Abstract
NSP and Cas family proteins form multidomain signaling platforms that mediate cell migration and invasion through a collection of distinct signaling motifs. Members of each family interact via their respective C-terminal domains, but the mechanism of this association has remained enigmatic. Here we present the crystal structures of the C-terminal domain from the human NSP protein BCAR3 and the complex of NSP3 with p130Cas. BCAR3 adopts the Cdc25-homology fold of Ras GTPase exchange factors, but exhibits a “closed” conformation incapable of enzymatic activity. The NSP3–p130Cas complex structure reveals that this closed conformation is instrumental for interaction of NSP proteins with a focal adhesion-targeting domain present in Cas proteins. This enzyme to adaptor conversion enables high affinity, yet promiscuous, interactions between NSP and Cas proteins and represents an unprecedented mechanistic paradigm linking cellular signaling networks.
INTRODUCTION
The integration of signals triggered by diverse stimuli is essential for cell communication in higher organisms and is primarily achieved through the assembly of multiprotein signaling nodes. The Crk-associated substrate (Cas) and novel SH2-containing protein (NSP) families form one such class of signaling nodes, and correspondingly harbor multiple protein-protein interaction domains and motifs that mediate association with an abundance of cell signaling factors. This allows them to function as mediators that integrate signals emanating from cell adhesion and environmental stimuli to promote cell migration, adhesion and invasion. As such, disregulation of signaling through Cas and NSP proteins is relevant to a spectrum of disease processes, particularly tumor progression and cancer metastasis.
The complexity of NSP-Cas functions is best exemplified by two prominent family members p130Cas and the NSP protein BCAR3, which are also known for their ability to confer antiestrogen resistance in breast cancer1,2. Firmly embedded in the Src-Crk signaling axis, p130Cas interacts with more than a dozen signaling factors besides Src family kinases and Crk, including FAK, PYK2, FRNK, RapGEF1, Aurora kinase A, PI3K, NMP4, NCK1 and SHIP2 (reviewed in ref 3). BCAR3 is associated with enhanced PI3K activity downstream of growth factor receptors and subsequent activation of Rac, Cdc42, AKT and PAK14. Besides this spectrum of signaling partners, p130Cas and BCAR3 also bind directly to each other5, thereby linking their respective signaling networks to form a platform that mediates migratory signaling. In addition to BCAR3, p130Cas is also capable of interacting with NSP family members NSP3 and NSP16,7. On the other hand, BCAR3 has been shown to interact with HEF1 (Cas-L) in addition to p130Cas8. Further combinations are created by association of the NSP family members NSP3 and NSP1 with the Cas proteins Cas-L and Efs, adding to the diversity of NSP–Cas signaling networks9–11. NSP and Cas family proteins are therefore capable of propagating class-specific but promiscuous combinatorial networks, which are each essential to allow their diverse functions in specialized cellular processes. As such the effect of aberrant NSP–Cas signaling is manifested in a range of disease processes. p130Cas and BCAR3 have long been known to exert a concerted effect in cancer12,13, while the NSP3 (SHEP1)–Cas-L signaling node has recently been found to be crucial for B-cell migration and maturation9,11. Additionally the NSP3–p130Cas module is reported to be essential in neuronal cells in for proper olfactory development, and when absent causes a phenotype reminiscent of the human developmental disorder Kallmann syndrome14. These examples underline the central importance of distinct NSP–Cas modules in cellular processes that require integration of adhesion and chemotactic stimuli.
While it is known that the respective C-terminal domains of Cas and NSP proteins are fundamental to their interaction (Supplementary Fig. 1a)7, it has remained unclear at the molecular level how different family members can interact in this class-specific yet promiscuous manner. Moreover the predicted nature of the C-terminal Cas and NSP domains suggest an unprecedented interaction mechanism. The NSP-binding portion of Cas proteins has been proposed to assume a focal-adhesion targeting (FAT) domain-like fold, while the C-terminal domain of NSP proteins shares weak homology with Cdc25-homology domains (Cdc25-hd) found in GTPase exchange factors15,16. However, the molecular mechanism of a direct Cdc25-hd:FAT interaction is currently without precedent, and the question of how this influences the putative activities of each domain while mechanically linking NSP and Cas signaling remains outstanding.
To shed light on the molecular features enabling the formation of Cas–NSP signaling modules, we have solved the crystal structures of the C-terminal region of unbound human BCAR3 and of the complex between human NSP3 and p130Cas. These structures in combination with our biochemical and biological analyses show that the Cdc25-homology domain present in NSP proteins has evolved as an adaptor domain based on an enzymatic fold used in RasGEFs. This unique NSP structure is specifically tailored to tightly bind the FAT domain of Cas proteins using a distinctive extended interface. Furthermore, the sophisticated and class-specific interaction patterns observed in the NSP3 and p130Cas interfaces are conserved among the members of both families. As a result, NSP and Cas proteins can promiscuously interact with each other to form various combinations of signaling modules that finely regulate cellular processes.
RESULTS
BCAR3 has an unconventional Cdc25-homology domain
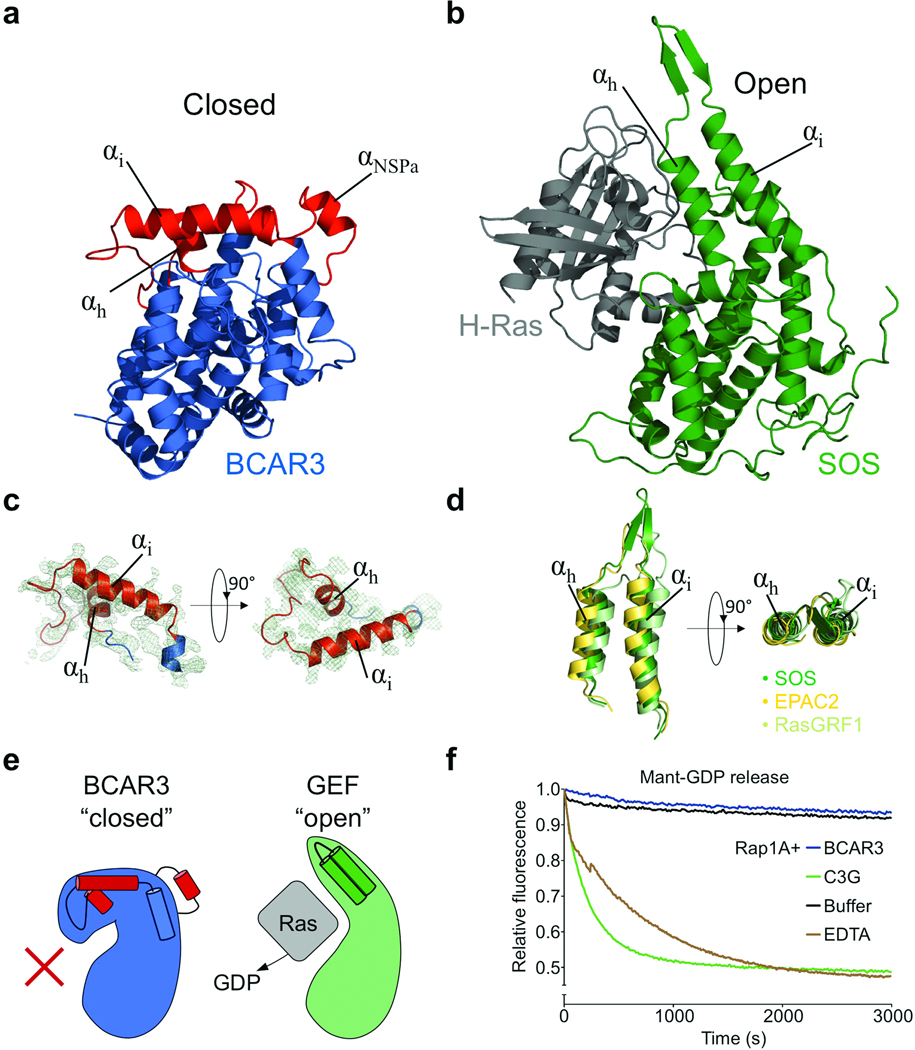
To elucidate the enigmatic nature of NSP protein signaling, we sought to obtain structural and mechanistic insight into the C-terminal domain of the best characterized family member, BCAR3. Thorough in vitro investigation of this domain, or any other NSP C-terminal domain, had previously been prevented by the poor protein chemical properties of this region. After testing various constructs, we succeeded in generating a soluble BCAR3 C-terminal domain (spanning residues 502–825) with properties suitable for biochemical and structural investigation. Extensive screening of crystallization conditions resulted in well-diffracting BCAR3 crystals, which allowed us to solve the structure of BCAR3 at a resolution of 2.4 Å (Fig. 1a). The crystal structure contains four molecules of BCAR3 in the asymmetric unit, which overlay with excellent fidelity (RMSD range 0.24–0.45 Å). The BCAR3 structure reveals an unexpected Ras-type GDP exchange factor architecture that sheds light on the contradictory nature of NSP proteins reported in the literature. BCAR3 adopts the overall fold of the Cdc25-homology domain structures solved to date with RMSD values between the BCAR3 core and SOS, EPAC2 or RasGRF1 of 2.18 Å, 2.35 Å and 2.19 Å respectively17–19 (Supplementary Fig. 1b). However, intriguingly BCAR3 shows dramatic deviations in two distinct regions that result in a novel ”closed” arrangement of the Cdc25-homology domain (Fig. 1, Supplementary Fig. 2). In detail, helices αh and αi (nomenclature derived from SOS17) each bend approximately 60° in orthogonal directions relative to their known GEF counterparts (Fig. 1c,d). Helix αh is also markedly shorter in BCAR3 than in other Cdc25-homology domains, and hydrophobic interactions by Leu713, Val714 and Leu716 cause the helix to be buried between αb and αi. Helix αi is broken into two portions (αi and αi*, Supplementary Fig. 2), with αi folding towards the C-terminus of αf in the Cdc25-homology domain core achieving the closed BCAR3 conformation (Fig. 1). The second markedly different region is comprised of residues 565–592. This “NSP-specific region” adopts a distinct conformation that includes helices αNSPa and αNSPb and is well conserved in all NSP proteins.
Figure 1.
The BCAR3 C-terminal domain resembles a Cdc25-homology domain but adopts a closed conformation incapable of canonical GEF function. (a) Structure of the BCAR3 Cas interaction domain. The C-terminal domain of the NSP BCAR3 overall adopts a Cdc25 homology domain fold (blue) yet crucial regions (red) are altered when compared with the prototypical Cdc25-homology domain of Ras GEFs. Also indicated in red is additional helix αNSPa in a region unique to NSP proteins. (b) SOS in complex with Ras. SOS (green) adopts an open conformation accommodating its target GTPase Ras (grey; PDBid:1bkd)17. (c) Structure of the closed “helical hairpin” in BCAR3 with the 2Fo-Fc electron density map (contoured to 1 σ) displayed from two orthogonal directions. (d) The helical hairpins of SOS, EPAC and RasGRF1. Orientations are identical to those used in panel c. (e) Schematic illustrating conformations of “closed” BCAR3 compared to a canonical “open” GEF conformation. The canonical GTPase interaction site is occluded in the “closed” Cdc25-homology domain of BCAR3 rendering it incapable of canonical GEF function. (f) The Cdc25-homology domain of BCAR3 shows no GEF activity in vitro, tested by comparing the release of mant-GDP from Rap1A, a putative GTPase target for BCAR3, in the presence of BCAR3, C3G, or EDTA.
Comparison with complexes of catalytically competent Cdc25-homology domains bound to their target GTPases shows that BCAR3 assumes a conformation incompatible with classical GEF function because its GTPase binding site is completely occluded (Fig. 1e and Supplementary Fig. 3a). This occlusion is mainly achieved by the closed conformation of helix αi and its preceding linker comprised of BCAR3 residues 718–736. In active “open” Cdc25-homology domains, helices αh and αi form a hairpin structure (Fig. 1) that is crucial for activity because it displaces the switch 1 region of target GTPases17,20. In fact, previous studies show that even slight conformational adjustments in this hairpin have substantial effects on exchange activity19,21. Thus, the dramatic changes observed in BCAR3 imply a drastic functional redirection of crucial GEF catalytic elements.
Closed BCAR3 lacks catalytic activity in vitro
To validate our structural observations and gain further insight into the functional properties of the BCAR3 C-terminal domain, we examined its ability to promote nucleotide exchange in vitro, which has previously been controversial in the literature5,16,22–24. Using BCAR3 502–825 allowed us to assess the ability of BCAR3 to induce GDP release from its putative target GTPase, Rap1A25. In contrast to positive controls with either EDTA or C3G (a bona fide GEF for Rap1A26), which effectively promote GDP release from Rap1A, no activity was observed for BCAR3 (Fig. 1f). Furthermore no interaction between the BCAR3 Cdc25-homology domain and Rap1A was detected in in vitro pulldowns or isothermal titration calorimetry experiments (data not shown). These results show a complete lack of catalytic activity for BCAR3, in accordance with the crystal structure, and reveal that BCAR3 represents a non-catalytic adaption of the Cdc25-homology domain fold.
Structure of the NSP3–p130Cas complex
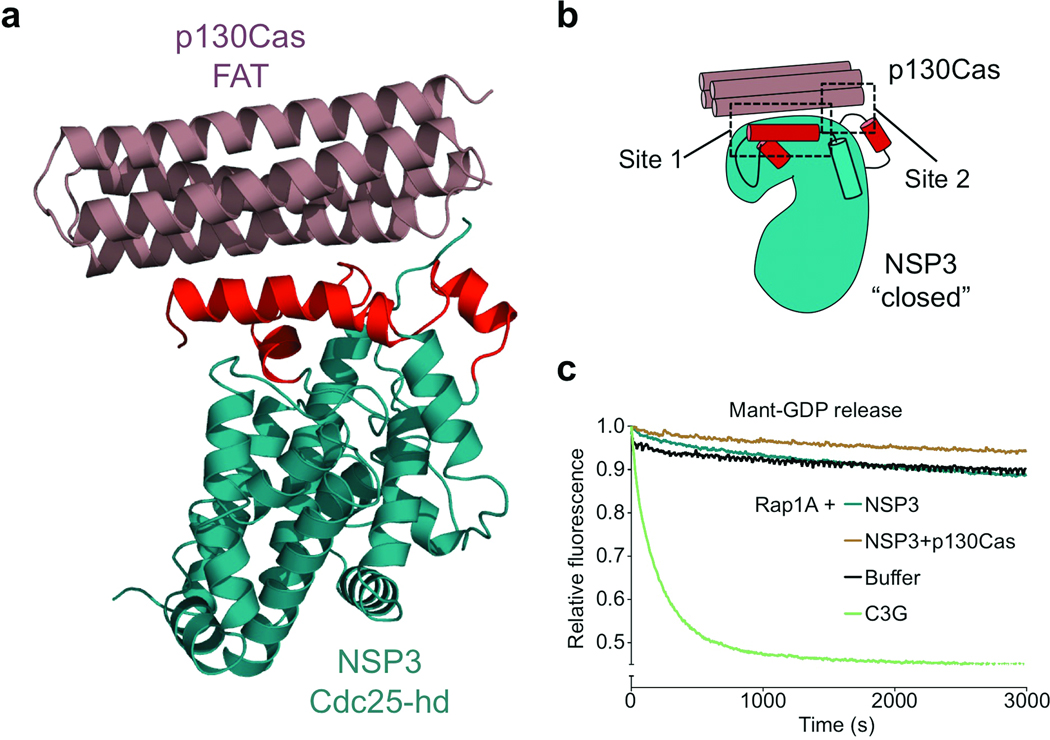
To further understand the function of the observed “closed” NSP conformation and ultimately understand the molecular basis of NSP–Cas signaling modules, we next sought to obtain the structure of an NSP–Cas interaction complex. To this end, we extended our investigations to the BCAR3 relative NSP3 (SHEP1) and the prototypical Cas family member p130Cas. We identified constructs encoding residues 382–703 of NSP3 and 645–870 of p130Cas, which allowed us to generate the individual protein domains as well as complexes of NSP3 or BCAR3 bound to p130Cas, all exhibiting excellent biochemical properties (Supplementary Fig. 3b,c). After extensive crystallization trials using both complexes, we were able to solve the crystal structure of NSP3 bound to p130Cas at a resolution of 2.5 Å (Fig. 2a). The asymmetric unit of the crystal contains two NSP3–p130Cas complexes (Supplementary Fig. 4), one of which is more clearly defined by electron density and is described hereafter. In line with our biochemical characterization (Supplementary Fig. 3b,c and 5a–c), the crystal structure shows that the two proteins form a 1:1 arrangement characterized by an extensive binding interface that buries a total of 1,192 Å2. This substantial interface is in good agreement with isothermal titration calorimetry results, which support a very tight (30 nM) interaction between p130Cas and the closely related NSP protein BCAR3 (Supplementary Fig. 5d).
Figure 2.
The NSP3–p130Cas complex. (a) Crystal structure of the NSP3–p130Cas complex – the closed conformation of the NSP3 Cdc25-homology domain is instrumental in Cas binding. NSP3 C-terminal domain (cyan) resembles a Cdc25-homology domain that adopts the same “closed” conformation observed for unbound BCAR3. The p130Cas C-terminal domain (brown) forms a four-helix bundle typical of focal adhesion targeting (FAT) domains. (b) Schematic representation of the NSP3–p130Cas interaction. The rearranged Cdc25-homology domain elements of NSP3 provide the two primary binding interactions (designated Site 1 and Site 2). (c) NSP3 GEF activity assay in the presence and absence of p130Cas C-terminal domain. NSP3 does not catalyze Rap1A nucleotide exchange either alone or in complex with p130Cas in a nucleotide exchange experiment similar to that shown in Fig. 1f.
In the final model of the NSP3–p130Cas complex (Fig. 2a), NSP3 residues 386–698 are well defined by electron density with the exception of the loop between residues 599 and 612, which was not included in the model. The C-terminal domain of p130Cas is defined by electron density from residues 739 to 872 and adopts the four-helix bundle fold characteristic of FAT domains. The structure of NSP3 closely resembles a Cdc25-homology domain fold, but intriguingly shares the closed conformation observed for unbound BCAR3. A comparative backbone RMSD of 0.97 Å between p130Cas-bound NSP3 and the unbound BCAR3 underscores their extremely close structural similarity (Supplementary Fig. 6a)
The closed NSP Cdc25-homology domain is instrumental for Cas binding
The structure of the NSP3–p130Cas complex reveals that the closed conformation observed for the NSP3 C-terminal domain is key in enabling a Cdc25-hd–FAT domain interaction to form a previously undescribed signaling tether. This unique binding interface utilized by NSP3 and p130Cas can be divided into two main binding motifs (Fig. 3, Supplementary Fig. 7a), which coincide with the two regions that deviate from the canonical Cdc25-homology domain fold of NSP3 (and BCAR3) and are strictly dependent on the closed conformation of the domain. In the first motif, which we dub Site 1, a hydrophobic strip in helix αi of NSP3 interacts with a highly complementary surface on p130Cas. Site 2 is generated by a distinct region between αNSPa and αb of NSP3 and synergizes with Site 1 to generate a tight and unique signaling interaction.
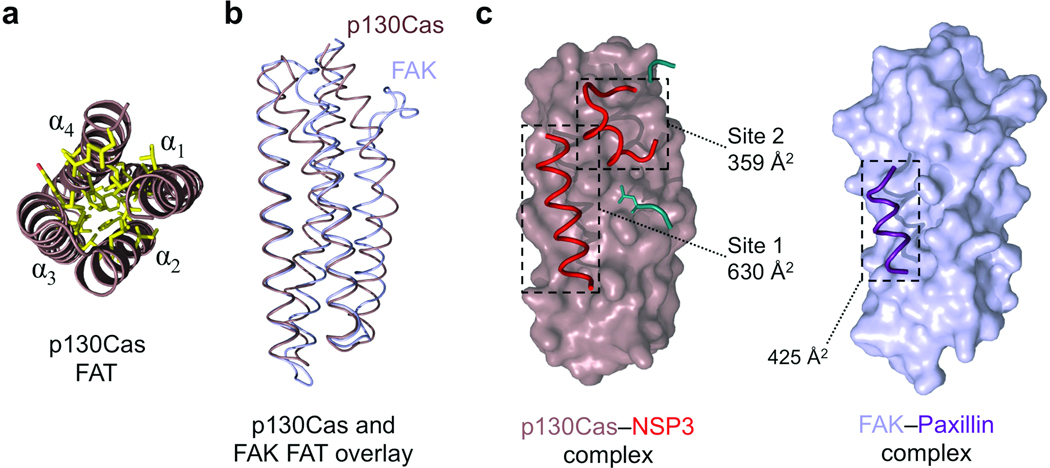
Figure 3.
The FAT domain of p130Cas is well defined and employs a novel extended binding mode. (a) The p130Cas FAT domain core. End-on view of the four helical bundle of p130Cas (brown) showing a close knit hydrophobic network (yellow) stabilizing the core of the helical arrangement. (b) Backbone overlay of p130Cas (brown) with FAK (light blue, from PDB 1ow7)30. (c) The interaction mode used by the FAT domain of p130Cas and NSP3 compared to FAK binding to the Paxillin LD-4 peptide (purple). NSP3 elements contributing to the Site 1 and Site 2 binding motifs are colored red. Buried surface areas for NSP3–p130Cas Site 1, Site 2, and FAK–Paxillin are indicated. Additional NSP3 residues participating in the Cas interaction are colored cyan, and add an extra 233 Å2 in buried surface area (not annotated).
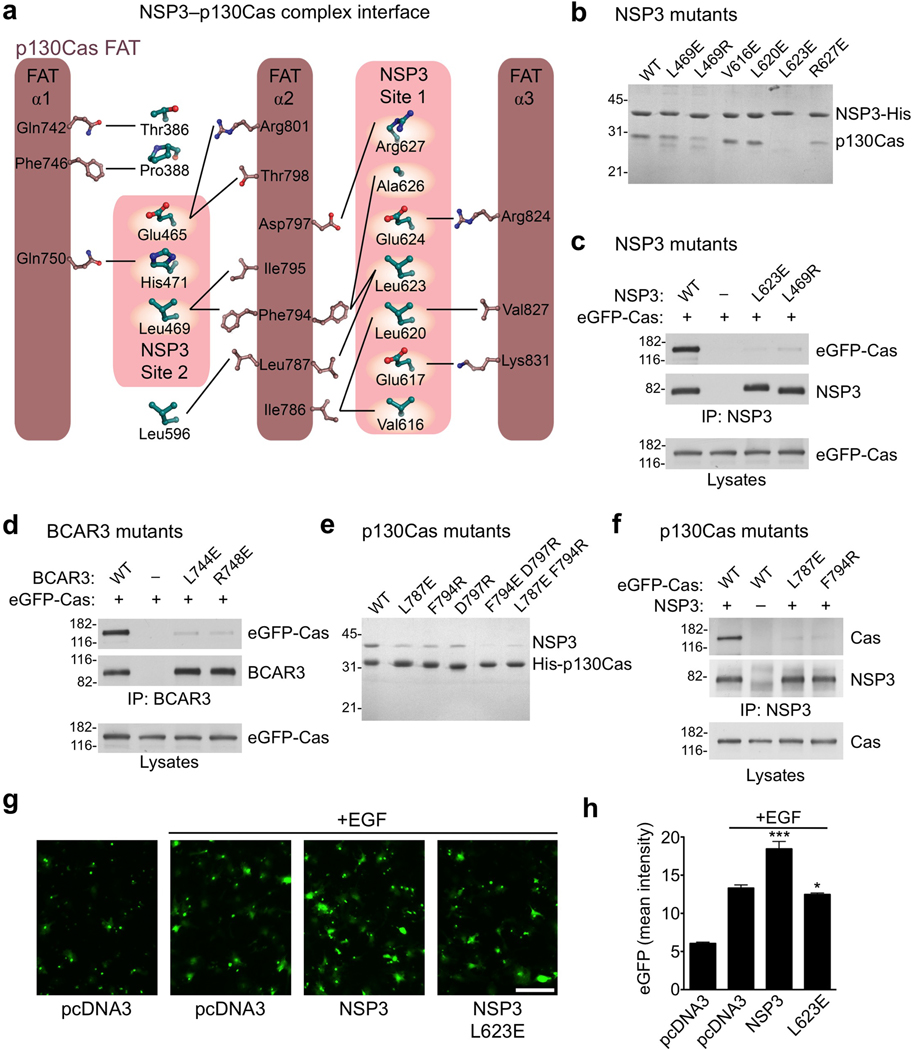
Specifically, in Site 1 Val616, Leu620 and Leu623 on helix αi packs into the hydrophobic groove between α2 and α3 of the p130Cas FAT domain (Fig. 4a, Supplementary Fig. 7a). These contacts are further augmented by charged interactions mediated by Glu617, Glu624 and Arg627 of NSP3. In site 2 helix αNSPb and subsequent residues Glu465 and Leu469-Pro470-His471 interact with a predominantly hydrophobic groove between α1 and α2 of the p130Cas FAT domain. Supplementing the two main interfaces is a peripheral association of residues 386–388 from the N-terminus of the NSP3 Cdc25-homology domain and Leu596 from helix αh of NSP3 (Fig. 3c), which contributes to a hydrophobic surface that links the Site 1 and Site 2 binding motifs.
Figure 4.
Analysis of NSP family-p130Cas interactions in vitro and in vivo. (a) Schematic representation of the interfaces between NSP3 and p130Cas. (b) A subset of well-expressed NSP3 mutants (Cdc25-homology domain) were purified as His6-tagged proteins and assayed for their ability to selectively co-purify untagged WT p130Cas C-terminal domain from E. coli lysates. (c) NSP3 (full-length) mutants L623E and L469R show almost complete loss of p130Cas (full-length) binding, as shown following co-immunoprecipitation with an anti-NSP3 antibody. (d) Residues L744E and R748E in BCAR3 (equivalent to NSP3 residues Leu623 and Arg627 respectively) were examined for interaction with Cas following immunoprecipitation with an anti-BCAR3 antibody. Their near complete loss of p130Cas association in COS cells is in agreement with a conserved mode of binding. (e) Co-purification of recombinant p130Cas mutants with NSP3, performed as shown in panel b, but using p130Cas with a His6–tag and untagged NSP3. (f) p130Cas mutants were examined for their ability to bind NSP3 in COS cells by immunopreciptation with an anti-NSP3 antibody. (g,h) NSP3–Cas interaction promotes chemotaxis towards an EGF chemoattractant in transwell migration assays. (g) Representative images of cells that relocated from the upper to the lower side of the Transwell filter (scale bar, 100 µm). (h) Histogram quantifying the mean eGFP intensity from transfected cells on the lower side of the filters. Error bars indicate the standard deviation from six microscope field measurements. (* P<0.05 and *** P<0.001)
As discussed for BCAR3, the closed NSP3 conformation renders key elements of the Cdc25-homology domain incapable of participating in a canonical interaction with a Ras GTPase. Furthermore, the presence of Cas in the complex provides even greater steric hindrance to such a hypothetical GTPase interaction. To further confirm the validity of this structural observation, we performed in vitro nucleotide exchange assays with both unbound NSP3 and NSP3 in complex with the p130Cas FAT domain. In addition to Rap1A, we included Rap2A as a further putative target GTPase for NSP327. The results do not show any detectable in vitro exchange activity for either unbound or Cas-bound NSP3 (Fig. 2c and Supplementary Fig. 6b). This is in accordance with our structural observations depicting a closed conformation of the NSP3 Cdc25-homology domain as essential for Cas binding.
Cas extends the paradigm of FAT domain interactions
Besides revealing a critical adaption in NSP proteins, the NSP3–p130Cas complex provides the first atomic resolution insight demonstrating that the C-terminal domains of Cas family proteins adopt a well-defined FAT domain type four-helix bundle. Extensive hydrophobic interactions in the core of the domain suggest a stable arrangement of these helices (Fig. 3a). Generally, FAT domains are best known to function as adaptor domains in focal adhesion signaling28, where they interact with binding partners by recruiting helical motifs to surface grooves between their constituent helices29–31. However, these interactions are characteristically weak29,32, whereas we observe a dissociation constant in the low nanomolar range for the binding of BCAR3 to the FAT domain of p130Cas and tight binding between NSP3 and p130Cas (Supplementary Fig. 5). While the tertiary structure of the C-terminal domain of p130Cas is nearly identical to that of other FAT domains (Fig. 3b), p130Cas achieves extremely tight binding to NSP3 by a novel adaptation of the FAT domain interaction mode (Fig. 3c) utilizing the bi-partite binding motifs described above. Firstly the NSP3–p130Cas interaction at Site 1 is accomplished through a binding groove between helices α2 and α3 of the p130Cas FAT domain. This interaction is reminiscent of the α2–α3 binding site used by FAK and Pyk2 to bind paxillin30,31, but p130Cas accommodates a distinctly longer helical stretch from NSP3 and buries a substantially greater surface area of 630 Å2 (Fig. 3c). The interaction at Site 2 on the other hand, has not been observed for other FAT-type four helical bundles. On p130Cas this interface is formed by helix α1 and the C-terminal portion of helix α2 of the FAT scaffold, and in concert with the extended Site 1 interface achieves the unusually tight binding of the p130Cas FAT domain to NSP proteins.
Biochemistry and biology of NSP–Cas complexes
To further verify our structural findings, we performed extensive biochemical and biological mutational studies. We designed structure-based mutations to disrupt binding elements while conserving the integrity of the protein domains (Fig. 4a). All NSP3, BCAR3 and p130Cas mutant domains showed excellent expression and solubility suitable for binding assays. The NSP3 R627E Site 1 mutation or L469R Site 2 mutation reduced complex formation with the p130Cas FAT domain in vitro, indicating that they substantially weaken the tight interaction (Fig. 4b, Supplementary Fig. 7b). In addition, the NSP3 L623E Site 1 mutation completely abrogated the interaction even at the substantial concentrations used in our in vitro association experiments (Fig. 4b). In COS cells the full-length NSP3 L623E and L469R mutants also efficiently disrupted the complex with full-length p130Cas when the two proteins were co-expressed (Fig. 4c). BCAR3 residues Leu744 and Arg748 correspond to Leu623 and Arg627 in NSP3, and when mutated also effectively disrupted the association with p130Cas in COS cells confirming that different NSP proteins utilize a conserved p130Cas binding mode (Fig. 4d). Mutation of Arg743 in mouse BCAR3, corresponding to Arg748 in human BCAR3, has also been independently confirmed to disrupt p130Cas binding in recent studies16,33. However, our in vitro results suggest that mutating Leu623 in NSP3 more effectively abrogates binding and therefore represents a more potent probe for dissecting NSP3–Cas signaling modules. The potency of Leu623 is in good agreement with its location in the hydrophobic core of the Site 1 interaction, in close proximity to the hydrophobic pair Phe794 and Leu787 of p130Cas (Fig. 4a). In contrast, the salt bridge that Arg627 forms with Asp797 of p130Cas is more peripheral to the interface.
We also examined interface mutations in p130Cas for their ability to counteract complex formation with NSP proteins. Based on structural insight, we chose Leu787, Phe794 and Asp797 as promising candidates for mutagenesis. Individual mutations of each of these residues (L787E, F794R and D797R) weakened the in vitro association of the p130Cas FAT domain with the NSP3 Cdc25-homology domain, while combinations of two mutations virtually abolished complex formation (Fig. 4e, Supplementary Fig. 7b). In addition, the single p130Cas L787E and F794R mutations markedly reduced association between full-length p130Cas and NSP3 co-expressed in COS cells, further confirming features of the NSP3–p130Cas complex in the cellular context (Fig. 4f).
To examine the functional effects of targeted disruption of the NSP3–p130Cas module on cell migration, we tested the NSP3 L623E mutant in a Transwell migration assay. NSP3 has previously been shown to promote COS cell migration towards EGF as a chemoattractant in a Cas-dependent manner27. The specific interface mutant L623E abrogated the NSP3-dependent enhancement of COS cell migration towards epidermal growth factor (EGF). Cells expressing this mutant showed migratory ability towards EGF similar to that of cells transfected with empty vector control (Fig. 4g,h). This emphasizes the role of NSP–Cas module formation in linking cell motility and growth factor signaling, while at the same time demonstrating the potency of structural interface mutations for investigation of these signaling nodes.
Promiscuous yet class-specific NSP–Cas interactions
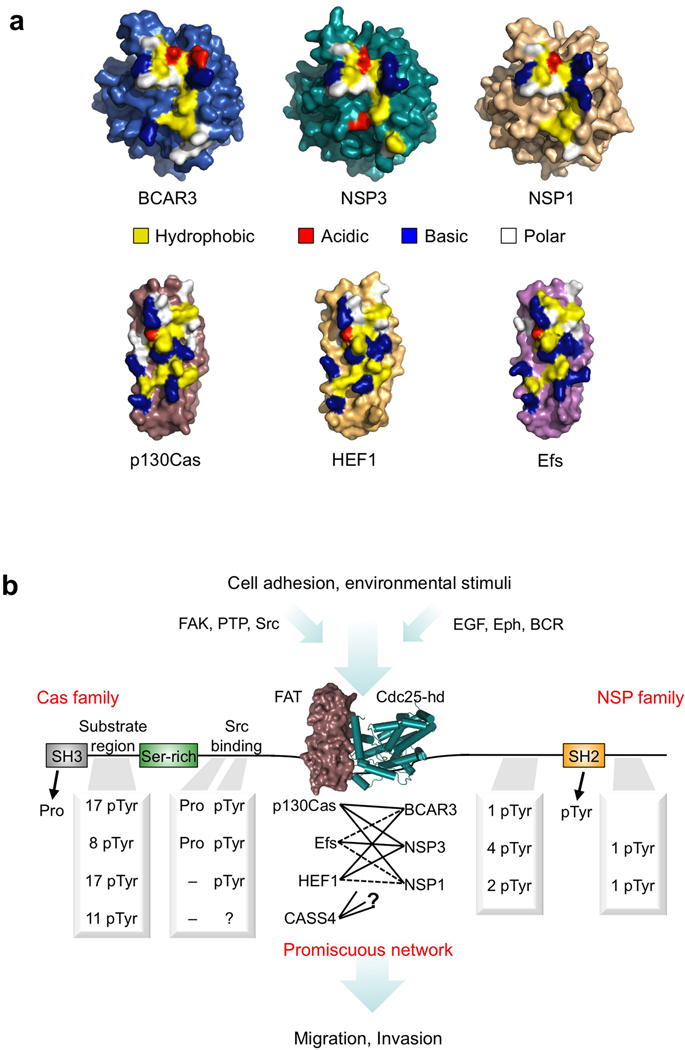
The structures of BCAR3, NSP3 and p130Cas provide not only the first insight into the C-terminal regions of NSP and Cas proteins and their interactions, but also a basis upon which to apply features depicted in the wider context of NSP and Cas family signaling. To this end, we created molecular models for the C-terminal domains of the remaining family members based on sequence profile alignments and the crystal structures solved in this work. Mapping of primary interface residues on these models shows that residues involved in NSP–Cas complex-formation are clearly conserved within each family (Fig. 5a). In line with this degree of conservation are residues that determine the core fold of NSP and Cas family C-terminal domains (Supplementary Fig. 8), whereas remaining surface features are considerably more divergent. This illustrates that all three NSP family proteins have pervasively repurposed an enzymatic fold as a recruitment module, while Cas family proteins share a C-terminal FAT domain that accommodates the extended and tight binding interface observed in the NSP3–p130Cas complex. This extended interface, built upon the Site 1 and Site 2 binding motifs, is elegantly specific to NSP–Cas interactions yet allows different combinations of high-affinity mechanical linkages that integrate heterogeneous signaling pathways between the two families of proteins.
Figure 5.
NSP–Cas modules: Class-specific yet promiscuous signaling nodes. (a) Surface representations of the BCAR3, NSP3 and p130Cas structures solved in this work, as well as models of NSP1, HEF1 and Efs. Key residues involved in the NSP–Cas interaction are colored in yellow, blue, red or white indicating hydrophobic, basic, acidic or polar amino acids respectively. (b) Schematic representation of NSP–Cas signaling nodes. Association between the C-terminal domains of Cas and NSP family members forms a crucial junction downstream of adhesion and environmental stimuli, bringing together pairs of multifaceted signaling proteins that regulate cellular migration and invasion. The C-terminal association is of high-affinity but promiscuous between family members, and allows different combinations of docking sites and signaling outputs based upon the specific NSP–Cas module formed. Phosphorylation sites (pTyr) are annotated based on (www.phosphosite.org), except for Efs, which is based on ref 10. Proline motifs (Pro) for Src family kinase binding were identified manually based upon the sequence of each Cas family protein. (Abbreviations used as follows: PTP, Protein tyrosine phosphatases; EGF, epidermal growth factor; Eph, Ephrin receptors; BCR, B-cell receptor)
DISCUSSION
NSP and Cas family proteins interact with each other to form multidomain signaling modules. Members from both families have been linked to pathological processes, such as breast cancer antiestrogen resistance2,34 or metastasis in melanoma and lung cancer35,36. Analysis of a NSP3 knockout mouse line also suggests an involvement in developmental disorders such as Kallmann syndrome14. Here we provide the first-in-class structures of the unbound BCAR3 Cdc25-homology domain and the complex of the NSP3 Cdc25-homology domain with the FAT domain of the scaffolding protein p130Cas, as well as comprehensive biochemical and cell-based analyses of these regions.
The Cdc25-homology domain is typically found in exchange factors for Ras-type small GTPases, which activate their target GTPases by catalyzing nucleotide exchange. Since the GTP-GDP dependent cycling of Ras proteins is essential for many aspects of normal cellular physiology and plays a critical role in diseases such as cancer, Cdc25-homology domains have been the focus of intense investigation. Several elegant structural studies have revealed the molecular mechanisms underlying nucleotide exchange by Cdc25-homology domains17,19,20, including their regulation by additional interacting domains18,37 and other proteins including Ras itself38. In contrast to these catalytically active Cdc25-homology domains, the BCAR3 Cdc25-homology domain adopts an unprecedented closed conformation with nearly all functional elements completely remodeled, rendering it virtually incapable of performing canonical exchange factor function. Moreover, the structure of the NSP3–p130Cas complex shows that the same closed conformation is essential for a tight interaction with the FAT domain of p130Cas. Residues at the interface between NSP3 and p130Cas partially overlap with the interface suggested in a recent report utilizing small-angle X-ray scattering (SAXS) to study the BCAR3–p130Cas complex16. However, this SAXS model assumes the BCAR3 Cdc25-homology domain to adopt a classical GEF-type fold similar to SOS, rather than the closed form now observed in the crystal structures of both isolated BCAR3 and NSP3 bound to p130Cas. The complete rearrangement of the helical hairpin in NSP proteins markedly modifies the Cdc25-homology domain and in effect creates an entirely new binding surface. Consequently, NSP proteins employ an aberrant enzymatic domain as an adaptor to link two multidomain signaling proteins.
Alignments of NSP proteins and Cas proteins reveal that both families (except the more distantly related CASS4) have preserved the key residues that are critical for their interaction (Fig. 5a and Supplementary Fig. 8). This analysis explains how NSP and Cas proteins are able to tightly associate in a promiscuous but class-specific manner11,12,39. As a consequence, different pairwise combinations of NSP and Cas proteins can exploit functional differences such as divergent phosphorylation sites characteristic of the particular NSP–Cas protein module3,6,10,27 (Fig. 5b). In such a manner the signaling outcome can be fine-tuned by the particular NSP–Cas signaling nodes present in a cell type. This class-specific promiscuity represents an effective structural mechanism to achieve sensitive regulation of cellular signaling networks by NSP–Cas modules.
In summary, the novel closed Cdc25-homology domain conformation of NSP proteins represents a unique adaption enabling tight interaction with the C-terminal FAT domain of Cas proteins. Besides revealing a new structural paradigm used by the NSP class of adaptor complexes, this work also provides a new basis for investigating the role of NSP–Cas signaling modules in cell migration, invasiveness and other key physiological and pathological processes.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
Supplementary Material
Table 1.
Data collection, phasing and refinement statistics
| BCAR3 Native | NSP3–p130Cas Native |
NSP3–p130Cas SeMet |
NSP3–p130Cas Thiomerosal |
|
|---|---|---|---|---|
| Data collection | ||||
| Space group | P212121 | I41 | I41 | I41 |
| Cell dimensions | ||||
| a, b, c (Å) | 50.23, 151.89, 196.50 | 171.88, 171.88, 78.27 | 172.04, 172.04, 79.16 | 171.72, 171.72, 77.76 |
| α, β, γ (°) | ||||
| Peak | Peak | |||
| Wavelength | 1.5418 | 1.075 | 0.9792 | 1.000 |
| Resolution (Å) | 19.85-2.4 | 29.5-2.5 | 29.6-2.9 | 50.0-2.6 |
| Rmerge | 0.096 (0.532) | 0.071 (0.844) | 0.056 (0.697) | 0.069 (0.552) |
| I / σI | 13.7 (3.0) | 22.7 (3.7) | 18.4 (2.3) | 35.2 (3.6) |
| Completeness (%) | 98.7 (96.3) | 99.8 (100.0) | 99.5 (98.0) | 97.1 (83.1) |
| Redundancy | 5.2 (4.6) | 14.9 (14.9) | 6.2 (6.1) | 14.7 (12.0) |
| Refinement | ||||
| Resolution (Å) | 19.75–2.4 | 29.5–2.5 | ||
| No. reflections | 56,380 | 37,296 | ||
| Rwork / Rfree | 17.4/24.4 | 19.6/26.6 | ||
| No. atoms | ||||
| Protein | 9,883 | 6,532 | ||
| Ligand/ion | 41 | 7 | ||
| Water | 373 | 59 | ||
| B-factors | ||||
| Protein | 42.4 | 100.4 (complex 1 72.9, complex 2 130.0) | ||
| Ligand/ion | 53.7 | 116 | ||
| Water | 37.5 | 69.6 | ||
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.008 | 0.008 | ||
| Bond angles (°) | 1.053 | 1.064 | ||
Values in parentheses are for highest-resolution shell.
ACKNOWLEDGEMENTS
From the Sanford|Burnham Medical Research Institute we thank Scott Snipas for protein sequencing, Dr. Andrey Bobkov for analytical ultracentrifugation and isothermal titration calorimetry and Dr. Guy Salvesen for critical discussion of the manuscript. We also thank the Hope for a Cure Foundation (www.hopeforacurefoundation.org) for donation of equipment and Dr. John Badger (DeltaG technologies) for assistance in model evaluation. This work was supported by NIH grants P01CA102583 to SJR and EBP, R01CA116099 and P01HD025938 to EBP and DOD-BCRP Fellowship BC100466 to PDM. Data collection at beamline X29 of the National Synchrotron Light Source was also supported by Biological and Environmental Research Department of Energy and the NIH National Center for Research Resources.
Footnotes
ACCESSION NUMBERS
The atomic coordinates of BCAR3 and the NSP3–p130Cas complex have been deposited in the Protein Data Bank with the accession codes 3t6a and 3t6g respectively.
AUTHOR CONTRIBUTIONS
P.D.M. grew crystals, solved the crystal structures, designed and carried out in vitro experiments and wrote the manuscript. Y.W. designed, performed and analyzed in vivo experiments, M.K.D. and J.J.L. expressed and purified proteins and grew initial NSP3–p130Cas crystals and H.R. carried out crystallographic data collection. S.J.R. and E.B.P. designed experiments, analyzed data and wrote the manuscript.
REFERENCES
- 1.Brinkman A, van der Flier S, Kok EM, Dorssers LC. BCAR1, a human homologue of the adapter protein p130Cas, and antiestrogen resistance in breast cancer cells. J Natl Cancer Inst. 2000;92:112–120. doi: 10.1093/jnci/92.2.112. [DOI] [PubMed] [Google Scholar]
- 2.van Agthoven T, et al. Identification of BCAR3 by a random search for genes involved in antiestrogen resistance of human breast cancer cells. Embo J. 1998;17:2799–2808. doi: 10.1093/emboj/17.10.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cabodi S, Del Pilar Camacho-Leal M, Di Stefano P, Defilippi P. Integrin signalling adaptors: not only figurants in the cancer story. Nat Rev Cancer. 2010;10:858–870. doi: 10.1038/nrc2967. [DOI] [PubMed] [Google Scholar]
- 4.Felekkis K, Quilliam LA, Lerner A. Characterization of AND-34 function and signaling. Meth Enzymol. 2006;407:55–63. doi: 10.1016/S0076-6879(05)07006-0. [DOI] [PubMed] [Google Scholar]
- 5.Gotoh T, Cai D, Tian X, Feig LA, Lerner A. p130Cas regulates the activity of AND-34, a novel Ral, Rap1, and R-Ras guanine nucleotide exchange factor. J Biol Chem. 2000;275:30118–30123. doi: 10.1074/jbc.M003074200. [DOI] [PubMed] [Google Scholar]
- 6.Lu Y, Brush J, Stewart TA. NSP1 defines a novel family of adaptor proteins linking integrin and tyrosine kinase receptors to the c-Jun N-terminal kinase/stress-activated protein kinase signaling pathway. The Journal of biological chemistry. 1999;274:10047–10052. doi: 10.1074/jbc.274.15.10047. [DOI] [PubMed] [Google Scholar]
- 7.Sakakibara A, Hattori S. Chat, a Cas/HEF1-associated adaptor protein that integrates multiple signaling pathways. J Biol Chem. 2000;275:6404–6410. doi: 10.1074/jbc.275.9.6404. [DOI] [PubMed] [Google Scholar]
- 8.Cai D, et al. The GDP exchange factor AND-34 is expressed in B cells, associates with HEF1, and activates Cdc42. J Immunol. 2003;170:969–978. doi: 10.4049/jimmunol.170.2.969. [DOI] [PubMed] [Google Scholar]
- 9.Al-Shami A, et al. The adaptor protein Sh2d3c is critical for marginal zone B cell development and function. The Journal of Immunology. 2010;185:327–334. doi: 10.4049/jimmunol.1000096. [DOI] [PubMed] [Google Scholar]
- 10.Alexandropoulos K, Regelmann AG. Regulation of T-lymphocyte physiology by the Chat-H/CasL adapter complex. Immunol Rev. 2009;232:160–174. doi: 10.1111/j.1600-065X.2009.00831.x. [DOI] [PubMed] [Google Scholar]
- 11.Browne CD, et al. SHEP1 partners with CasL to promote marginal zone B-cell maturation. Proceedings of the National Academy of Sciences of the United States of America. 2010 doi: 10.1073/pnas.1007558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schrecengost RS, Riggins RB, Thomas KS, Guerrero MS, Bouton AH. Breast cancer antiestrogen resistance-3 expression regulates breast cancer cell migration through promotion of p130Cas membrane localization and membrane ruffling. Cancer Res. 2007;67:6174–6182. doi: 10.1158/0008-5472.CAN-06-3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuh NR, Guerrero MS, Schrecengost RS, Bouton AH. BCAR3 regulates Src/p130 Cas association, Src kinase activity, and breast cancer adhesion signaling. J Biol Chem. 2010;285:2309–2317. doi: 10.1074/jbc.M109.046631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, et al. The SRC homology 2 domain protein Shep1 plays an important role in the penetration of olfactory sensory axons into the forebrain. J Neurosci. 2010;30:13201–13210. doi: 10.1523/JNEUROSCI.3289-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai D, Clayton LK, Smolyar A, Lerner A. AND-34, a novel p130Cas-binding thymic stromal cell protein regulated by adhesion and inflammatory cytokines. J Immunol. 1999;163:2104–2112. [PubMed] [Google Scholar]
- 16.Garron M-L, et al. Structural insights into the association between BCAR3 and Cas family members, an atypical complex implicated in anti-oestrogen resistance. J Mol Biol. 2009;386:190–203. doi: 10.1016/j.jmb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 17.Boriack-Sjodin PA, Margarit SM, Bar-Sagi D, Kuriyan J. The structural basis of the activation of Ras by Sos. Nature. 1998;394:337–343. doi: 10.1038/28548. [DOI] [PubMed] [Google Scholar]
- 18.Rehmann H, Das J, Knipscheer P, Wittinghofer A, Bos JL. Structure of the cyclic-AMP-responsive exchange factor Epac2 in its auto-inhibited state. Nature. 2006;439:625–628. doi: 10.1038/nature04468. [DOI] [PubMed] [Google Scholar]
- 19.Freedman TS, et al. A Ras-induced conformational switch in the Ras activator Son of sevenless. Proc Natl Acad Sci USA. 2006;103:16692–16697. doi: 10.1073/pnas.0608127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rehmann H, et al. Structure of Epac2 in complex with a cyclic AMP analogue and RAP1B. Nature. 2008;455:124–127. doi: 10.1038/nature07187. [DOI] [PubMed] [Google Scholar]
- 21.Freedman TS, et al. Differences in flexibility underlie functional differences in the Ras activators son of sevenless and Ras guanine nucleotide releasing factor 1. Structure. 2009;17:41–53. doi: 10.1016/j.str.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bos JL, de Rooij J, Reedquist KA. Rap1 signalling: adhering to new models. Nat Rev Mol Cell Biol. 2001;2:369–377. doi: 10.1038/35073073. [DOI] [PubMed] [Google Scholar]
- 23.Dodelet VC, Pazzagli C, Zisch AH, Hauser CA, Pasquale EB. A novel signaling intermediate, SHEP1, directly couples Eph receptors to R-Ras and Rap1A. J Biol Chem. 1999;274:31941–31946. doi: 10.1074/jbc.274.45.31941. [DOI] [PubMed] [Google Scholar]
- 24.Sakakibara A, Ohba Y, Kurokawa K, Matsuda M, Hattori S. Novel function of Chat in controlling cell adhesion via Cas-Crk-C3G-pathway-mediated Rap1 activation. J Cell Sci. 2002;115:4915–4924. doi: 10.1242/jcs.00207. [DOI] [PubMed] [Google Scholar]
- 25.Riggins RB, Quilliam LA, Bouton AH. Synergistic promotion of c-Src activation and cell migration by Cas and AND-34/BCAR3. J Biol Chem. 2003;278:28264–28273. doi: 10.1074/jbc.M303535200. [DOI] [PubMed] [Google Scholar]
- 26.van den Berghe N, Cool RH, Horn G, Wittinghofer A. Biochemical characterization of C3G: an exchange factor that discriminates between Rap1 and Rap2 and is not inhibited by Rap1A(S17N) Oncogene. 1997;15:845–850. doi: 10.1038/sj.onc.1201407. [DOI] [PubMed] [Google Scholar]
- 27.Dail M, et al. SHEP1 function in cell migration is impaired by a single amino acid mutation that disrupts association with the scaffolding protein cas but not with Ras GTPases. J Biol Chem. 2004;279:41892–41902. doi: 10.1074/jbc.M402929200. [DOI] [PubMed] [Google Scholar]
- 28.Deakin NO, Turner CE. Paxillin comes of age. J Cell Sci. 2008;121:2435–2444. doi: 10.1242/jcs.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertolucci CM, Guibao CD, Zheng J. Structural features of the focal adhesion kinase-paxillin complex give insight into the dynamics of focal adhesion assembly. Protein Sci. 2005;14:644–652. doi: 10.1110/ps.041107205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoellerer MK, et al. Molecular recognition of paxillin LD motifs by the focal adhesion targeting domain. Structure. 2003;11:1207–1217. doi: 10.1016/j.str.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 31.Lulo J, Yuzawa S, Schlessinger J. Crystal structures of free and ligand-bound focal adhesion targeting domain of Pyk2. Biochemical and biophysical research communications. 2009;383:347–352. doi: 10.1016/j.bbrc.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 32.Thomas JW, et al. The role of focal adhesion kinase binding in the regulation of tyrosine phosphorylation of paxillin. J Biol Chem. 1999;274:36684–36692. doi: 10.1074/jbc.274.51.36684. [DOI] [PubMed] [Google Scholar]
- 33.Vanden Borre P, Near RI, Makkinje A, Mostoslavsky G, Lerner A. BCAR3/AND-34 can signal independent of complex formation with CAS family members or the presence of p130Cas. Cellular Signalling. 2011 doi: 10.1016/j.cellsig.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Agthoven T, et al. Functional identification of genes causing estrogen independence of human breast cancer cells. Breast Cancer Res Treat. 2009;114:23–30. doi: 10.1007/s10549-008-9969-5. [DOI] [PubMed] [Google Scholar]
- 35.Ji H, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 36.Kim M, et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125:1269–1281. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 37.Sondermann H, et al. Structural analysis of autoinhibition in the Ras activator Son of sevenless. Cell. 2004;119:393–405. doi: 10.1016/j.cell.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 38.Margarit SM, et al. Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell. 2003;112:685–695. doi: 10.1016/s0092-8674(03)00149-1. [DOI] [PubMed] [Google Scholar]
- 39.Roselli S, Wallez Y, Wang L, Vervoort V, Pasquale EB. The SH2 domain protein Shep1 regulates the in vivo signaling function of the scaffolding protein Cas. Cell Signal. 2010 doi: 10.1016/j.cellsig.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.