Abstract
Male germline stem cells (GSCs) in Drosophila melanogaster divide asymmetrically by orienting mitotic spindle with respect to the niche, a microenvironment that specifies stem cell identity. The spindle orientation is prepared during interphase through stereotypical positioning of the centrosomes. We recently demonstrated that GSCs possess a checkpoint (“the centrosome orientation checkpoint”) that monitors correct centrosome orientation prior to mitosis to ensure an oriented spindle and thus asymmetric outcome of the division. Here, we show that Par-1, a serine/threonine kinase that regulates polarity in many systems, is involved in this checkpoint. Par-1 shows a cell cycle-dependent localization to the spectrosome, a germline-specific, endoplasmic reticulum-like organelle. Furthermore, the localization of cyclin A, which is normally localized to the spectrosome, is perturbed in par-1 mutant GSCs. Interestingly, overexpression of mutant cyclin A that does not localize to the spectrosome and mutation in hts, a core component of the spectrosome, both lead to defects in the centrosome orientation checkpoint. We propose that the regulation of cyclin A localization via Par-1 function plays a critical role in the centrosome orientation checkpoint.
Keywords: stem cell, stem cell niche, centrosome, spindle orientation, asymmetric stem cell division, Drosophila, cyclin A, spectrosome
Introduction
Many adult stem cells divide asymmetrically, producing one self-renewed stem cell and one differentiating cell, thereby contributing to tissue homeostasis (Morrison and Kimble, 2006). Disruption of this balance can lead to tumorigenesis/tissue hyperplasia (due to excess stem cell self-renewal) or tissue degeneration/aging (due to excess differentiation or lack of self-renewal). It has been increasingly recognized that many adult stem cells reside in a special microenvironment, or niche, which provides essential signals for stem cell maintenance, identity, and proliferation (Morrison and Spradling, 2008). In such a niche, stem cells often orient their mitotic spindles to determine the outcome of stem cell division: stem cells divide either symmetrically to increase stem cell number or asymmetrically to maintain stem cell number (Yamashita, 2010). Spindle orientation perpendicular to the niche component maintains one daughter of the stem cell division within the niche and displaces the other outside the niche, leading to an asymmetric outcome of the stem cell division.
Drosophila male germline stem cells (GSCs) reside in a defined microenvironment at the apical tip of the testis. The hub cells as well as somatic cyst stem cells (CySCs) are critical constituents of the GSC niche (Kiger et al., 2001; Leatherman and Dinardo, 2008, 2010; Tulina and Matunis, 2001), and the attachment of GSCs to the hub cells is the key to maintaining GSCs within the niche (Voog et al., 2008). Drosophila male GSCs divide asymmetrically by orienting their mitotic spindle perpendicularly toward the hub, so that one daughter of the division is attached to the hub while the other is displaced from the hub (Yamashita et al., 2003). Spindle orientation is set up during interphase through stereotypical positioning of the mother and daughter centrosomes: the mother centrosome is always closely associated with the hub-GSC interface throughout the cell cycle, while the daughter centrosome is replicated next to the mother centrosome and migrates to the opposite side of the cell during interphase (Yamashita et al., 2003; Yamashita et al., 2007). Stereotypical centrosome behavior in preparation for division orientation has been described in Drosophila neuroblasts (Rebollo et al., 2007; Rusan and Peifer, 2007) and mouse radial glia progenitor cells (Wang et al., 2009), suggesting that centrosome positioning is an evolutionarily conserved mechanism for asymmetric stem cell division.
We recently showed that GSCs without stereotypical centrosome positioning (referred as to misoriented GSCs; Fig. 1A) exhibit delayed cell cycle progression (Cheng et al., 2008). Misoriented GSCs are defined as those in which neither of the two centrosomes is located adjacent to the hub cells. GSCs resume cell division once the centrosome orientation is corrected, suggesting the presence of a surveillance mechanism (hereafter referred to as the centrosome orientation checkpoint) to monitor correct centrosome orientation to ensure asymmetric stem cell division. Indeed, we recently demonstrated the presence of such a checkpoint by showing that GSCs mutant for the centrosomin (cnn) gene or those overexpressing a dominant-negative form of E-cadherin fail to delay the cell cycle even when centrosomes are misoriented (Inaba et al., 2010). The surveillance mechanism that coordinates the position of the spindle and cell cycle progression is best understood as the spindle position checkpoint (SPOC) in budding yeast (Burke, 2009). The SPOC inhibits the mitotic exit network (MEN) when the spindle is mispositioned within the mother cell. Although essentially all multicellular organisms must undergo asymmetric cell divisions, which often require correct spindle orientation, the mechanisms equivalent to the SPOC have never been characterized outside yeast.
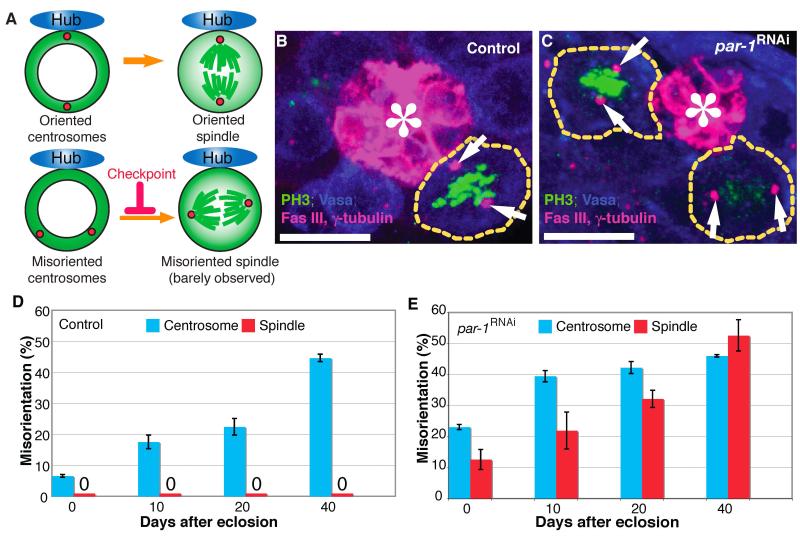
Fig. 1. Par-1 is required for the centrosome orientation checkpoint in male germline stem cells (GSCs).
(A) Model of the centrosome orientation checkpoint. Upon centrosome misorientation, GSCs undergo cell cycle arrest/delay. Centrosome misorientation is defined as neither of the two centrosomes being closely associated with the hub-GSC interface during interphase. Spindle misorientation is defined as neither of the two spindle poles being closely associated with the hub-GSC interface during mitosis. (B and C) Examples of oriented spindles in control GSCs and misoriented spindles in par-1RNAi GSCs (nos-gal4>UAS-Par-1 RNAi). Misoriented spindles in metaphase and anaphase are visible in C. Arrows indicate spindle poles. Green: PH3, phosphorylated histone H3 (mitotic chromosomes). Blue: Vasa (germ cells). Red: Fas III, fasciclin III (hub, *) and γ-tubulin (centrosome). The scale bar is 10 μm. (D and E) The frequency of centrosome (blue) and spindle (red) misorientation in control (D) and par-1RNAi (E) flies with age. Data are shown as means ± standard deviation (S.D.)
Here, we show that Par-1 is a critical component of the centrosome orientation checkpoint in Drosophila male GSCs. Par-1 was first isolated among par (partition-defective) genes that are required for the asymmetric division of Caenorhabditis elegans zygotes (Kemphues et al., 1988). Subsequently, a series of studies established the evolutionarily conserved roles of par genes in cell polarity in various contexts, including epithelial cells and Drosophila oocytes [reviewed in (St Johnston and Ahringer, 2010)]. The data suggest that the function of Par-1 in the centrosome orientation checkpoint is to regulate the localization of cyclin A. We show that localization of cyclin A, which is known to be dispensable for mitosis during embryonic development (Dienemann and Sprenger, 2004), is critical for the centrosome orientation checkpoint. We propose that cyclin A localization is a critical target of the centrosome orientation checkpoint, linking cellular asymmetry to cell cycle progression.
Materials and methods
Fly husbandry and strains
All fly stocks were raised on standard Bloomington medium at 25°C. The following fly stocks were used: Par-1-GFP (generated by the Flytrap project (Buszczak et al., 2007; Kelso et al., 2004; Morin et al., 2001) and obtained from the Spradling laboratory); nos-gal4 (Van Doren et al., 1998); Shaggy-GFP; UAS-HA-Cyclin A, and UAS-HA-NLS-Cyclin A (Dienemann and Sprenger, 2004) [plasmids were obtained from Dr. F. Sprenger and transgenic flies were obtained using standard P-element transformation (Rubin and Spradling, 1983)]; par-1K05603, Df(2R)BSC22, hts01103, and Df(2R)BSC26 (obtained from the Bloomington stock center); par-1w3 and par-106323 (obtained from Daniel St. Johnston (Shulman et al., 2000)); UAS-par-1RNAi (obtained from Dr. B. Lu (Zhang et al., 2007); UAS-DEFL (obtained from Hiroki Oda (Oda and Tsukita, 1999)). For the construction of Cyclin AΔC, stop codons were introduced at M488 and E489 residues by site-directed mutagenesis using the UAS-HA-Cyclin A plasmid as a template, resulting in the truncation of 44 amino acids from the C terminus. The resulting plasmid was used to generate transgenic flies harboring UAS-Cyclin AΔC using standard P-element transformation.
Immunofluorescence microscopy
Immunofluorescence staining was performed as described previously (Cheng et al., 2008). For cyclin A staining, testes were dissected into phosphate-buffered saline (PBS), transferred into chilled (−20°C) 90% ethanol and 3.7% formaldehyde fixative for 10 min, and then washed in PBS plus Tween 20 (PBST) for 30 min. The primary antibodies used were: mouse anti-γ-tubulin (1:100; GTU-88; Sigma), mouse anti-fasciclin III [1:20; developed by C. Goodman and obtained from the Developmental Studies Hybridoma Bank (DSHB)], mouse anti-Adducin-like (1:20, developed by H. D. Lipshitz and obtained from DSHB), mouse anti-Cyclin A (1:10, developed by C.F. Lehner and obtained from DSHB), rabbit anti-Thr3-phosphorylated Histone H3 (1:200; Upstate), goat anti-Vasa (1:100; dC-13; Santa Cruz Biotechnology), rabbit anti-Vasa (1:100; Santa Cruz Biotechnology), rat anti-HA (1:2000; clone 3F10; Roche). Images were taken using a Leica TCS SP5 confocal microscope with a 63× oil immersion objective (NA = 1.4) and processed using Adobe Photoshop software.
Western blotting and immunoprecipitation
Testes (60 pairs/sample) were dissected into PBS at room temperature within 20–30 min. Testes were then dissolved in lithium dodecyl sulfate (LDS) sample buffer (NuPAGE; Invitrogen) supplemented with 2% sodium dodecyl sulfate (SDS) and protease inhibitor cocktail (EDTA-free; Roche). Samples were separated on NuPAGE Bis-Tris gels (4–12%; Invitrogen) and transferred onto polyvinylidene fluoride (PVDF) membranes (Immobilon-P; Millipore). Blots were blocked in PBS containing 5% nonfat milk and 0.1% Triton X-100, followed by incubation with primary antibodies diluted in PBS containing 5% nonfat milk and 0.1% Triton X-100. Blots were then washed with PBS containing 0.1% Triton X-100, followed by incubation with secondary antibody. After washing with PBS, detection was performed using an enhanced chemiluminescence system (Amersham). For immunoprecipitation, testes were dissected into PBS at room temperature within 20–30 min, rinsed with buffer I [25mM HEPES, 80 mM KCl, 5 mM MgCl2, 1 mM ethylene glycol tetraacetic acid (EGTA), 10 mM β-glycerophosphate, 10 mM NaF, 1 mM NaVO3, 0.2 M sucrose (pH 7.4)], frozen in liquid nitrogen, and stored at −80°C until usage. A total of 800 testes/sample were homogenized and solubilized with lysis buffer [buffer I supplemented with 0.1% NP40 and protease inhibitor cocktail (EDTA-free; Roche)] for 8 h at 4°C. Supernatants (after centrifugation at 13,000 rpm for 30 min at 4°C using a tabletop centrifuge) were then incubated with anti-GFP antibody (rabbit; ab290; Abcam) and protein A agarose (Roche) overnight at 4°C. The beads were then washed five times with lysis buffer. Bound proteins were dissolved in SDS loading buffer and analyzed by western blotting as described above. Primary antibodies were anti-Cyclin A (mouse; A12-s; DSHB; 1:5), anti-β-Actin (mouse monoclonal; clone AC-15; Sigma; 1:2500), anti-GFP (chicken; GFP-1-2-; Aves labs; 1:10,000). Secondary antibodies were horseradish peroxidase-conjugated anti-mouse, anti-rabbit, and anti-chicken (Jackson ImmunoResearch Laboratories; 1:4000).
Results
Par-1 is required for centrosome and spindle orientation in Drosophila male GSCs
Our previous studies suggested the presence of a novel checkpoint that monitors correct centrosome orientation within GSCs prior to commitment to mitosis (Fig. 1A)(Cheng et al., 2008; Inaba et al., 2010). Upon centrosome misorientation in interphase, wild-type GSCs are arrested in the cell cycle for a prolonged time period, but enter mitosis upon reorientation (Cheng et al., 2008). When this checkpoint is perturbed, GSCs enter mitosis with misoriented centrosomes, leading to misoriented spindles (Inaba et al., 2010) (Fig. 1A). Therefore, the presence of spindle misorientation indicates a failure in the centrosome orientation checkpoint.
Par-1 controls cellular polarity in many systems, including C. elegans early embryos (Cheeks et al., 2004; Kemphues et al., 1988) and Drosophila oocytes (Benton et al., 2002; Benton and St Johnston, 2003; Huynh et al., 2001; Shulman et al., 2000). We tested whether Par-1 might play a role in centrosome and/or spindle orientation in male GSCs using previously characterized UAS-par-1 RNAi (Zhang et al., 2007) as well as viable allelic combinations (Shulman et al., 2000). In GSCs from young wild-type flies, the centrosome was oriented throughout the cell cycle with minimal (~5%) centrosome misorientation, and misoriented spindles were never observed (Fig. 1B, D). Although centrosome misorientation significantly increased with age in wild-type flies, spindle misorientation never increased (Fig. 1D) because GSCs possess the centrosome orientation checkpoint (Inaba et al., 2010) (Cheng et al., 2008). We have previously shown that the increase in centrosome misorientation with age is due to dedifferentiation of spermatogonia to GSC identity (Cheng et al., 2008). In this study, increased centrosome misorientation in aged flies was utilized as a “sensitized” background to highlight the presence of misoriented spindles due to a defect in the centrosome orientation checkpoint: in wild-type GSCs, the misoriented spindle was never observed even in testes from old flies, whereas mutant GSCs with defective centrosome orientation checkpoint would show spindle misorientation, in particular in the aged flies, which have a high frequency of centrosome misorientation.
Unlike wild-type GSCs, GSCs expressing par-1 RNAi [nos-gal4>UAS-par-1 RNAi (Zhang et al., 2007); hereafter referred to as par-1RNAi] had a high frequency of centrosome misorientation and frequently underwent mitosis with misoriented spindles (Fig. 1C, E). The requirement of Par-1 for correct centrosome and spindle orientation was confirmed using viable alleles of par-1 mutants (Table 1). The spindle misorientation observed in par-1 mutant GSCs is not merely due to increased centrosome misorientation during interphase. For example, 20-day-old wild-type testes contained approximately 20% of GSCs with misoriented centrosomes (Fig. 1D), but no misoriented spindles. By contrast, on day 0, approximately 20% of par-1RNAi GSCs exhibited centrosome misorientation similar to 20-day-old wild-type flies, but unlike in wild-type flies, this was associated with over 10% spindle misorientation (Fig. 1E), suggesting that par-1RNAi GSCs are defective in preventing mitosis upon centrosome misorientation. It should be noted that, in par-1RNAi GSCs, the centrosome misorientation reached plateau as the flies age. This is presumably because of the scoring criteria, by which misoriented centrosomes/spindles (neither of two centrosomes being away from the hub) cannot go beyond 50-60% centrosome misorientation: GSCs are scored as oriented when one of two centrosomes is close to the hub-GSC interface. Since the hub-GSC interface is ~25% of the entire GSC cortex, the probability that a randomly oriented centrosome is observed close to the hub-GSC interface would be ~25%. Thus, the probability that a GSC with two centrosomes is scored as misoriented would be up to ~60% (100%-25%-25%+12.5%=62.5%) even when centrosome orientation is random (Cheng et al., 2008).
Table 1.
Frequencies of misoriented centrosomes and spindles observed in allelic combinations of par-1 mutants (day 0)
| Genotype | Centrosome misorientation (n = GSCs counted) |
Spindle misorientation (n = mitotic GSCs counted) |
|---|---|---|
| nos-gal4>UAS-Par-1 RNAi* | 23.1% (n = 355) | 12.6% (n = 262) |
| TM3; UAS-Par-1 RNAi*, ** | 6.6% (n = 318) | 0% (n = 153) |
| par-1K06323/par-1k05603 | 36.4% (n = 143) | 25% (n = 32) |
| par-1K06323/CyO or par-1k05603/CyO*** | 12.4% (n = 105) | 0% (n = 38) |
| par-1w3/par-1K06323 | 41.6% (n = 101) | 29.6% (n = 27) |
| par-1w3 /CyO or par-1K06323/CyO*** | 14.1% (n = 177) | 0% (n = 35) |
| Df(2R)BSC22/par-1K06323 | 46.6% (n = 118) | 26.3% (n = 19) |
| Df(2R)BSC22/CyO or par- 1K06323/SM6a*** |
11.9% (n = 109) | 0% (n = 30) |
Same data as shown in Fig. 1.
Progeny with balancer chromosome from the same cross was used as the “no gal4 driver” control.
Progeny with balancer chromosome from the same cross was used as the heterozygous control.
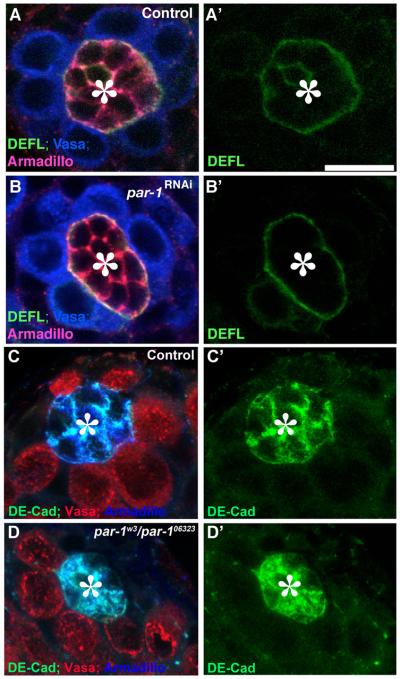
It is possible that Par-1 is required for the establishment of GSC polarity, but not for the centrosome orientation checkpoint. If par-1 mutant GSCs have no polarity, then the observed misoriented spindle might simply reflect the lack of polarity instead of the lack of the checkpoint function. However, this possibility is unlikely for the following reasons: 1) par-1RNAi and par-1w3/par-1k06323 GSCs showed increased centrosome misorientation with age, although it was always higher than in the control. This shows that par-1RNAi GSCs are “more oriented” at day 0 compared with later time points, suggesting that par-1RNAi GSCs maintain polarity themselves at least to some extent. 2) While par-1RNAi and par-1w3/par-1k06323 GSCs display a significant frequency of spindle misorientation, the frequency is still lower than that of centrosome misorientation. This suggests that par-1 mutant GSCs retain residual activity toward correct centrosome orientation prior to commitment to mitosis, although it is only partial and often leads to spindle misorientation. 3) par-1RNAi and par-1w3/par-1k06323 GSCs maintain the polarity with respect to the hub, judging from the fact that proteins known to be polarized within GSCs [DE-cadherin, Armadillo/β-catenin (Yamashita et al., 2003)] maintained their correct, polarized localization (Fig. 2). Therefore, it is unlikely that spindle misorientation observed in par-1 mutant GSCs is due to the lack of GSC polarity with respect to the hub cells.
Fig. 2. par-1mutant GSCs maintain the polarity toward the hub cells.
DE-cadherin as well as Armadillo/β-catenin correctly localizes in control GSCs (A, C) as well as in par-1RNAi GSCs (B) and par-1w3/par-106323 GSCs. (A, B) Green: DEFL (E-cad-GFP expressed in germ cells). Blue: Vasa (germ cells). Red: Armadillo. (C, D) Green: DE-Cad. Blue: Armadillo. Red: Vasa. The hub is indicated by the asterisk (*). The scale bar is 10 μm.
It is also possible that par-1 mutant GSCs are defective in general cell cycle regulation, and enter mitosis faster than normal GSCs irrespective of their centrosome orientation, leaving little time for GSCs to correct the centrosome orientation. However, this is also unlikely because the mitotic index of par-1RNAi GSCs is comparable to that of control GSCs [~0.18 mitotic GSCs/testis in control (n = 859) vs. ~0.16 mitotic GSCs/testis in par-1RNAi (n = 1689)], suggesting that the spindle misorientation observed in par-1RNAi GSCs is not a secondary effect of accelerated cell cycle progression. Taken together, these results suggest that Par-1 is required for centrosome orientation and its checkpoint in GSCs.
Par-1 and cyclin A colocalize to the spectrosome in a cell-cycle dependent manner
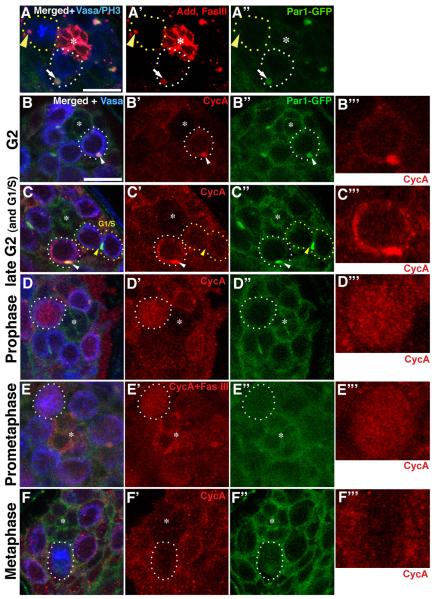
To better understand the function of Par-1 in the GSC centrosome orientation checkpoint, we first investigated cell cycle-dependent changes in Par-1 localization. Par-1 has been reported to localize to the spectrosome/fusome (Cox et al., 2001; Huynh et al., 2001; Lighthouse et al., 2008) (also shown in Supplementary Fig. S1), a germline-specific, endoplasmic reticulum (ER)-like organelle (Snapp et al., 2004). The spectrosome is a spherical structure that is found in GSCs, while the fusome is a branched version of the spectrosome that runs through the cytoplasm of interconnected spermatogonia at later stages (de Cuevas et al., 1997). As reported, Par-1 was localized to the spectrosome of GSCs throughout most of the cell cycle, as indicated by its colocalization with Adducin-like/Hu li tai shao (Hts), a major component of the spectrosome/fusome (Lin et al., 1994) (Fig. 3A, white arrow). However, this spectrosomal localization was diminished during mitosis, although the spectrosome itself remained intact during this time (Fig. 3A, yellow arrowhead).
Fig. 3. Localization of Par-1 and cyclin A during the GSC cell cycle.
(A) Par-1 localizes to the spectrosome (marked by Adducin-like) in interphase (white-dotted circle and arrow) but is released from it in mitosis (yellow-dotted circle and arrowhead). (B–F) Localization of Par-1 and cyclin A during the cell cycle. Cyclin A (CycA) localizes to the spectrosome in early G2 (B), while it is not detectable in G1 (when the GSC is still connected to the gonialblast, marked as “G1”in C). Cyclin A accumulates in the cytoplasm in addition to the spectrosome in late G2 (C, white-dotted circle). Cyclin A is released from the spectrosome in prophase (D) and prometaphase (E) when the spectrosomal localization of Par-1 is minimal. Cyclin A is degraded by metaphase (F). B”’-F”’ show higher magnification images of cyclin A channel of selected GSCs without markings. Green: Par1-GFP. Blue: PH3 (mitotic chromosome) and Vasa (germ cells). Red: Adducin-like and Fas III in (A), cyclin A in (B, C, D, F), and cyclin A and Fas III in (E). The scale bar is 10 μm and the hub is marked by the asterisk (*).
Similar to Par-1, cyclin A has been reported to localize to the spectrosome/fusome (Lilly et al., 2000). In the female germline, cyclin A regulates the number of transit-amplifying divisions. As reported, cyclin A localized to the spectrosome/fusome in the male germline (Supplementary Fig. S2A). We found that cyclin A also underwent dynamic changes in localization during the cell cycle. In earlier stages of the cell cycle, when the GSC is still connected to the gonialblast, cyclin A was barely detectable (Fig. 3C, marked as G1/S). Presumably, this period corresponds to G1/S phase, when the cyclin A level is known to be very low (Fry and Yamano, 2006). As the cell cycle progresses, separation of GSC and GB is completed and simultaneously the cyclin A level gradually increases; this cyclin A was colocalized with the spectrosome (Fig. 3B). As the cyclin A level becomes even higher in G2 phase, cyclin A was observed in the spectrosome as well as in the cytoplasm (Fig. 3C). Cyclin A was subsequently released into the cytoplasm and nucleus at around the onset of mitosis—i.e., prophase and prometaphase (Fig. 3D, E; see also Supplementary Fig. S2C, D). Prophase was judged based on the fact that the nuclear envelope was not yet broken down but chromosome condensation was detectable with phosphorylated histone H3 staining (Supplementary Fig. S2C). Vasa staining also helped identifying the mitotic phases, since Vasa localizes to the cytoplasm, with prominent association around the nuclear envelope before it breaks down, then evenly distributed within the cell after the nuclear envelope breakdown. During prophase, the level of cyclin A in the nucleus was comparable to that in the cytoplasm, clearly demonstrating that cyclin A was now released into the nucleus. Prometaphase was judged based on the fact that the nuclear envelope was broken down but the metaphase plate was not yet formed (Fig. 3E). The release of cyclin A into the cytoplasm and nucleus during early mitosis was not due to structural perturbation of the spectrosome, because the spectrosome structure (positive for Adducin-like, a major structural component of the spectrosome/fusome) was maintained throughout the cell cycle, including mitosis (Fig. 3A). The entire depth of the GSCs was scanned to determine whether any residual spectrosomal localization of cyclin A exists, but we did not detect such localization of cyclin A during mitosis. Cyclin A was quickly degraded by metaphase (Fig. 3F). This is consistent with previous reports in many cell types demonstrating that cyclin A degradation precedes anaphase onset (Fry and Yamano, 2006; Tin Su, 2001). Interestingly, we noted that high cytoplasmic/nuclear levels of cyclin A were always associated with a minimal (or undetectable) spectrosomal level of Par-1 (Fig. 3D”, E”). Localization change of cyclin A during mitosis is also detailed in Supplementary Fig. S2. Together, we concluded that Par-1 and cyclin A show dynamic localization changes during the cell cycle, with the release of cyclin A from the spectrosome coinciding with a diminished amount of Par-1 at the spectrosome.
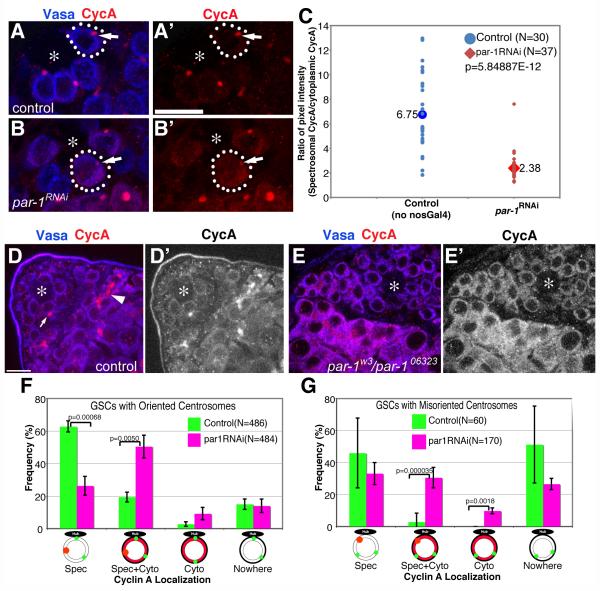
Cyclin A localization is perturbed in par-1 mutant GSCs
To address the potential functional relationship between cyclin A and Par-1, we examined cyclin A localization in par-1 mutant GSCs. First, we noted that cyclin A protein was more frequently localized to the cytoplasm in par-1RNAi and par-1w3/par-1k06323 mutant GSCs (Fig. 4B, E) than in control GSCs (Fig. 4A, D). Because spectrosomal cyclin A was not completely abolished in par-1RNAi GSCs (Fig. 4B, arrow), we compared the ratio of cytoplasmic cyclin A to spectrosomal cyclin A in control vs. par-1RNAi GSCs by randomly choosing GSCs with any detectable cyclin A (i.e., not in metaphase-telophase or G1/early S phase; Fig. 4C). For this analysis, the ratio was measured for every single GSC from the chosen frames to avoid any bias: for example, in Fig. 4B, the unmarked GSC on the top right of marked GSC appears to have more prominent spectrosomal cyclin A. Such cells were also counted to obtain the data shown in Fig. 4C. The frequency of GSCs with any detectable cyclin A was not significantly different in control vs. par-1RNAi GSCs. While control GSCs showed a high concentration of cyclin A in the spectrosome (spectrosome/cytoplasm ratio was 6.75), par-1RNAi GSCs showed a concentration of spectrosomal cyclin A that was only 2.38 times higher than that of cytoplasmic cyclin A. The cytoplasmic localization of cyclin A was somewhat clearer in par-1w3/par-1k06323 mutant testes (Fig. 4D, E). The reduction in the spectrosomal cyclin A is likely due to cyclin A relocalization to the cytoplasm rather than reduced cyclin A protein levels because cyclin A protein levels were not significantly changed in par-1 mutant testes (Supplementary Fig. S3A). These results strongly suggest that Par-1 is required for cyclin A localization to the spectrosome. A similar tendency for cyclin A to localize to the cytoplasm was also observed in spermatogonial cells (Fig. 4E). It is unlikely that the spectrosome integrity is disrupted in general in par-1 mutant GSCs and the defective cyclin A localization is only secondary to such a structural defect of the spectrosome: Adducin-like/Hts and Shaggy (Morin et al., 2001), two known spectrosomal components, were correctly localized to the spectrosome (Supplementary Fig. S4), suggesting that cyclin A is a specific spectrosomal component that is affected in par-1 mutant GSCs.
Fig. 4. Cyclin A localization is perturbed in par-1RNAi mutant GSCs.
(A and B) Cyclin A localization in control (A) vs. par-1RNAi (B) GSCs. Arrows indicate the spectrosomes. Red: Cyclin A. Blue: Vasa (germ cells). The scale bar is 10 μm and the hub is marked by the asterisk (*). (C) The ratio of pixel intensity of spectrosomal cyclin A/cytoplasmic cyclin A in control vs. par-1RNAi GSCs. The ratio of spectrosomal/cytoplasmic cyclin A was determined by measuring the pixel intensity of cyclin A staining in the spectrosomal/cytoplasmic area, from which the background (hub) pixel intensity was subtracted. While control GSCs showed 6.75 times the spectrosomal concentration of cyclin A, par-1RNAi GSCs showed only 2.38 times the concentration of cyclin A. (D and E) Cyclin A localization in control (D) vs. par-1w3/par-106323 (E) GSCs. (F and G) Summary of cyclin A localization in control vs. par-1RNAi GSCs when centrosomes are oriented (F) or misoriented (G). Data are shown as means ± S.D.
To assess defective cyclin A localization in par-1 mutant GSCs in more detail, we scored the frequency of the cyclin A localization pattern in control vs. par-1RNAi GSCs. First, we focused on GSCs with oriented centrosomes in control vs. par-1RNAi GSCs. In control GSCs with correctly oriented centrosomes, cyclin A was confined to the spectrosome in more than 60% of GSCs (“Spec”, Fig. 4F). By contrast, such spectrosomal localization of cyclin A was observed only in ~25% of par-1RNAi GSCs (“Spec”, Fig. 4F). In control GSCs, approximately 20% had cytoplasmic cyclin A (“Spec + Cyto” and “Cyto”, Fig. 4F), presumably reflecting the cell cycle stage (i.e., late G2). In par-1RNAi GSCs, however, approximately 60% of GSCs showed cytoplasmic cyclin A (“Spec + Cyto” and “Cyto”, Fig. 4F), which is significantly higher than that of the control.
Next, we compared cyclin A localization in control vs. par-1RNAi GSCs when centrosomes were misoriented (Fig. 4G). When centrosomes were misoriented in wild-type GSCs, the frequency of GSCs with cytoplasmic cyclin A was dramatically reduced (less than 3%; “Spec + Cyto” and “Cyto”, Fig. 4G), suggesting that these GSCs were not approaching late G2/mitosis. It should be noted that many (~50%) control GSCs with misoriented centrosomes had no detectable cyclin A (Fig. 4G), implying that either cyclin A protein levels were also being regulated in response to centrosome misorientation or GSCs were being arrested at the stage before cyclin A accumulation (see Discussion for detail). In contrast to control GSCs, par-1RNAi GSCs had a high frequency of cytoplasmic cyclin A, even when centrosomes were misoriented (~40% of misoriented GSCs; “Spec + Cyto” and “Cyto”, Fig. 4G). It is unlikely that the change in cyclin A localization in par-1RNAi GSCs is due to defective cell cycle progression, arresting GSCs at particular cell cycle stage when cyclin A localizes to the cytoplasm, because, as noted above, the mitotic index of control vs. par-1RNAi GSCs was similar. Taken together, these findings suggest that Par-1 is required for correct cyclin A localization to the spectrosome during interphase.
Expression of cyclin A mutants that do not localize to the spectrosome leads to a defective centrosome orientation checkpoint
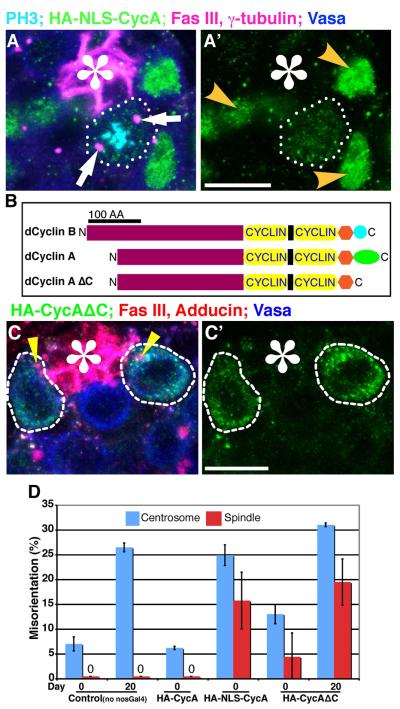
The above data are consistent with the hypothesis that Par-1 prevents precocious mitosis by preventing translocation of cyclin A from the spectrosome to the cytoplasm and nucleus. This hypothesis predicts that cyclin A that is not confined to the spectrosome would promote mitosis irrespective of centrosome orientation. To address this possibility, we first examined the effect of expression of cyclin A with a nuclear localization signal (NLS-Cyclin A) (Dienemann and Sprenger, 2004). It was reported that cyclin A localization is dispensable for mitotic progression during early embryogenesis (Dienemann and Sprenger, 2004). When NLS-Cyclin A was overexpressed in GSCs (nos-Gal4>UAS-HA-NLS-Cyclin A), GSCs frequently underwent mitosis with misoriented spindles (Fig. 5A, D and Supplementary Table S1). Importantly, expressing wild-type cyclin A (HA-Cyclin A) caused no defect in centrosome or spindle orientation, similar to control flies (Fig. 5D. Since there was no difference between control and those expressing HA-Cyclin A, the day 20 data point for HA-Cyclin A is not shown). These results suggest that the centrosome orientation checkpoint is abrogated upon expression of NLS-Cyclin A.
Fig. 5. Cyclin A localization is critical for the centrosome orientation checkpoint.
(A) An example of a misoriented spindle in GSC-overexpressing NLS-Cyclin A (nos-gal4>HA-NLS-Cyclin A). Arrows indicate spindle poles of the misoriented spindle. Nuclear localization is clear in interphase GSCs (yellow arrowheads). Green: HA. Blue: Vasa(germ cells). Red: Fas III (hub, *) and γ-tubulin (centrosome). The scale bar is 10 μm. (B) Schematic of cyclin A and cyclin B structures showing high similarity except for the C-terminal region. “Cyclin” indicates the cyclin folds. (C) Cyclin AΔC does not localize to the spectrosome (indicated by yellow arrowheads) and is in the cytoplasm. Green: HA. Blue: Vasa (germ cells). Red: Fas III (hub, *) and Adducin-like (spectrosome). The scale bar is 10 μm. (D) Summary of misoriented centrosomes and spindles upon expression of NLS-HA-Cyclin A or HA-Cyclin AΔC. The centrosome and spindle orientation phenotype in GSCs expressing HA-Cyclin AΔC becomes more obvious at age day 20, while the phenotype of GSCs expressing HA-NLS-CycA is already obvious at age day 0, and GSCs expressing HA-CycA was comparable with the control at any time point (day 20 not shown). Data are shown as means ± S.D.
However, it is possible that nuclear-localized cyclin A is accelerating the GSC cell cycle so that GSCs do not have enough time to correct misoriented centrosomes, resulting in misoriented spindles. Thus, to further investigate the function of spectrosomal localization of cyclin A, we first determined a region of cyclin A protein that is required for correct spectrosome localization. We found that the 44 amino acids at the C-terminal region of cyclin A are essential for spectrosome localization. This C-terminal region is not conserved in cyclin B protein (Fig. 5B), which does not localize to the spectrosome, despite the high homology between cyclins A and B. When expressed in germ cells, cyclin A without the C-terminal region (Cyclin AΔC) localized to the cytoplasm (nos-gal4>UAS-HA-Cyclin AΔC; Fig. 5C) and, strikingly, resulted in mitosis with misoriented spindles in GSCs upon overexpression (Fig. 5D), indicating that GSCs expressing Cyclin AΔC are defective in the centrosome orientation checkpoint. These results suggest the importance of cyclin A localization in the centrosome orientation checkpoint. Interestingly, NLS-Cyclin A was more potent in inducing interphase centrosome misorientation as well as mitotic spindle misorientation than Cyclin AΔC, possibly because of its constitutive localization to the nucleus. In contrast, centrosome and spindle misorientation was moderate upon expression of Cyclin AΔC; however, misorientation became more apparent at 20 days of age. The difference might be explained by that NLS-Cyclin A indeed accelerates GSC mitosis to some extent, while Cyclin AΔC only abrogates the centrosome orientation checkpoint without driving GSCs into mitosis. Importantly, both HA-NLS-Cyclin A and HA-Cyclin AΔC were degraded at the right timing (Supplementary Fig. S5), suggesting that altered timing of degradation due to their changed localization is not the reason for inducing mitosis with misoriented spindles. Taken together, these data suggest that Cyclin A localization is critical in the centrosome orientation checkpoint.
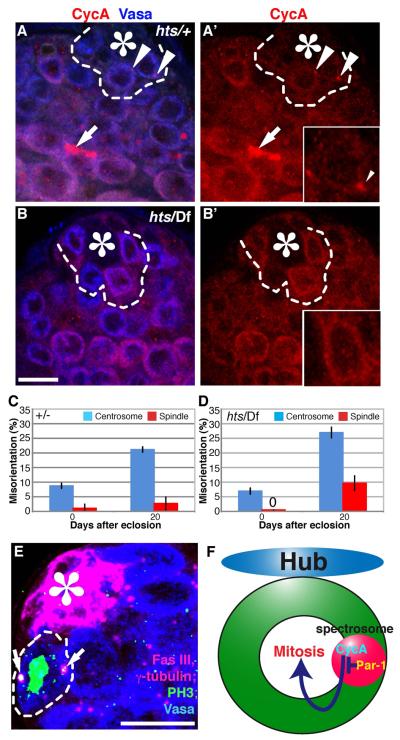
The hts mutant is defective in the centrosome orientation checkpoint
The above data indicate the importance of the spectrosome in the centrosome orientation checkpoint. To address the functional importance of the spectrosome structure in the centrosome orientation checkpoint, we examined the hu li tai shao (hts) mutant. Hts, or Adducin-like, is an integral component of the spectrosome/fusome, and hts mutants lack any detectable spectrosome/fusome structure (Lin et al., 1994). Indeed, cyclin A was completely cytoplasmic in hts mutants (Fig. 6A, B). Strikingly, hts mutants showed elevated spindle misorientation at 20 days of age, without a significant increase in centrosome misorientation compared with the control (Fig. 6C, D, E). Spindle misorientation was not observed in young hts mutant flies, presumably due to a low frequency of centrosome misorientation at this age. This implies that spectrosome is not required for correct centrosome orientation but only for the centrosome orientation checkpoint. Taken together, these data collectively suggest that the spectrosome functions to confine cyclin A and that cyclin A localization to the spectrosome is a critical step in the centrosome orientation checkpoint.
Fig. 6. hts mutation affects the centrosome orientation checkpoint.
(A and B) Cyclin A localization in control (A) vs. hts01103/Df(2R)BSC26 (B) testes. Cyclin A localizes to the cytoplasm in hts mutants. Arrowhead in (A) indicates the spectrosome, while arrow indicates the fusome. GSCs are indicated by dotted lines. Red: Cyclin A. Blue: Vasa (germ cells). The scale bar is 10 μm and the hub is marked by the asterisk (*). The high magnification images of cyclin A channel for selected GSCs are shown (insets). (C ) Centrosome and spindle misorientation in control (heterozygous siblings from the cross of hts/CyO × Df(2R)BSC26/CyO) and hts mutant flies at ages day 0 and day 20. hts mutants are defective in the centrosome orientation checkpoint but not in centrosome orientation. Data are shown as means ± S.D. (D) Example of a misoriented spindle in hts01103/Df(2R)BSC26 testis. Green: PH3 (mitotic chromosomes). Red: Fas III (hub, *) and γ-tubulin (centrosome). Blue: Vasa (germ cells). (E) Model: Par-1 confines cyclin A in the spectrosome, thereby delaying mitotic progression.
Dsas-4, a centriolar component, is not required for the centrosome orientation checkpoint
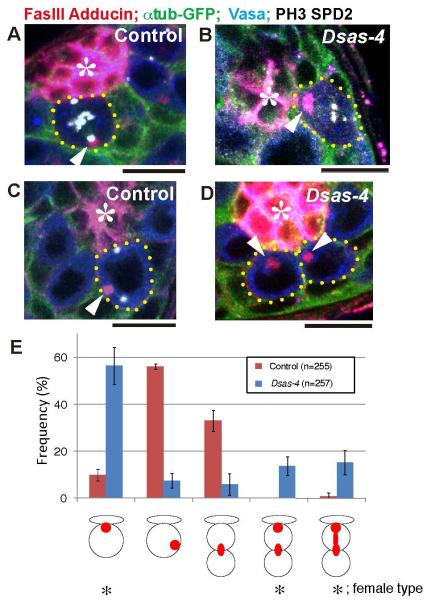
Recently, it was reported that mutants of Dsas-4 do not exhibit misoriented spindle (Riparbelli and Callaini, 2011). Dsas-4 is a core centriole component and its mutant is completely devoid of the centrosomes. It should be noted that the centrosome orientation checkpoint that we propose based on our data is the mechanism that monitors the position of the centrosome, and Dsas-4 mutant does not contain the very structure to be monitored. However, the lack of the spindle misorientation in Dsas-4 mutant raised a question on the importance of the centrosome in spindle orientation of male GSC division. Thus, we examined the spindle orientation in Dsas-4 mutant in detail. First, we confirmed that GSC spindle is correctly oriented toward the hub cells in Dsas-4 mutant as in wild type (Fig. 7A, B). Interestingly, in wild type GSCs, we consistently observed that the spectrosome is associated with the distal spindle pole (Fig. 7A, arrowhead), opposite to the female, where the spectrosome is associated with the proximal (apical) spindle pole (Deng and Lin, 1997). In female GSCs, the apically localized spectrosome is required to anchor the spindle pole and to orient the mitotic spindle. In Dsas-4 mutant male GSCs, the spectrosome was consistently observed at the apical spindle pole, close to the hub cells (Fig. 7B, arrowhead), which is the opposite to the wild type male GSCs, and similar to the wild type female GSCs. Closer inspection revealed that the spectrosome localization pattern in Dsas-4 mutant male GSCs is more similar to that of wild type female GSCs, rather than wild type male GSCs. In interphase wild type male GSCs, the spectrosome is localized at a random place (Fig. 7C, arrowhed) (Yamashita et al., 2003). However, in Dsas-4 mutant male GSCs, the spectrosome was consistently associated with the hub-GSC interface (Fig. 7D, arrowhead). The spectrosome localization pattern in wild type vs. Dsas-4 mutant GSCs is summarized in Fig. 7E. Taken together, these data suggest that, in the complete absence of the centrosomes, the spectrosome apparently functions as a “back up” mechanism to orient mitotic spindle, and able to orient mitotic spindle correctly in Dsas-4 mutant male GSCs.
Fig. 7. Proximal pole of the oriented spindle is associated with the spectrosome in Dsas-4 mutant.
(A) In control male GSCs, spindle is oriented and the spectrosome is associated with the distal spindle pole. Arrowheads indicate the spectrosome-associated spindle pole. Red: Fas III (hub, *) and Adducin-like (spectrosome). Green: GFP-α-tubulin. Blue: Vasa (germ cells). White: phospho-histone H3 (PH3) and Spd-2 (centrosome). Bar: 10μm. (B) In the Dsas-4 mutant GSCs, spindle is correctly oriented, but the spectrosome is associated with the proximal spindle pole. (C) In interphase wild type GSCs, the spectrosome (arrowhead) is not associated with the hub-GSC interface. (D) In interphase Dsas-4 mutant GSCs, the spectrosome (arrowheads) is consistently associated with the hub-GSC interface. (E) The summary of spectrosome localization pattern in control vs. Dsas-4 mutant GSCs. Female type spectrosome localization is indicated by asterisks.
Discussion
Here we show that Par-1 acts as a component of the centrosome orientation checkpoint, probably through its ability to influence cyclin A localization. This checkpoint ensures the asymmetric outcome of GSC division by delaying cell cycle progression when centrosomes are not correctly oriented. Such a checkpoint would provide an additional layer of accuracy in oriented stem cell division. Our study highlights the importance of cyclin A localization in the centrosome orientation checkpoint. Intriguingly, it was reported that in cultured mammalian cells, cyclin A is confined to the endoplasmic reticulum (ER) via its interaction with a protein called SCAPER (Tsang et al., 2007). The spectrosome/fusome has been shown to be a part of the ER (Snapp et al., 2004), therefore, regulation of cyclin A through its localization is likely evolutionarily conserved.
The fact that the wild type misoriented GSCs tend to have lower/non-detectable cyclin A levels (Fig. 4) suggests that GSCs degrade cyclin A or that the arrest point of the centrosome orientation checkpoint is before cyclin A accumulation. It is possible that distinct mechanisms stall the cell cycle, depending on when the centrosome misorientation is sensed. For example, when the centrosome misorientation is detected earlier in the cell cycle (i.e., before cyclin A accumulation), the cell cycle would be stalled before cyclin A protein synthesis/accumulation. In contrast, when the centrosome misorientation is detected later in the cell cycle (i.e., after cyclin A accumulation), the cell cycle would be stalled by preventing translocation of cyclin A from the spectrosome to the cytoplasm/nucleus. Further studies are required to dissect the detailed mechanisms that monitor centrosome orientation, possibly depending on the cell cycle stage.
It is currently unclear how Par-1 might regulate cyclin A localization in response to centrosome misorientation. Direct interaction between Par-1 and cyclin A was not detected in immunoprecipitation experiments (Supplementary Fig. S3B), thus the molecular mechanism by which Par-1 regulates cyclin A localization to the spectrosome/fusome remains to be determined. It is formally possible that cyclin A mislocalization and the defective checkpoint response are two unrelated consequences of par-1 mutation. However, considering that the expression of cyclin A mutant proteins defective in spectrosome localization is sufficient to perturb the centrosome orientation checkpoint, we favor the possibility that cyclin A is indeed part of a Par-1-dependent checkpoint response to centrosome misorientation. Future identification of proteins that recruit/anchor cyclin A to the spectrosome will provide further insight into this process.
We have shown that the mother centrosome is consistently located at the hub-GSC interface, while the daughter centrosome migrates to the opposite side (Yamashita et al., 2007). Whether the centrosome orientation checkpoint monitors the correct positioning of the mother centrosome or any centrosome is currently unknown. However, given that dedifferentiated GSCs, which must have lost their “original” mother centrosome (generated earlier during development) when they committed to differentiation, still retain the centrosome orientation checkpoint (Cheng et al., 2008), the centrosome orientation checkpoint does not appear to monitor the presence of “original” mother centrosomes. It is still possible that the centrosome orientation checkpoint monitors the presence of “mature” centrosomes (not necessarily from earlier in development, but >2 cell cycle-old centrosomes) at the hub-GSC interface. Interestingly, it was recently shown that the daughter centrosome is consistently inherited by stem cells during the divisions of Drosophila neuroblast (Conduit and Raff, 2010; Januschke et al., 2011). Given the precise inheritance of mother or daughter centrosomes depending on the context/stem cell system, it is tempting to speculate that the centrosome orientation checkpoint monitors the presence of the mother centrosome in male GSCs, and possibly an equivalent mechanism monitors the daughter centrosome inheritance in neuroblasts.
In developing embryos, cyclin A localization was reported to be dispensable for its activity (Dienemann and Sprenger, 2004). Even the plasma membrane-bound form of cyclin A was shown to be able to fulfill its function to promote mitosis. Indeed, the mutant forms of cyclin A protein used in this study (NLS-CycA and Cyclin AΔC) are “functional” in that they can promote the cell cycle progression into mitosis. Instead, we propose that these cyclin A mutant proteins cannot be subjected to a negative regulation by Par-1. It is possible that the embryonic cell cycle has minimal negative regulation as in embryonic stem cells, while male GSCs have an additional regulatory step (i.e., the centrosome orientation checkpoint) that negatively regulates mitotic entry.
The lack of spindle misorientation in Dsas-4 mutant male GSCs is intriguing. In the complete absence of the centrosome, the spindle was correctly oriented in dividing GSCs (Riparbelli and Callaini, 2011) (Fig. 7B), while defective centrosome function in cnn mutant leads to abrogation of the centrosome orientation checkpoint (Inaba et al., 2010). Dsas-4 mutant male GSCs apparently orient the mitotic spindle via anchorage of spindle pole to the apically-localized spectrosome, which is highly reminiscent to spindle orientation mechanism in female GSCs (Deng and Lin, 1997). The prediction would be that the spindle orientation is randomized in Dsas-4 hts double mutant male GSCs, which lacks both the centrosome and spectrosome. Unfortunately, the analysis of the double mutant was technically very challenging; Dsas-4 single mutant flies die as pharate adult, and the survival of the double mutant was worse. Furthermore, we were never able to observe any mitotic GSCs from those pharate adult double mutants that we managed to recover and analyze. Thus, future studies will be required to test this prediction.
Our study illuminates the importance of stem cell-specific regulators of the general cell cycle machinery such as cyclin A. We propose that stem cells have developed elaborate mechanisms to ensure an asymmetric outcome of the stem cell division, the failure of which can lead to tumorigenesis or tissue degeneration.
Supplementary Material
Highlights.
Identified Par-1 kinase as a component of the centrosome orientation checkpoint
Cyclin A is a downstream target of the centrosome orientation checkpoint
Cyclin A localization is essential for the centrosome orientation checkpoint
Spectrosome as a critical organelle for the centrosome orientation checkpoint
Acknowledgments
We are grateful to Bingwei Lu, Frank Sprenger, Daniel St. Johnston, Hiroki Oda, the FlyTrap project, the Bloomington Stock Center, the Developmental Studies Hybridoma Bank for reagents, and members of the Yamashita laboratory for helpful discussions. This research was supported by a University of Michigan start-up fund, the March of Dimes Basil O’Conner Starter Scholar Research Award, the Searle Scholar Program, NIH R01GM086481 (to Y.M.Y.), and the University of Michigan Center for Organogenesis postdoctoral fellowship (to H.Y.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Benton R, Palacios IM, Johnston DS. Drosophila 14-3-3/PAR-5 Is an Essential Mediator of PAR-1 Function in Axis Formation. Dev Cell. 2002;3:659–671. doi: 10.1016/s1534-5807(02)00320-9. [DOI] [PubMed] [Google Scholar]
- Benton R, St Johnston D. Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 2003;115:691–704. doi: 10.1016/s0092-8674(03)00938-3. [DOI] [PubMed] [Google Scholar]
- Burke DJ. Interpreting spatial information and regulating mitosis in response to spindle orientation. Genes Dev. 2009;23:1613–1618. doi: 10.1101/gad.1826409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, Skora AD, Nystul TG, Ohlstein B, Allen A, Wilhelm JE, Murphy TD, Levis RW, Matunis E, Srivali N, Hoskins RA, Spradling AC. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 2007;175:1505–1531. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeks RJ, Canman JC, Gabriel WN, Meyer N, Strome S, Goldstein B. C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Curr Biol. 2004;14:851–862. doi: 10.1016/j.cub.2004.05.022. [DOI] [PubMed] [Google Scholar]
- Cheng J, Turkel N, Hemati N, Fuller MT, Hunt AJ, Yamashita YM. Centrosome misorientation reduces stem cell division during ageing. Nature. 2008;456:599–604. doi: 10.1038/nature07386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit PT, Raff JW. Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in drosophila neuroblasts. Curr Biol. 2010;20:2187–2192. doi: 10.1016/j.cub.2010.11.055. [DOI] [PubMed] [Google Scholar]
- Cox DN, Lu B, Sun TQ, Williams LT, Jan YN. Drosophila par-1 is required for oocyte differentiation and microtubule organization. Curr Biol. 2001;11:75–87. doi: 10.1016/s0960-9822(01)00027-6. [DOI] [PubMed] [Google Scholar]
- de Cuevas M, Lilly MA, Spradling AC. Germline cyst formation in Drosophila. Annu Rev Genet. 1997;31:405–428. doi: 10.1146/annurev.genet.31.1.405. [DOI] [PubMed] [Google Scholar]
- Deng W, Lin H. Spectrosomes and fusomes anchor mitotic spindles during asymmetric germ cell divisions and facilitate the formation of a polarized microtubule array for oocyte specification in Drosophila. Dev Biol. 1997;189:79–94. doi: 10.1006/dbio.1997.8669. [DOI] [PubMed] [Google Scholar]
- Dienemann A, Sprenger F. Requirements of cyclin a for mitosis are independent of its subcellular localization. Curr Biol. 2004;14:1117–1123. doi: 10.1016/j.cub.2004.06.024. [DOI] [PubMed] [Google Scholar]
- Fry AM, Yamano H. APC/C-mediated degradation in early mitosis: how to avoid spindle assembly checkpoint inhibition. Cell Cycle. 2006;5:1487–1491. doi: 10.4161/cc.5.14.3003. [DOI] [PubMed] [Google Scholar]
- Huynh JR, Shulman JM, Benton R, St Johnston D. PAR-1 is required for the maintenance of oocyte fate in Drosophila. Development. 2001;128:1201–1209. doi: 10.1242/dev.128.7.1201. [DOI] [PubMed] [Google Scholar]
- Inaba M, Yuan H, Salzmann V, Fuller MT, Yamashita YM. E-cadherin is required for centrosome and spindle orientation in Drosophila male germline stem cells. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0012473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J, Llamazares S, Reina J, Gonzalez C. Drosophila neuroblasts retain the daughter centrosome. Nat Commun. 2011;2:243. doi: 10.1038/ncomms1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelso RJ, Buszczak M, Quinones AT, Castiblanco C, Mazzalupo S, Cooley L. Flytrap, a database documenting a GFP protein-trap insertion screen in Drosophila melanogaster. Nucleic Acids Res. 2004;32:D418–420. doi: 10.1093/nar/gkh014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294:2542–2545. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- Leatherman JL, Dinardo S. Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell. 2008;3:44–54. doi: 10.1016/j.stem.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherman JL, Dinardo S. Germline self-renewal requires cyst stem cells and stat regulates niche adhesion in Drosophila testes. Nat Cell Biol. 2010;12:806–811. doi: 10.1038/ncb2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lighthouse DV, Buszczak M, Spradling AC. New components of the Drosophila fusome suggest it plays novel roles in signaling and transport. Dev Biol. 2008;317:59–71. doi: 10.1016/j.ydbio.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly MA, de Cuevas M, Spradling AC. Cyclin A associates with the fusome during germline cyst formation in the Drosophila ovary. Dev Biol. 2000;218:53–63. doi: 10.1006/dbio.1999.9570. [DOI] [PubMed] [Google Scholar]
- Lin H, Yue L, Spradling AC. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development. 1994;120:947–956. doi: 10.1242/dev.120.4.947. [DOI] [PubMed] [Google Scholar]
- Morin X, Daneman R, Zavortink M, Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc Natl Acad Sci U S A. 2001;98:15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Tsukita S. Nonchordate classic cadherins have a structurally and functionally unique domain that is absent from chordate classic cadherins. Dev Biol. 1999;216:406–422. doi: 10.1006/dbio.1999.9494. [DOI] [PubMed] [Google Scholar]
- Rebollo E, Sampaio P, Januschke J, Llamazares S, Varmark H, Gonzalez C. Functionally Unequal Centrosomes Drive Spindle Orientation in Asymmetrically Dividing Drosophila Neural Stem Cells. Dev Cell. 2007;12:467–474. doi: 10.1016/j.devcel.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Riparbelli MG, Callaini G. Male gametogenesis without centrioles. Dev Biol. 2011;349:427–439. doi: 10.1016/j.ydbio.2010.10.021. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Vectors for P element-mediated gene transfer in Drosophila. Nucleic Acids Res. 1983;11:6341–6351. doi: 10.1093/nar/11.18.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan NM, Peifer M. A role for a novel centrosome cycle in asymmetric cell division. J Cell Biol. 2007;177:13–20. doi: 10.1083/jcb.200612140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman JM, Benton R, St Johnston D. The Drosophila homolog of C. elegans PAR-1 organizes the oocyte cytoskeleton and directs oskar mRNA localization to the posterior pole. Cell. 2000;101:377–388. doi: 10.1016/s0092-8674(00)80848-x. [DOI] [PubMed] [Google Scholar]
- Snapp EL, Iida T, Frescas D, Lippincott-Schwartz J, Lilly MA. The fusome mediates intercellular endoplasmic reticulum connectivity in Drosophila ovarian cysts. Mol Biol Cell. 2004;15:4512–4521. doi: 10.1091/mbc.E04-06-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D, Ahringer J. Cell polarity in eggs and epithelia: parallels and diversity. Cell. 2010;141:757–774. doi: 10.1016/j.cell.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Tin Su T. Cell cycle: how, when and why cells get rid of cyclin A. Curr Biol. 2001;11:R467–469. doi: 10.1016/s0960-9822(01)00283-4. [DOI] [PubMed] [Google Scholar]
- Tsang WY, Wang L, Chen Z, Sanchez I, Dynlacht BD. SCAPER, a novel cyclin A-interacting protein that regulates cell cycle progression. J Cell Biol. 2007;178:621–633. doi: 10.1083/jcb.200701166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulina N, Matunis E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science. 2001;294:2546–2549. doi: 10.1126/science.1066700. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8:243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- Voog J, D’Alterio C, Jones DL. Multipotent somatic stem cells contribute to the stem cell niche in the Drosophila testis. Nature. 2008;454:1132–1136. doi: 10.1038/nature07173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tsai JW, Imai JH, Lian WN, Vallee RB, Shi SH. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM. Cell adhesion in regulation of asymmetric stem cell division. Curr Opin Cell Biol. 2010;22:605–610. doi: 10.1016/j.ceb.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM, Jones DL, Fuller MT. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 2003;301:1547–1550. doi: 10.1126/science.1087795. [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Guo H, Kwan H, Wang JW, Kosek J, Lu B. PAR-1 kinase phosphorylates Dlg and regulates its postsynaptic targeting at the Drosophila neuromuscular junction. Neuron. 2007;53:201–215. doi: 10.1016/j.neuron.2006.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.