Abstract
The target-decoy approach to estimating and controlling false discovery rate (FDR) has become a de facto standard in shotgun proteomics, and it has been applied at both the peptide-to-spectrum match (PSM) and protein levels. Current bioinformatics methods control either the PSM- or the protein-level FDR, but not both. In order to obtain the most reliable information from their data, users must employ one method when the number of tandem mass spectra exceeds the number of proteins in the database and another method when the reverse is true. Here we propose a simple variation of the standard target-decoy strategy that estimates and controls PSM and protein FDRs simultaneously, regardless of the relative numbers of spectra and proteins. We demonstrate that even if the final goal is a list of PSMs with a fixed low FDR and not a list of protein identifications, the proposed two-dimensional strategy offers advantages over a pure PSM-level strategy.
Keywords: Mass spectrometry, target decoy strategy, false discovery rate, peptide identification, protein identification
1. Introduction
Shotgun or “bottom-up” proteomics analyzes complex protein mixtures by digesting the proteins with a protease such as trypsin and then identifying the resulting peptides using tandem mass spectrometry (MS/MS) and protein database-search software such as SEQUEST1 or Mascot2. The reliability and reproducibility of peptide identifications have improved greatly in the past four years due to the widespread adoption of the target-decoy approach to false discovery rate (FDR) estimation3, 4. The basic target-decoy strategy augments the actual or “target” protein database with a matched set of “decoy” protein sequences, and then searches the combined database to produce a list of peptide-to-spectrum matches (PSMs). Variants of the basic approach use separate target and decoy databases5–7 or estimate local rather than global FDR8. In all cases, decoy PSMs simulate false target PSMs, and hence can be used to estimate the number of false target PSMs meeting some set of acceptance criteria. Acceptance criteria vary widely, and we distinguish two classes of methods: “Peptide-centric” methods filter the list of PSMs to an acceptably low FDR before integrating PSMs by protein, and “protein-centric” methods integrate first and filter later, primarily by protein score.
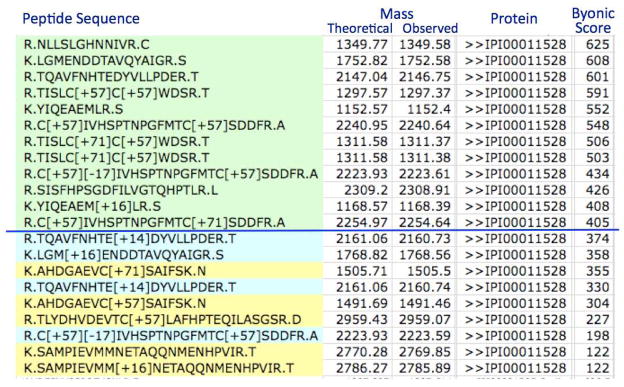
Peptide-centric methods often give poor performance at the protein level. Even a low rate of false PSMs may map to a high rate of false protein identifications, because false PSMs tend to scatter all over the protein database rather than cluster on a small number of proteins. On the other hand, strict filtering at the PSM level to avoid false protein identifications may discard a large number of PSMs that a human expert would accept as true due to their peptide or protein identity. Figure 1 illustrates the problem using as an example all the PSMs to a single protein in a single Byonic9 search. The PSMs shown in green have Byonic matching scores above the 1% FDR cutoff and would thus be accepted by 1% FDR filtering, but the PSMs in blue and yellow would be discarded. The protein IPI00011528 is one of only ~260 confidently identified proteins in a search of the Aurum data set10 of ~10,000 tandem mass spectra against a database containing ~100,000 target and decoy proteins, and hence most human experts would accept the yellow identifications simply because they match an identified protein. The expert would a fortiori accept the blue identifications because they match not just an identified protein, but also identified peptides (green).
Figure 1. Spectrum identifications matching one protein.
In this screenshot of Combyne output, only the spectrum-to-peptide matches (PSMs) shown in green score above the 1% FDR threshold, but a human expert would probably accept the lower-scoring PSMs (yellow and blue) because they match a high-scoring protein. A human expert might favor the blue PSMs over the yellow PSMs because not only do they match a high-scoring protein, but they match high-scoring peptides within that protein.
Protein-centric methods may give poor performance at the PSM level, but this problem tends to be less obvious. Combyne11, ProValt12, and MAYU13 are protein assembly tools that take unfiltered or lightly filtered PSMs as input and estimate protein FDR using the target-decoy strategy. None of these tools, however, estimates the FDR of PSMs from accepted proteins, and this PSM-level FDR can vary from almost negligible when database proteins outnumber spectra by 10 to 1 as in the Aurum example, to unacceptably large when spectra outnumber proteins by 10 to 1 or more, as is often the case with large data sets or small protein databases. False matches to true proteins may give misleading information about protein sequence, abundance, and modifications.
In order to handle both PSM and protein levels, researchers have also devised methods that take protein-level features into account when deciding which PSMs to accept. ProteinProphet14 uses the “number of sibling peptides” to rescore PSMs, meaning the number of other matched peptides in the protein(s) containing the match. The first published version of Percolator15 used similar protein-level features. Protein-level features improve the separation of true and false PSMs for most search results, yet they interact adversely with the target-decoy strategy and cause it to underestimate the PSM-level FDR. By boosting the score of a PSM to a confidently identified protein, which is much more likely to be a target than a decoy, these methods violate the central assumption of the target-decoy strategy: at any given score threshold, decoy matches are just as likely as false target matches.4 Although the score boost occurs after the database search, it introduces a bias similar to one incurred by multi-stage search strategies that expand the search space around high-scoring proteins16 or “hot” peptides17 by adding modifications and/or nonspecific digestion. For reasons related to this bias, the second version of Percolator18 dropped all protein-level features, and Panoramics19, a more recent tool akin to ProteinProphet, avoided the use of sibling peptides from the outset; however, these tools also give up the discriminatory power of protein-level information.
Here we propose a simpler and more general solution to the peptide/protein quandary: a two-dimensional target-decoy method called ProteinsFirst that can control protein- and PSM-level FDR simultaneously. Using this method, researchers can estimate the PSM FDR after protein assembly. The estimate is nearly unbiased, and the small remaining bias is in the conservative direction, meaning that the method tends to overestimate the PSM FDR.
2. Methods
We start by describing ProteinsFirst at a generic level, so that it can be used with any proteomics search program (Mascot, SEQUEST, etc.) and any protein assembly program (ProteinProphet, MAYU, etc.). We then describe computational experiments on ProteinsFirst, using Byonic as the search engine and Combyne as the protein assembly program. We also benchmark against Percolator, a freely available tool for PSM filtering. We emphasize that ProteinsFirst is a “script” for processing protein and PSM lists computed by other bioinformatics tools. ProteinsFirst cuts the lists to simultaneously control protein and PSM FDRs. It discards PSMs to proteins too low on the protein list and adds PSMs to certain decoy proteins, but does not otherwise rerank the lists.
2.1. ProteinsFirst Algorithm
We assume that each target protein has a corresponding decoy of the same length. We could, for example, create corresponding decoys by reversing each target protein, that is, reading the sequence from C- to N-terminus3. In Section 2.2 we describe a more refined method for creating corresponding decoys. The ProteinsFirst algorithm performs the following steps, with steps 5 and 6 giving the crucial difference between ProteinsFirst and a standard protein-centric approach:
Search the concatenated target-decoy database using a proteomics search engine such as Mascot, SEQUEST, or Byonic.
Rank proteins by protein score based upon the results of the search.
Assign each PSM (target or decoy) to the highest rank protein that contains the peptide.
Discard all PSMs from proteins with protein score below some threshold Tprot. Estimate the protein FDR in the list of target proteins by (# decoy proteins)/(# target proteins)
Add PSMs from the corresponding decoys of all target proteins with score above Tprot.
Sort the PSMs (target and decoy) by score and discard all PSMs below some threshold Tpep. Estimate the PSM FDR in the list of target PSMs by (# decoy PSMs)/(# of target PSMs).
In the algorithm just sketched, the protein FDR is controlled by parameter Tprot and the spectrum FDR by both Tprot and Tpep. If we set Tprot so low that it admits all proteins, ProteinsFirst reduces to a standard peptide-centric approach, and if we set Tpep so low that it admits all peptides, ProteinsFirst reduces to a standard protein-centric approach. As in the usual target-decoy strategy, there is some play in the selection of these two parameters; for a conservative bias we could cut the protein and PSM lists when the estimated FDR first crosses the acceptable level. For step 2 we can use any protein ranking method that treats target and decoy proteins identically. We could rank proteins simply by the number of distinct peptides above some score threshold (the “two-peptide rule”), or use a more sophisticated protein-scoring tool such as ProteinProphet, ProValt, Combyne, or MAYU, all of which make efforts to decide exactly which members of a set of homologous proteins are present. ProteinsFirst is robust to variations in homolog acceptance, because it adds PSMs from the corresponding decoys of all target proteins with score above Tprot, regardless of whether these proteins are actually present.
2.2. Computational Experiments
Proteomics Data Set
For our computational experiments we used a data set named 20080618_Jurkat_0p5ul_top10_run1 from Genentech (South San Francisco, CA), now publicly available on Tranche/Proteome Commons. As described previously,20 Jurkat cells were lysed using 8M urea/50 mM Tris (pH 7.5) in the presence of protease and phosphatase inhibitors (Roche), then separated by SDS-PAGE. Proteins of mass at least 70 kDa were digested in gel with trypsin, extracted, and dried. This sample had no cysteine treatment. Approximately 0.5 μg of digested peptides was injected for analysis on a Thermo LTQ Orbitrap with Orbitrap single-MS scans (resolution 60,000) and LTQ MS/MS scans performed on the top 10 peaks per full scan. The data set contains 5853 MS/MS spectra, with assigned charges from +2 to +6, with 3683 +2, 1782 +3, 356 +4, 30 +5, and 2 +6.
Software
We used our own prototype search engine, Byonic, and configured it for a semitryptic search with the following variable (optional) modifications and a limit of two modifications per peptide: oxidized methionine (M[+16]), pyro-glu cyclizations (N-terminal E[−18] and Q[−17]), and deamidated asparagine and glutamine (N[+1] and Q[+1]). We used mass tolerances of 20 ppm for precursor mass measurements and 0.5 Da for fragment mass measurements; we also allowed “off-by-one” errors in the precursor mass, that is, the recorded mass was not the monoisotopic mass but rather the mass of the peptide with one 13C.
We used our own protein assembly tool, Combyne, to assign PSMs to proteins and rank proteins by score. As described previously, Combyne assigns confidences (p-values) to PSMs using Byonic score, Byonic delta (the difference between the top score and the second-best score), Byonic z-score (the number of standard deviations between the top score and the mean score of random peptides), cleavage specificity, number of modifications, and peptide length; this p-value step does not use any protein-level features. (In this study, Combyne’s p-value step did not use “corroborations” between PSMs,11 for example, boosting the score of a PSM with a semitryptic peptide that is a substring of a tryptic peptide, because corroborations integrate over more than one PSM.) After its p-value step, Combyne computes protein p-values, ranks proteins, and assigns PSMs to proteins, as in steps 2 – 4 of ProteinsFirst. We implemented ProteinsFirst inside Combyne (a C program) by adding steps 5 and 6. It is important to note that ProteinsFirst uses Combyne’s PSM and protein p-values as arbitrary scores, and not as measures of statistical significance; all that matters is the order of the scores.
Methods
For comparison of ProteinsFirst with peptide-centric approaches to separating true from false PSMs, we benchmarked five computational methods. All five methods used the same Byonic search results, but then processed the PSMs in different ways. In all cases, FDR was estimated by summing down a sorted list of PSMs or proteins and estimating FDR by (# of decoy identifications)/(# of target identifications), and cutting when the FDR first crosses the acceptable level (set to 1% in this experiment). The methods are as follows:
(Pep) Run Combyne’s p-value step and sort PSMs by Combyne’s p-value.
(Prot) Use Combyne to rank proteins and accept all PSMs to accepted proteins.
(ProtFirst) Run ProteinsFirst using Combyne to rank proteins and Combyne’s p-value as the PSM score.
(PercPep) Run Percolator (Version 1.17, built November 30, 2010) and sort PSMs by Percolator’s posterior error probability. We set up Percolator to use almost the same statistical features as Combyne: Byonic score, delta, and z-score, along with cleavage specificity, precursor mass error, and peptide length. (We used precursor mass error in Percolator but not Combyne, because Combyne cannot be easily reconfigured to use new features.)
(PercProt) This method is the same as PercPep, except that PercProt used one protein-level feature: the number of sibling peptides. More precisely, for a PSM with peptide p, this feature is the number of distinct peptides appearing in PSMs from the protein(s) that contain p.
Protein Databases
In order to test ProteinsFirst at various ratios of spectra to proteins, we used three protein databases of varying sizes. The large database is the Uniprot Human database without isoforms (downloaded June 2, 2011), containing 20,334 target proteins. The medium database is a subset of the large database, containing 1012 target proteins, about one-fourth (259) of which score above the top-ranked decoy. The small database is a subset of the medium database, containing 347 target proteins, most of which score above the top-ranked decoy.
For each database protein we generated a corresponding decoy by fixing the initial residue and the final R or K within each fully tryptic peptide and reversing all the remaining residues. Thus the target protein
>IPI00003947 Ig lambda chain V-II region
QSALTQPRSVSGSPGHSVTISCIGTSSNVGDYKYVSWYQQHPGKAPKLIIYEVSSR
PSGVPDRFSGSKSGNTASLTISGLQAEDEADYYCCSYIGSYVFGTGTKVIVLG
gives rise to the “reverse”
>Decoy IPI00003947 Ig lambda chain V-II region
QPQTLASRSYDGVNSSTGICSITVSHGPSGSVKYGPHQQYWSVKAPKLSSVEYIIR
PDPVGSRFSGSKSTGTGFVYSGIYSCCYYDAEDEAQLGSITLSATNGKVGLVI.
This method of making decoys ensures that each fully tryptic target peptide has a decoy counterpart, exactly matched in mass, amino acid content, and initial and final residues. Initial and final residues have an influence on the overall search due to terminal-residue effects such as pyro-glu modifications and reduced cleavage before proline.
3. Results
We first give a mathematical proof that under certain natural assumptions ProteinsFirst gives an estimate of the FDR with only a small conservative bias. We then give empirical results on the Jurkat data set, comparing low-FDR PSM lists computed by ProteinsFirst with lists computed using two peptide-centric algorithms and one protein-centric algorithm. We remind the reader that the FDR in a list of identifications is the number of false identifications divided by the total number of identifications.
3.1. Theoretical Results
Assume c is a candidate peptide from a protein database, s is an MS/MS spectrum, and Spep(s, c) is a real-valued score that measures the quality of the match between c and s. We are normally interested in only the top-scoring peptide p for each spectrum,
and for simplicity we assume that there is only one top-scoring peptide p (which may appear in more than one protein). We model a PSM as a tuple (s, p, Spep(s, p), P, Sprot(P), t), where s is an MS/MS spectrum, p is the top-scoring peptide, Spep(s, p) is the score of peptide p, P is the top-scoring protein that contains peptide p, Sprot(P) is the protein score for P, and t is a Boolean indicator for whether P is a target or a decoy. We make no assumptions about the meanings of peptide and protein scores, except that a higher score is considered better (more likely to be true) than a lower score. We use Prob[·] to denote the probability of an event, and E[·] to denote the expectation of a random variable.
We now distinguish three types of PSMs: true matches in which p is indeed the peptide that produced spectrum s and P is a target protein, false (target) matches in which p is not the correct peptide and P is a target protein, and decoy matches in which P is a decoy protein (and we call p a decoy peptide). We consider Spep(s, c), Spep(s, p) (= maxc { Spep(s, c) }), and Sprot(P) to be random variables. The hypotheses underlying the target-decoy strategy are: (A1) For any spectrum s, the peptide score Spep(s, c) is independently and identically distributed (i.i.d.) for all false candidates, regardless of whether c is a false target or a decoy, and (A2) each spectrum s is scored against the same number of decoy and false target candidate peptides. (Many spectra will be compared against one more decoy than false target, since there is also a true target, so (A2) is not strictly true.) Then the random variable maxc{ Spep(s, c) | c is a decoy } has the same distribution as maxc{ Spep(s, c) | c is a false target }, since each is the maximum of the same number of i.i.d. random variables.
Now assume we have a collection of spectra, s1, s2, …, sn, with top-scoring peptides p1, p2, …, pn. For any given i, 1 ≤ i ≤ n, and any score threshold Tpep, Prob [Spep(si, pi) ≥ Tpep | pi is a decoy] = Prob [Spep(si, pi) ≥ Tpep | pi is a false target]. Summing from i=1 to n,
Notice that this statement holds even if different spectra give different distributions of random scores, for example, some spectra tend to give high scores and others low scores. It also holds even if there are dependencies among the spectra, for example, si+1 is known to contain the same peptide as si, because expectations of random variables sum regardless of independence.
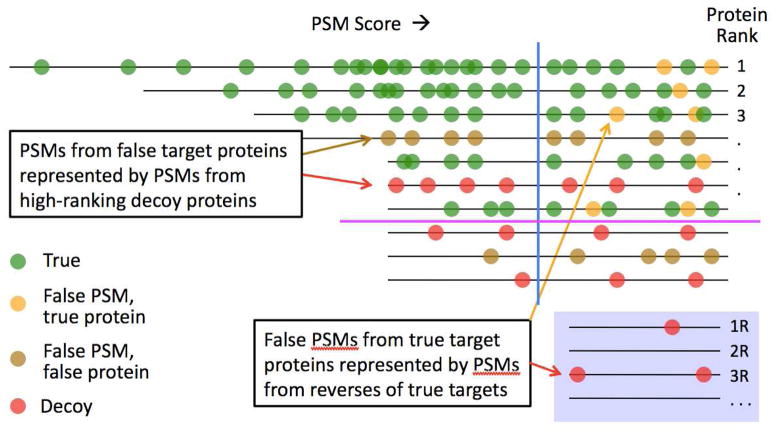
To analyze ProteinsFirst, we divide target proteins into two types: a false target protein P has no true matches, and a true protein has at least one true match. We then divide the false target PSMs into two types as shown in Figure 2: false matches to false target proteins and false matches to true proteins. Assume as above that we have a collection of spectra, s1, s2, …, sn, with top-scoring peptides p1, p2, …, pn from proteins P1, P2, …, Pn. We first consider the matches to high-scoring false target proteins. For any given i, 1 ≤ i ≤ n, any protein score threshold Tprot, and peptide score threshold Tpep, we assume the following, which can be informally stated in words as (A3) the protein-ranking method is not biased against decoy proteins:
Figure 2. Algorithm for multilevel FDR estimation.
PSMs are arranged in a 2D plot by PSM score and protein rank. There are three kinds of false PSMs: false matches to true proteins (yellow), false matches to false target proteins (brown), and decoy matches (red). If we discard PSMs based on score alone (blue line), we lose many true PSMs (green). If we discard PSMs based on protein first (purple line), we lose few true PSMs. After discarding PSMs to low-ranking proteins, we add PSMs to the matched decoys (“reverses”) of high-ranking target proteins (those above the purple line), denoted 1R, 2R, …, in order to estimate the overall FDR.
In most protein-ranking methods, we do not have exact equality above, because some target proteins are true, so that if the protein database is equally balanced between decoys and targets, it has more decoys than false targets.13 This bias will generally be very small, since the user will typically run the algorithm with a small protein FDR. If the user allows high FDR in ProteinsFirst by setting Tprot so low that Step 4 admits all proteins, each decoy PSM will be duplicated, overestimating the PSM FDR by a factor of two.
We next consider the false matches to true proteins. For any given i, 1 ≤ i ≤ n, any protein score threshold Tprot, and any peptide score threshold Tpep, the following holds true because step 5 of ProteinsFirst explicitly adds all PSMs to the matched decoys of high-scoring target proteins.
We again do not generally have exact equality, because step 5 adds all PSMs to the matched decoys of high-scoring target proteins Pi, whether or not Pi is true. Again this bias will be small if ProteinsFirst is run with a small protein FDR.
Now summing from i=1 to n,
This statement holds even if different spectra give different distributions of random scores, and even if there are dependencies among the spectra.
3.2. Experimental Results
Table 1 gives the results for the five methods described above. All methods except the protein-centric method Prot are set to give 1% PSM FDR as measured by the target/decoy strategy. Prot does not estimate PSM FDR, so it was set to give 1% protein FDR. We see that the use of a protein-level feature (number of sibling peptides), improved Percolator’s sensitivity (# of PSMs) by less than 1% for the Byonic searches on the small and medium protein databases, but by 18% for the large protein database. As argued in the introduction, however, the 18% number may be a slight overestimate due to the adverse interaction of the target/decoy strategy and protein-level features. (The overestimate is necessarily slight, because as we discuss below the total number of false matches to the top proteins in the Byonic search against the large database is small.) We see that protein-level features are most helpful in the case that the number of spectra is smaller than the number of proteins in the protein database. Similarly, we see that ProteinsFirst improved Pep’s sensitivity only on the large-database search, and only by a modest 7% more PSMs.
Table 1. Comparison of ProteinsFirst with four other methods of filtering PSMs.
PercPep, PercProt, and Pep are peptide-centric approaches that give PSM lists with estimated 1% PSM FDR; Prot is a protein-centric approach that gives estimated 1% protein FDR; and ProtFirst is our new two-dimensional approach that gives estimated 1% FDR at both the PSM and protein levels. PercPep uses Percolator and peptide-level features. PercProt adds one protein-level feature and gains sensitivity on the large-database search, but for this method the target/decoy strategy gives an optimistically biased FDR estimate. Pep uses essentially the same peptide-level features as PercPep, but a different algorithm for combining these features. Prot uses the same peptide-level features as PercPep and Pep to rescore PSMs, but it goes on to rank proteins and then accepts all PSMs to top-ranked proteins; this method controls protein FDR but not PSM FDR. ProtFirst uses the same peptide-level features as PercPep, Pep, and Prot, and uses ProteinsFirst to limit both protein- and PSM-level FDR to 1%. For each algorithm, we give the number of target PSMs accepted, the number of unique target proteins hit by the PSMs, and the number of unique decoy proteins hit by the PSMs.
| Algorithm | Small Protein Database # PSMs, # Targets, # Decoys |
Medium Protein Database # PSMs, # Targets, # Decoys |
Large Protein Database # PSMs, # Targets, # Decoys |
|---|---|---|---|
| PercPep | 3552, 295, 39 | 3378, 321, 36 | 3019, 316, 14 |
| PercProt | 3574, 289, 36 | 3406, 302, 36 | 3576, 294, 29 |
| Pep | 3894, 288, 16 | 3835, 306, 20 | 3332, 285, 19 |
| Prot | 4236, 270, 3 | 3826, 286, 3 | 3577, 270, 3 |
| ProtFirst | 3857, 270, 3 | 3759, 286, 3 | 3577, 270, 3 |
Although the PSM FDR is held at 1% for the experiments with PercPep, PercProt, Pep, and ProteinsFirst, the protein FDR rate varies widely. PercPep, PercProt, and Pep give relatively high protein FDRs, ranging from 4.4% (= 14/316) up to 13% (= 39/295); whereas, ProteinsFirst holds the protein FDR rate at 1%.
Prot is a pure protein-centric approach that ranks proteins using Combyne, sets the protein FDR to 1%, and then accepts all PSMs to proteins on the accepted protein list. Prot is identical to steps 1 – 4 of ProteinsFirst. (Prot is almost identical to ProteinsFirst with Tpep set very low. The only difference is that ProteinsFirst with low Tpep overestimates PSM FDR.) As we see in Table 1, Prot gives identical results to ProteinsFirst, because in this case ProteinsFirst accepted all PSMs to accepted proteins, and still reached only an estimated 0.8% PSM FDR. The large-database search has a 1 to 7 ratio (= 5853/(2 × 20,334)) of spectra to database proteins. For the medium database, however, which has ratio 2.9 to 1 (= 5853/(2 × 1012)), Prot accepts 3826 PSMs instead of the 3759 accepted by ProteinsFirst. As in ProteinsFirst, we can estimate the number of false PSMs among these 3826 by the number of PSMs to matched decoys of the accepted proteins, and obtain a PSM FDR of 2.4% (= 90/3826). For the small-database search, which has ratio 8.4 to 1 (= 5853/(2 × 397)), Prot accepts 4236 PSMs with an estimated PSM FDR of 6.9% (= 293/4236), which may be unacceptably large and lead to mistaken inferences.
Table 1 also shows that Pep, which used Combyne to rescore Byonic results, generally outperformed PercPep, which used Percolator. PercPep gave 9% to 12% fewer PSMs at 1% FDR (3552, 3378, and 3019 versus 3894, 3835, and 3332). PercPep also accepted somewhat more PSMs hitting decoy proteins, evidence that PercPep had a higher protein-level FDR in its list of accepted PSMs, even though neither PercPep nor Pep attempts to control protein-level FDR.
4. Discussion
We first discuss the difference between Combyne and Percolator before turning attention to our primary topic, ProteinsFirst. In our experiments Combyne and Percolator used almost the same statistical features for rescoring, but they differed in machine-learning methods. Combyne models feature probability distributions, one-parameter exponential distributions, only for false PSMs, rather than for both true and false PSMs, and combines feature probabilities into a p-value simply by assuming independence. Percolator uses semi-supervised learning, with decoys providing a source of labeled false PSMs, and a discriminative classifier based on support-vector machines (SVMs) to compute posterior error probabilities and q-values. There are too many differences between the two approaches to determine the source of the difference in performance. We do notice, however, that there is a fairly large difference in model complexity. An SVM classifier depends upon an unspecified number of variables (the PSM feature vectors chosen as support vectors), even in the case that there are few features, whereas Combyne uses only six parameters regardless of the size of the data set.
We again note that one protein-level feature (the number of sibling peptides) gave a substantial improvement in Percolator’s results on the search against the large protein database; this is evidence that peptide-centric approaches that avoid protein-level features altogether are relatively weak methods. ProteinsFirst provides a way to use an integrated protein-level feature (protein rank) without biasing the PSM FDR estimate.
ProteinsFirst is a natural extension of the target/decoy strategy that can simultaneously measure and control the false discovery rate at two different levels: the PSM level and the protein level. ProteinsFirst rests on essentially the same theoretical underpinnings as the usual target/decoy strategy, namely that decoy PSMs faithfully model false target PSMs. ProteinsFirst, however, relies on a more detailed parallel between targets and decoys than is necessary for estimating only the PSM FDR. The usual target/decoy strategy gives an unbiased estimate of PSM FDR so long as the decoy database faithfully models the target peptides. ProteinsFirst, however, simultaneously estimates protein- and PSM-level FDR, and hence the decoys must match the targets in protein lengths and numbers as well as in peptide-level statistics. Fortuitously, most groups are already using decoy databases appropriate for ProteinsFirst.
One additional caution applies to ProteinsFirst and indeed to all protein-level FDR estimation. In very simple samples, for example, an excised gel band, almost all the true PSMs may hit a single protein. In this case, ProteinsFirst’s protein FDR estimate suffers from undersampling: if there is only one true protein, then admitting two proteins gives a protein FDR of 50%. ProteinsFirst’s PSM FDR estimate will also suffer from undersampling, but not as severely, and indeed will be essentially the same as the estimate from the usual peptide-centric target/decoy strategy.
Except for the problem of undersampling in simple samples, ProteinsFirst can be applied regardless of the relative numbers of tandem mass spectra in the data set and proteins in the protein database, so that users need not decide whether a peptide- or a protein-centric method is more appropriate. ProteinsFirst gives a lower and more predictable protein FDR than the peptide-centric methods benchmarked here, and this advantage holds across a wide range of spectrum to protein ratios. ProteinsFirst gives a more predictable PSM FDR than protein-centric methods, and in the case of a high spectrum to protein ratio, say 2 to 1 or larger, ProteinsFirst’s PSM FDR is apt to be much lower as well, because in this case a significant number of false PSMs hit true proteins.
We have described ProteinsFirst for the basic target-decoy strategy, but the algorithm can be adapted to variant target-decoy strategies. The algorithm can also be generalized to other multilevel identification problems. The abstract requirements are an observed base level (e.g., PSMs) along with any number of higher levels (e.g., peptides, proteins, protein families or organisms), all of which can be modeled simultaneously by an appropriately designed system of decoys. If higher-level identifications are informative about the correctness of base-level identifications, then a multidimensional algorithm should outperform the simplistic approach of accepting/rejecting base level identifications without consideration of the higher levels.
Acknowledgments
MB was supported in part by NIH grant R21GM094557 and YJK by an NSF CRA Computing Innovations postdoctoral fellowship.
References
- 1.Eng J, McCormack AL, Yates JR. An approach to correlate tandem mass spectra data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 2.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 3.Moore RE, Young MK, Lee TD. Qscore: an algorithm for evaluating SEQUEST database search results. Journal of the American Society for Mass Spectrometry. 2002;13:378–386. doi: 10.1016/S1044-0305(02)00352-5. [DOI] [PubMed] [Google Scholar]
- 4.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 5.Navarro P, Vazquez J. A refined method to calculate false discovery rates for peptide identification using decoy databases. Journal of proteome research. 2009;8:1792–1796. doi: 10.1021/pr800362h. [DOI] [PubMed] [Google Scholar]
- 6.Wang G, Wu WW, Zhang Z, Masilamani S, Shen RF. Decoy methods for assessing false positives and false discovery rates in shotgun proteomics. Analytical chemistry. 2009;81:146–159. doi: 10.1021/ac801664q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen C, et al. On the estimation of false positives in peptide identifications using decoy search strategy. Proteomics. 2009;9:194–204. doi: 10.1002/pmic.200800330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang WH, Shilov IV, Seymour SL. Nonlinear fitting method for determining local false discovery rates from decoy database searches. Journal of proteome research. 2008;7:3661–3667. doi: 10.1021/pr070492f. [DOI] [PubMed] [Google Scholar]
- 9.Bern M, Cai Y, Goldberg D. Lookup peaks: a hybrid of de novo sequencing and database search for protein identification by tandem mass spectrometry. Analytical chemistry. 2007;79:1393–1400. doi: 10.1021/ac0617013. [DOI] [PubMed] [Google Scholar]
- 10.Falkner JA, et al. Validated MALDI-TOF/TOF mass spectra for protein standards. Journal of the American Society for Mass Spectrometry. 2007;18:850–855. doi: 10.1016/j.jasms.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Bern M, Goldberg D. Improved ranking functions for protein and modification-site identifications. J Comput Biol. 2008;15:705–719. doi: 10.1089/cmb.2007.0119. [DOI] [PubMed] [Google Scholar]
- 12.Weatherly DB, et al. A Heuristic method for assigning a false-discovery rate for protein identifications from Mascot database search results. Mol Cell Proteomics. 2005;4:762–772. doi: 10.1074/mcp.M400215-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Reiter L, et al. Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol Cell Proteomics. 2009;8:2405–2417. doi: 10.1074/mcp.M900317-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Mol Cell Proteomics. 2005;4:1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 16.Everett LJ, Bierl C, Master SR. Unbiased statistical analysis for multi-stage proteomic search strategies. Journal of proteome research. 9:700–707. doi: 10.1021/pr900256v. [DOI] [PubMed] [Google Scholar]
- 17.Bern M, Kil YJ. Comment on "unbiased statistical analysis for multi-stage proteomic search strategies". Journal of proteome research. 10:2123–2127. doi: 10.1021/pr101143m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spivak M, Weston J, Bottou L, Kall L, Noble WS. Improvements to the percolator algorithm for Peptide identification from shotgun proteomics data sets. Journal of proteome research. 2009;8:3737–3745. doi: 10.1021/pr801109k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng J, Naiman DQ, Cooper B. Probability model for assessing proteins assembled from peptide sequences inferred from tandem mass spectrometry data. Analytical chemistry. 2007;79:3901–3911. doi: 10.1021/ac070202e. [DOI] [PubMed] [Google Scholar]
- 20.Kil YJ, Becker C, Sandoval W, Goldberg D, Bern M. Preview: a program for surveying shotgun proteomics tandem mass spectrometry data. Analytical chemistry. 83:5259–5267. doi: 10.1021/ac200609a. [DOI] [PMC free article] [PubMed] [Google Scholar]