hepatitis C patients.
PPARβ has received a lot of attention in the research of metabolic diseases recently. Activation of mouse PPARβ increases fatty acid β-oxidation, which is accompanied by marked reduction of lipid droplets in skeletal muscle (31). Hepatic-restricted PPARβ activation produces hepatic glycogen and increase in monounsaturated fatty acid production as well as up-regulation of glucose utilization (32). Our results showed hepatic PPARβ mRNA was decreased and GLUT2 mRNA level was increased in hepatitis C patients suggesting an imbalance in using fat and sugar as energy sources in HCV-infected livers. Co-regulator NCOA3 mRNA level is increased in HCV-infected livers, which is consistent with the finding that deletion of NCOA3 in mice prevents high fat diet-induced steatosis and inflammation (33). However, whether the modest change of NCOA3 mRNA level found in patients are biologically significant remains to be validated.
In vitro, HCV core proteins NS2 and NS5 induce hepatic lipid accumulation by activating SREBP-1c and PPARγ (34-36). However, SREBP-1c mRNA level reduced in HCV-infected livers. Our unpublished data also show reduced expression of SREBP-1c in HCV core protein transgenic mice. The expression of SREBP-1c is negatively regulated by SHP and CAR and positively regulated by PPARβ (Fig. 4). The up-regulated SHP and CAR as well as down-regulated PPARβ found in our HCV-infected patients could explain reduced SREBP-1c expression. Reduced expression of SREBP-1c could also be due to overloading of fat in the hepatocyte caused by up-regulated expression of FATP2 and FATP5, and thus potentially is an adaptive response.
SHP plays a central role in lipid and glucose metabolism by inhibiting SREBP-1c, MTP, NTCP, and CYP7A1 expression and increasing G6P and PEPCK expression (13, 37). In HCV-infected patients, increased hepatic SHP, MTP, NTCP, and CYP7A1 mRNA was observed, and FXR, G6P, and PEPCK mRNA levels did not change. This finding suggests that the FXR-SHP-CYP7A1 regulatory loop is totally compromised in HCV-infected liver. The observed changes could be due to HCV infection. Alternatively, those changes could be adaptive host responses in order to minimize liver injury. MTP is essential for hepatic lipoprotein assembly and secretion, and VLDL is important for HCV secretion from the infected cells (23). In addition, bile acid via FXR promotes genotype 1 hepatitis C virus replication (38, 39). Thus, all these alterations are related to HCV life cycle.
Activation of CAR ameliorates hyperglycemia by suppressing glucose production and stimulating glucose uptake and usage in the liver and improves steatosis by inhibiting hepatic lipogenesis and inducing β oxidation (40). In our studied hepatitis C patients, CAR was significantly up-regulated and this was accompanied by decreased SREBP-1c and increased GLUT2 expression. This finding suggests that CAR may play a significant role in lipid and glucose metabolism in the HCV-infected livers.
In ethanol-fed mice, hepatic PPARα-mediated signaling is decreased (41-43). In addition, AMPK activity and fatty acid synthesis related genes are down-regulated (44). In HCV-infected patients who had a history of drinking, our results showed that PPARα and RXRα expression levels were increased with concomitant up regulation of their target genes involved in fatty acid oxidation and hepatic uptake and intracellular trafficking. Species difference may account for the differential findings (57). There are both current and non-current drinkers in group B, but no significant difference could be found in gene expression between the two groups (Table S2). This suggests the possibility of active drinking in “non-current drinkers”. In addition, the gene expression alteration does not seem to be caused by differences in disease severity because there was no difference in liver panel, severity of fibrosis, and inflammation in these two cohorts (Table S3). Although PPARα and RXRα and their target genes were up-regulated in patients with a history of alcohol drinking, the genes involved in anti-oxidant and inflammatory pathways did not change their expression level significantly (Fig. S2B. This result does not support the hypothesis that alcohol and HCV synergized through increasing PPARα activity, lipid peroxidation, and oxidative stress and thus liver injury. Other mechanisms have been proposed to explain the synergism of HCV infection and alcohol intake. For example, alcohol impairs the intracellular innate immune response in human hepatocytes and promotes HCV infection and replication (45).
Multivariate analysis showed independent association between the hepatic mRNA levels of FAS, FGF21, and IL-10 with HCV RNA. All these genes are regulated by nuclear receptors or co-regulators. FAS expression is regulated by SREBP-1c, which is regulated by LXR and PPARγ (13, 24, 25). FGF21 is involved in fat oxidation or lipolysis in the liver and is a target of PPARα (46-48). IL-10 is an anti-inflammatory cytokine and down-regulates Th1 effector mechanisms. The expression of IL-10 in hepatocytes is increased by fatty acids and such regulation is mediated by PPAR-γ coactivator 1α (PGC-1α) (49). The relationship between FAS and HCV replication has been studied in a subgenomic replicon system (50) and in the JFH1 infectious system (51). Inhibition of fatty acid synthase (FAS) by cerulenin or C75 blocks HCV replication. There have been no reports on the relationship between FGF21 and HCV replication. Our result for the first time showed a negative independent correlation between hepatic FGF21 mRNA level and HCV RNA level. In an HCV replicon study, PPARα agonist inhibits HCV replication in Huh7/Rep-Feo cells (52). PPARα agonist reduces serum HCV RNA titers in patients (53, 54). Whether this effect is mediated by FGF21 warrants further study because FGF21 is a PPARα target.
The current study has a few limitations. First, the sample size is not large. Larger sample size would increase the confidence of findings. Second, expression was detected at the mRNA level because most livers were obtained from biopsy. However, most of the studied genes are regulated by nuclear receptors at the transcriptional levels. Third, the number of studied genes was limited. We prioritized the assays by studying the expression of genes that have obvious roles in lipid, bile acid, and carbohydrate homeostasis. The studied cofactors are the most common ones for their receptive nuclear receptors. The ultimate approach is microarray using a large sample size. Fourth, the direct comparison between normal liver and chronic hepatitis C patients with a drinking history was not done due to lack of matched gender. However, the change caused by HCV and by alcohol may mask each other's effects. For example, PPARα mRNA was decreased in HCV-infected livers in comparison with normal livers. However, it was increased in HCV patients who had a history of alcohol drinking in comparison with those who were HCV-infected, but did not have a drinking history. Thus, it is likely that PPARα expression level is not different between control and HCV/alcohol group, however the change to PPARα is significant and the interaction between HCV and alcohol drinking deserves attention.
Supplementary Material
The expressions of nuclear receptors and their downstream target genes were studied in 23 HCV-infected livers and 15 donor livers. (A) Nuclear receptors. (B) Fatty acid synthesis pathway. (C) Fatty acid oxidation pathway. (D) Antioxidant pathway. Mean ± SEM are shown. p>0.05, comparisons between the two groups.
The expressions of nuclear receptors and their downstream target genes were studied hepatitis C male patients with (N=21) and without (13) a history of drinking. (A) Nuclear receptor co-regulators. *: p <0.05. (B) Antioxidant and inflammatory response pathway. (C) Fatty acid synthesis pathway. (D) VLDL secretion pathway. (E) Nuclear receptors. Mean ± SEM are shown. p >0.05 in all comparisons.
Fasting plasma glucose level (GLU) in hepatitis C patients with (HC+AL) and without (HC) a history of alcohol consumption. Mean ± SEM are shown. *: p <0.05 in the comparison between patients with and without a history of drinking.
Acknowledgments
The authors thank patients, physicians, nurses, Natali Navarro Cazarez, and Carly Thoma-Perry for their contribution to the KU Liver Center Tissue Bank. We also thank Zoe Raglow for her assistance in preparation of this paper.
Financial Support: This study is supported by NIH grants CA 53596, DK092100, and P20RR021940.
Abbreviations
- ACADS
acyl-CoA dehydrogenase
- ACC
acyl-coA carboxylase
- ACOX
acyl-CoA oxidase
- ALP
alkaline phosphatase
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- CAR
constitutive androstane receptor
- CHOL
total cholesterol
- CPT-1
carnitine palmitoyl transferase 1
- CYP4A11
cytochrome P450, family 4, subfamily A, polypeptide 11
- CYP7A1
cytochrome P450, family 7, subfamily A, polypeptide 1
- CYP2E1
cytochrome P450, family 2, subfamily E, polypeptide 1
- FABP
fatty acid binding protein
- FAE
Fatty acyl-CoA elongase
- FAS
fatty acid synthase
- CD36
CD36 molecule
- FATP
fatty acid transport protein
- FGF21
fibroblast growth factor 21
- FXR
farnesoid X receptor
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- GLUT
facilitated glucose transporter
- G6P
glucose-6-phosphatase
- HADH
hydroxyacyl-CoA dehydrogenase
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- IFN
interferon
- IL
interleukin
- LDLR
low-density lipoprotein receptor
- LRH-1
liver receptor homolog-1
- LXR
liver X receptor
- MTP
microsomal triglyceride transfer protein
- NCOA
nuclear receptor coactivator
- NCOR
nuclear receptor corepressor
- NOS2
nitric oxide synthase 2
- NTCP
Na+/taurocholate cotransporter
- PCR
polymerase chain reaction
- PEPCK
phosphoenolpyruvate carboxykinase
- PGC-1α
peroxisome proliferator activated receptor-gamma coactivator 1α
- PPAR
peroxisome proliferator activated receptor
- RAR
retinoic acid receptor
- REV-Erbβ
nuclear receptor subfamily 1, group D, member 2
- RIG1
Retinoid-inducible gene 1 protein
- RNA
ribonucleic acid
- RXR
retinoid X receptor
- SCD1
stearyl-coA dehydrogenase
- SHP
small heterodimer partner
- SREBP
steroid regulatory element-binding protein
- SVR
sustained virological response
- TBILI
total bilirubin
- TNF
tumor necrosis factor
- TRIG
triglyceride
- VLDL
very low-density lipoprotein
References
- 1.Kim WR. The burden of hepatitis C in the United States. Hepatology. 2002;36(5 Suppl 1):S30–4. doi: 10.1053/jhep.2002.36791. [DOI] [PubMed] [Google Scholar]
- 2.Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358(9286):958–65. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 3.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Goncales FL, Jr, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347(13):975–82. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 4.McHutchison JG, Lawitz EJ, Shiffman ML, Muir AJ, Galler GW, McCone J, et al. Peginterferon alfa-2b or alfa-2a with ribavirin for treatment of hepatitis C infection. N Engl J Med. 2009;361(6):580–93. doi: 10.1056/NEJMoa0808010. [DOI] [PubMed] [Google Scholar]
- 5.Poordad F, McCone J, Jr, Bacon BR, Bruno S, Manns MP, Sulkowski MS, et al. Boceprevir for untreated chronic HCV genotype 1 infection. N Engl J Med. 2011;364(13):1195–206. doi: 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bacon BR, Gordon SC, Lawitz E, Marcellin P, Vierling JM, Zeuzem S, et al. Boceprevir for previously treated chronic HCV genotype 1 infection. N Engl J Med. 2011;364(13):1207–17. doi: 10.1056/NEJMoa1009482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McHutchison JG, Everson GT, Gordon SC, Jacobson IM, Sulkowski M, Kauffman R, et al. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N Engl J Med. 2009;360(18):1827–38. doi: 10.1056/NEJMoa0806104. [DOI] [PubMed] [Google Scholar]
- 8.McHutchison JG, Manns MP, Muir AJ, Terrault NA, Jacobson IM, Afdhal NH, et al. Telaprevir for previously treated chronic HCV infection. N Engl J Med. 2010;362(14):1292–303. doi: 10.1056/NEJMoa0908014. [DOI] [PubMed] [Google Scholar]
- 9.Andre P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, et al. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol. 2002;76(14):6919–28. doi: 10.1128/JVI.76.14.6919-6928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asselah T, Rubbia-Brandt L, Marcellin P, Negro F. Steatosis in chronic hepatitis C: why does it really matter? Gut. 2006;55(1):123–30. doi: 10.1136/gut.2005.069757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czaja AJ, Carpenter HA, Santrach PJ, Moore SB. Host- and disease-specific factors affecting steatosis in chronic hepatitis C. J Hepatol. 1998;29(2):198–206. doi: 10.1016/s0168-8278(98)80004-4. [DOI] [PubMed] [Google Scholar]
- 12.Mirandola S, Realdon S, Iqbal J, Gerotto M, Dal Pero F, Bortoletto G, et al. Liver microsomal triglyceride transfer protein is involved in hepatitis C liver steatosis. Gastroenterology. 2006;130(6):1661–9. doi: 10.1053/j.gastro.2006.02.035. [DOI] [PubMed] [Google Scholar]
- 13.Wagner M, Zollner G, Trauner M. Nuclear receptors in liver disease. Hepatology. 2011;53(3):1023–34. doi: 10.1002/hep.24148. [DOI] [PubMed] [Google Scholar]
- 14.Wang K, Wan YJ. Nuclear receptors and inflammatory diseases. Exp Biol Med (Maywood) 2008;233(5):496–506. doi: 10.3181/0708-MR-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raglow Z, Thoma-Perry C, Gilroy R, Wan YJ. The interaction between HCV and nuclear receptor-mediated pathways. Pharmacol Ther. 2011 doi: 10.1016/j.pharmthera.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trauner M, Halilbasic E. Nuclear receptors as new perspective for the management of liver diseases. Gastroenterology. 2011;140(4):1120–5. e1–12. doi: 10.1053/j.gastro.2011.02.044. [DOI] [PubMed] [Google Scholar]
- 17.Peters MG, Terrault NA. Alcohol use and hepatitis C. Hepatology. 2002;36(5 Suppl 1):S220–5. doi: 10.1053/jhep.2002.36811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Safdar K, Schiff ER. Alcohol and hepatitis C. Semin Liver Dis. 2004;24(3):305–15. doi: 10.1055/s-2004-832942. [DOI] [PubMed] [Google Scholar]
- 19.Gyamfi MA, Wan YJ. Pathogenesis of alcoholic liver disease: the role of nuclear receptors. Exp Biol Med (Maywood) 2010;235(5):547–60. doi: 10.1258/ebm.2009.009249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mello T, Polvani S, Galli A. Peroxisome proliferator-activated receptor and retinoic x receptor in alcoholic liver disease. PPAR Res. 2009;2009:748174. doi: 10.1155/2009/748174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishak K, Baptista A, Bianchi L, Callea F, De Groote J, Gudat F, et al. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22(6):696–9. doi: 10.1016/0168-8278(95)80226-6. [DOI] [PubMed] [Google Scholar]
- 22.Agnello V, Abel G, Elfahal M, Knight GB, Zhang QX. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci U S A. 1999;96(22):12766–71. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Negro F. Abnormalities of lipid metabolism in hepatitis C virus infection. Gut. 2010;59(9):1279–87. doi: 10.1136/gut.2009.192732. [DOI] [PubMed] [Google Scholar]
- 24.Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, et al. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14(22):2819–30. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joseph SB, Laffitte BA, Patel PH, Watson MA, Matsukuma KE, Walczak R, et al. Direct and indirect mechanisms for regulation of fatty acid synthase gene expression by liver X receptors. J Biol Chem. 2002;277(13):11019–25. doi: 10.1074/jbc.M111041200. [DOI] [PubMed] [Google Scholar]
- 26.Burris TP. Nuclear hormone receptors for heme: REV-ERBalpha and REV-ERBbeta are ligand-regulated components of the mammalian clock. Mol Endocrinol. 2008;22(7):1509–20. doi: 10.1210/me.2007-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearce KH, Iannone MA, Simmons CA, Gray JG. Discovery of novel nuclear receptor modulating ligands: an integral role for peptide interaction profiling. Drug Discov Today. 2004;9(17):741–51. doi: 10.1016/S1359-6446(04)03201-5. [DOI] [PubMed] [Google Scholar]
- 28.Wan YJ, An D, Cai Y, Repa JJ, Hung-Po Chen T, Flores M, et al. Hepatocyte-specific mutation establishes retinoid X receptor alpha as a heterodimeric integrator of multiple physiological processes in the liver. Mol Cell Biol. 2000;20(12):4436–44. doi: 10.1128/mcb.20.12.4436-4444.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wan YJ, Cai Y, Lungo W, Fu P, Locker J, French S, et al. Peroxisome proliferator-activated receptor alpha-mediated pathways are altered in hepatocyte-specific retinoid X receptor alpha-deficient mice. J Biol Chem. 2000;275(36):28285–90. doi: 10.1074/jbc.M000934200. [DOI] [PubMed] [Google Scholar]
- 30.Dharancy S, Malapel M, Perlemuter G, Roskams T, Cheng Y, Dubuquoy L, et al. Impaired expression of the peroxisome proliferator-activated receptor alpha during hepatitis C virus infection. Gastroenterology. 2005;128(2):334–42. doi: 10.1053/j.gastro.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka T, Yamamoto J, Iwasaki S, Asaba H, Hamura H, Ikeda Y, et al. Activation of peroxisome proliferator-activated receptor delta induces fatty acid beta-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc Natl Acad Sci U S A. 2003;100(26):15924–9. doi: 10.1073/pnas.0306981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu S, Hatano B, Zhao M, Yen CC, Kang K, Reilly SM, et al. Role of peroxisome proliferator-activated receptor {delta}/{beta} in hepatic metabolic regulation. J Biol Chem. 2011;286(2):1237–47. doi: 10.1074/jbc.M110.138115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma X, Xu L, Wang S, Cui B, Li X, Xu J, et al. Deletion of steroid receptor coactivator-3 gene ameliorates hepatic steatosis. J Hepatol. 2010 doi: 10.1016/j.jhep.2010.11.022. [DOI] [PubMed] [Google Scholar]
- 34.Kim KH, Hong SP, Kim K, Park MJ, Kim KJ, Cheong J. HCV core protein induces hepatic lipid accumulation by activating SREBP1 and PPARgamma. Biochem Biophys Res Commun. 2007;355(4):883–8. doi: 10.1016/j.bbrc.2007.02.044. [DOI] [PubMed] [Google Scholar]
- 35.Kim K, Kim KH, Ha E, Park JY, Sakamoto N, Cheong J. Hepatitis C virus NS5A protein increases hepatic lipid accumulation via induction of activation and expression of PPARgamma. FEBS Lett. 2009;583(17):2720–6. doi: 10.1016/j.febslet.2009.07.034. [DOI] [PubMed] [Google Scholar]
- 36.Oem JK, Jackel-Cram C, Li YP, Zhou Y, Zhong J, Shimano H, et al. Activation of sterol regulatory element-binding protein 1c and fatty acid synthase transcription by hepatitis C virus non-structural protein 2. J Gen Virol. 2008;89(Pt 5):1225–30. doi: 10.1099/vir.0.83491-0. [DOI] [PubMed] [Google Scholar]
- 37.Trauner M. A little orphan runs to fat: the orphan receptor small heterodimer partner as a key player in the regulation of hepatic lipid metabolism. Hepatology. 2007;46(1):1–5. doi: 10.1002/hep.21801. [DOI] [PubMed] [Google Scholar]
- 38.Scholtes C, Diaz O, Icard V, Kaul A, Bartenschlager R, Lotteau V, et al. Enhancement of genotype 1 hepatitis C virus replication by bile acids through FXR. J Hepatol. 2008;48(2):192–9. doi: 10.1016/j.jhep.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 39.Chang KO, George DW. Bile acids promote the expression of hepatitis C virus in replicon-harboring cells. J Virol. 2007;81(18):9633–40. doi: 10.1128/JVI.00795-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong B, Saha PK, Huang W, Chen W, Abu-Elheiga LA, Wakil SJ, et al. Activation of nuclear receptor CAR ameliorates diabetes and fatty liver disease. Proc Natl Acad Sci U S A. 2009;106(44):18831–6. doi: 10.1073/pnas.0909731106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wan YJ, Morimoto M, Thurman RG, Bojes HK, French SW. Expression of the peroxisome proliferator-activated receptor gene is decreased in experimental alcoholic liver disease. Life Sci. 1995;56(5):307–17. doi: 10.1016/0024-3205(94)00953-8. [DOI] [PubMed] [Google Scholar]
- 42.Nakajima T, Kamijo Y, Tanaka N, Sugiyama E, Tanaka E, Kiyosawa K, et al. Peroxisome proliferator-activated receptor alpha protects against alcohol-induced liver damage. Hepatology. 2004;40(4):972–80. doi: 10.1002/hep.20399. [DOI] [PubMed] [Google Scholar]
- 43.Fischer M, You M, Matsumoto M, Crabb DW. Peroxisome proliferator-activated receptor alpha (PPARalpha) agonist treatment reverses PPARalpha dysfunction and abnormalities in hepatic lipid metabolism in ethanol-fed mice. J Biol Chem. 2003;278(30):27997–8004. doi: 10.1074/jbc.M302140200. [DOI] [PubMed] [Google Scholar]
- 44.Garcia-Villafranca J, Guillen A, Castro J. Ethanol consumption impairs regulation of fatty acid metabolism by decreasing the activity of AMP-activated protein kinase in rat liver. Biochimie. 2008;90(3):460–6. doi: 10.1016/j.biochi.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 45.Ye L, Wang S, Wang X, Zhou Y, Li J, Persidsky Y, et al. Alcohol impairs interferon signaling and enhances full cycle hepatitis C virus JFH-1 infection of human hepatocytes. Drug Alcohol Depend. 2010;112(1-2):107–16. doi: 10.1016/j.drugalcdep.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Inagaki T, Dutchak P, Zhao G, Ding X, Gautron L, Parameswara V, et al. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab. 2007;5(6):415–25. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 47.Reitman ML. FGF21: a missing link in the biology of fasting. Cell Metab. 2007;5(6):405–7. doi: 10.1016/j.cmet.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 48.Christodoulides C, Dyson P, Sprecher D, Tsintzas K, Karpe F. Circulating fibroblast growth factor 21 is induced by peroxisome proliferator-activated receptor agonists but not ketosis in man. J Clin Endocrinol Metab. 2009;94(9):3594–601. doi: 10.1210/jc.2009-0111. [DOI] [PubMed] [Google Scholar]
- 49.Morari J, Torsoni AS, Anhe GF, Roman EA, Cintra DE, Ward LS, et al. The role of proliferator-activated receptor gamma coactivator-1alpha in the fatty-acid-dependent transcriptional control of interleukin-10 in hepatic cells of rodents. Metabolism. 2010;59(2):215–23. doi: 10.1016/j.metabol.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 50.Su AI, Pezacki JP, Wodicka L, Brideau AD, Supekova L, Thimme R, et al. Genomic analysis of the host response to hepatitis C virus infection. Proc Natl Acad Sci U S A. 2002;99(24):15669–74. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang W, Hood BL, Chadwick SL, Liu S, Watkins SC, Luo G, et al. Fatty acid synthase is up-regulated during hepatitis C virus infection and regulates hepatitis C virus entry and production. Hepatology. 2008;48(5):1396–403. doi: 10.1002/hep.22508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishimura-Sakurai Y, Sakamoto N, Mogushi K, Nagaie S, Nakagawa M, Itsui Y, et al. Comparison of HCV-associated gene expression and cell signaling pathways in cells with or without HCV replicon and in replicon-cured cells. J Gastroenterol. 2010;45(5):523–36. doi: 10.1007/s00535-009-0162-3. [DOI] [PubMed] [Google Scholar]
- 53.Fujita N, Kaito M, Tanaka H, Horiike S, Adachi Y. Reduction of serum HCV RNA titer by bezafibrate therapy in patients with chronic hepatitis C. Am J Gastroenterol. 2004;99(11):2280. doi: 10.1111/j.1572-0241.2004.40695_3.x. [DOI] [PubMed] [Google Scholar]
- 54.Fujita N, Kaito M, Kai M, Sugimoto R, Tanaka H, Horiike S, et al. Effects of bezafibrate in patients with chronic hepatitis C virus infection: combination with interferon and ribavirin. J Viral Hepat. 2006;13(7):441–8. doi: 10.1111/j.1365-2893.2005.00718.x. [DOI] [PubMed] [Google Scholar]
- 55.Boulias K, Katrakili N, Bamberg K, Underhill P, Greenfield A, Talianidis I. Regulation of hepatic metabolic pathways by the orphan nuclear receptor SHP. EMBO J. 2005;24(14):2624–33. doi: 10.1038/sj.emboj.7600728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The expressions of nuclear receptors and their downstream target genes were studied in 23 HCV-infected livers and 15 donor livers. (A) Nuclear receptors. (B) Fatty acid synthesis pathway. (C) Fatty acid oxidation pathway. (D) Antioxidant pathway. Mean ± SEM are shown. p>0.05, comparisons between the two groups.
The expressions of nuclear receptors and their downstream target genes were studied hepatitis C male patients with (N=21) and without (13) a history of drinking. (A) Nuclear receptor co-regulators. *: p <0.05. (B) Antioxidant and inflammatory response pathway. (C) Fatty acid synthesis pathway. (D) VLDL secretion pathway. (E) Nuclear receptors. Mean ± SEM are shown. p >0.05 in all comparisons.
Fasting plasma glucose level (GLU) in hepatitis C patients with (HC+AL) and without (HC) a history of alcohol consumption. Mean ± SEM are shown. *: p <0.05 in the comparison between patients with and without a history of drinking.