Abstract
Typhoid fever is a persistent infection caused by host-adapted Salmonella strains adept at circumventing immune-mediated host defences. Given the importance of T cells in protection, the culling of activated CD4+ T cells after primary infection has been proposed as a potential immune evasion strategy used by this pathogen. We demonstrate that the purging of activated antigen-specific CD4+ T cells after virulent Salmonella infection requires SPI-2 encoded virulence determinants, and is not restricted only to cells with specificity to Salmonella-expressed antigens, but extends to CD4+ T cells primed to expand by co-infection with recombinant Listeria monocytogenes. Unexpectedly, however, the loss of activated CD4+ T cells during Salmonella infection demonstrated using a monoclonal population of adoptively transferred CD4+ T cells was not reproduced among the endogenous repertoire of antigen-specific CD4+ T cells identified with MHC class II tetramer. Analysis of T-cell receptor variable segment usage revealed the selective loss and reciprocal enrichment of defined CD4+ T-cell subsets after Salmonella co-infection that is associated with the purging of antigen-specific cells with the highest intensity of tetramer staining. Hence, virulent Salmonella triggers the selective culling of high avidity activated CD4+ T-cell subsets, which re-shapes the repertoire of antigen-specific T cells that persist later after infection.
Keywords: bacteria/bacterial infection, CD4/helper T cells, infection, T-cell receptor, tetramers
Introduction
Typhoid fever is a systemic infection caused by Salmonella strains that are highly adapted for persistence in humans, which include Salmonella enterica serotype Typhi and serotype Paratyphi.1,2 By extension, infection in mice with the related Salmonella enterica serotype Typhimurium (S. typhimurium) strain results in systemic persistent infection that recapitulates many clinical features of typhoid fever in humans.3,4 Hence, Salmonella have developed species-specific survival strategies that allow for persistent infection, and S. typhimurium infection in mice is a representative model for dissecting the Salmonella-specific immune evasion strategies required for persistent infection.5,6
T cells play important roles in optimal host defence after primary Salmonella infection in both humans and mice.7–11 For example, in patients with typhoid fever, the presence of T-cell-mediated immune responses is associated with faster recovery rates and a reduction in disease-related sequelae.11 Similarly, mice with T-cell defects are more susceptible because even attenuated Salmonella mutants can cause fatal infections.12–14 These protective effects can be more specifically attributed to the CD4+ T-cell subset because ablation of these cells significantly worsens Salmonella infection outcomes.6,13,15,16 By extension, Salmonella-expressed virulence determinants inhibit the priming of naive CD4+ T cells, which coincides with their blunted activation within the first 2–3 weeks after infection.6,17–21 Hence, circumventing the activation of protective CD4+ T cells represents an important survival strategy used by Salmonella for establishing and maintaining persistent infection.
In addition to impeding their activation, the active elimination of pathogen-specific CD4+ T cells after their expansion has been proposed as an additional immune evasion tactic used by Salmonella.22 However, the applicability of these findings to immune evasion by virulent Salmonella is uncertain because only attenuated mutants required to bypass the innate susceptibility of Nramp1-susceptible mice were analysed. By contrast, CD4+ T cells with the same specificity were found to progressively expand through day 37 after virulent Salmonella infection in Nramp1-resistant mice when MHC class II tetramers were used to track the response.6 In this regard, given the cross-regulation between Nramp1 and the expression of Salmonella virulence determinants,23 further investigation on the in vivo dynamics of Salmonella-specific CD4+ T cells primed by virulent infection in Nramp1-resistant mice seems imperative. Furthermore, whether the reported culling of activated CD4+ T cells by Salmonella is restricted only to cells with specificity for Salmonella-expressed antigen or more generalizably to other activated T-cell subsets is unknown. To address these questions, we tracked the dynamics of antigen-specific CD4+ T cells after infection with virulent Salmonella in Nramp1-resistant mice, which results in persistent instead of acute fatal infection.24,25 Using isogenic Salmonella mutants, we dissect the Salmonella intrinsic features that dictate the purging of antigen-specific CD4+ T cells after infection. We present results that demonstrate the selective culling of high avidity activated CD4+ T cells during persistent Salmonella infection.
Materials and methods
Mice
C57BL/6 (Nramp1-susceptible) and 129/Sv (Nramp1-resistant) mice were purchased from the National Cancer Institute, and B6.129 F1 mice were generated by intercrossing C57BL/6 female mice with 129/Sv male mice as described previously.19,26 CD45.1+ (Ly5.1+, Ptprca+/+) were purchased from the Jackson Laboratory. SM1 T-cell receptor (TCR) transgenic mice with CD4+ T cells specific for the I-Ab FliC427–441 peptide have been described elsewhere,27 and were maintained on a CD45.1+ Rag-1-deficient background. For adoptive transfer, 5 × 104 CD4+ T cells from SM1 TCR transgenic mice or 1 × 107 to 2 × 107 CD4+ T cells from CD45.1+ T cells were intravenously transferred 1 day before infection. All mice were maintained in specific pathogen-free facilities, and used when between 6 and 8 weeks of age according to University of Minnesota IACUC approved protocols.
Bacteria and infections
Wild-type S. typhimurium (SL1344) and the isogenic aflagellated mutant (BC490), the Salmonella pathogenicity island 2 (SPI-2) deficient (HH109) and corresponding isogenic wild-type (SL12023) strains have each been described elsewhere.22,27–30 For Salmonella infection, each strain was back-diluted to log-phase growth in brain–heart infusion medium at 37°, washed and diluted with sterile saline, and injected intravenously through the lateral tail vein (104 colony-forming units in 200 μl per mouse). To eradicate Salmonella, enrofloxacin was added to the drinking water (2 mg/ml) beginning on day 5 after infection as described previously.31 Recombinant Listeria monocytogenes (Lm) that are attenuated because of targeted defects in actA (required for intracellular and intercellular spread) and engineered to express the FliC427−441 (Lm-FliC) or 2W1S52–68 (Lm-2W1S) peptides as L. monocytogenes endogenous antigens have each been described previously.32,33 For L. monocytogenes infection, each strain was grown in brain–heart infusion medium supplemented with chloramphenicol (20 μg/ml) at 37°, washed and diluted with sterile saline, and injected intravenously (5 × 106 colony-forming units in 200 μl per mouse).
Reagents for flow cytometry and MHC tetramer staining
Fluorophore-conjugated antibodies and other reagents for flow cytometry were purchased from BD Biosciences (San Jose, CA) or eBioscience (San Diego, CA). MHC class II I-Ab 2W1S52–68 or FliC427–441 tetramers, and techniques for staining and enrichment of antigen-specific CD4+ T cells using these reagents have been described.6,32,34 The TCR variable segment usage among tetramer-positive and bulk CD4+ T cells was analysed after staining with FITC-conjugated antibodies to mouse Vβ2, Vβ3, Vβ4, Vβ5.1/5.2, Vβ6, Vβ7, Vβ8.1/8.2, Vβ8.3, Vβ9, Vβ10b, Vβ11, Vβ12, Vβ13, Vβ14 and Vβ17a (BD Biosciences).
Statistics
The percentage of cells, total cell numbers, and tetramer staining intensity for cells recovered between groups of mice were compared using Student's t-test (GraphPad, Prism software, La Jolla, CA) with P<0·05 taken as statistical significance.
Results
Antigen-specific CD4+ T cells expand, but are not sustained after Salmonella infection
To identify the dynamics whereby virulent Salmonella primes antigen-specific CD4+ T cells, we enumerated the expansion and contraction kinetics of Salmonella-specific CD4+ T cells following infection with the virulent wild-type strain SL1344. To bypass the innate susceptibility of mice containing only the susceptible allele of Nramp1, we used B6.129 F1 mice that are simultaneously resistant and develop persistent infection with virulent Salmonella and do not reject adoptively transferred cells from TCR transgenic mice on the C57BL/6 background.26 CD4+ T cells from CD45.1+ SM1 TCR transgenic mice with specificity for the Salmonella FliC427–441 I-Ab peptide were adoptively transferred into B6.129 F1 mice to track the Salmonella-specific CD4+ T-cell response early (day 5; when the bacterial burden is progressively increasing) and later (day 21; when the bacterial burden peaks and begins to decline).6,31 We found that FliC-specific CD45.1+ CD4+ T cells expand within the first 5 days after virulent Salmonella infection, indicating the efficient priming of these antigen-specific CD4+ T cells (Fig. 1). However, over the next 2 weeks, FliC-specific CD45.1+ CD4+ T cells were eliminated and by day 21 after infection they were below the limits of detection (< 200 cells) and < 0·1% compared with levels on day 5 (Fig. 1). Interestingly, these results illustrating the expansion and rapid elimination of adoptively transferred FliC-specific CD4+ T cells by virulent Salmonella are in sharp contrast to the progressive expansion from day 5 to day 37 for the relatively small population of endogenous CD4+ T cells with the same specificity identified with MHC class II tetramer,6 but reproduce the previously reported culling of activated adoptively transferred FliC-specific CD4+ T cells by avirulent Salmonella after infection in Nramp1-susceptible mice.22
Figure 1.
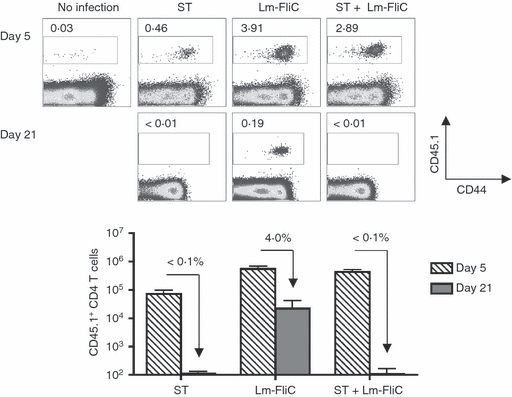
FliC-specific CD4+ T cells are primed, but not sustained after Salmonella infection. Percent (top) and total numbers (bottom) of FliC-specific CD45.1+ among CD4+ T cells at the indicated time-points after infection with wild-type Salmonella (ST), recombinant Listeria monocytogenes expressing the FliC427–441 peptide (Lm-FliC), or co-infection with both bacterial strains. The percent FliC-specific CD45.1+ CD4+ T cells remaining on day 21 compared with day 5 are indicated for each infection condition. These results are representative of three independent experiments each with similar results containing 6–10 mice per group. Bar, 1 SE.
To establish if the short-lived nature of adoptively transferred FliC-specific CD4+ T cells was unique to Salmonella infection, we also compared the expansion and contraction kinetics of these cells after infection with recombinant L. monocytogenes engineered to express the FliC427–441 peptide (Lm-FliC) as an endogenous antigen.33 By contrast to the near complete elimination of these cells that occurs after Salmonella infection, FliC-specific CD45.1+ CD4+ cells primed by Lm-FliC were maintained, and by day 21 after infection were found at ∼ 5% levels compared with day 5 (Fig. 1). Hence, Salmonella-specific features not shared by recombinant L. monocytogenes cause the elimination of expanded pathogen-specific CD4+ T cells later after infection.
To begin interrogating the apparent discordance between infection with Salmonella and recombinant L. monocytogenes in priming sustained pathogen-specific CD4+ T cells, we enumerated the dynamics of FliC-specific CD4+ T cells in mice after co-infection with Salmonella and Lm-FliC compared with mice infected with each separately. Despite the robust initial expansion of FliC-specific CD4+ T cells that occurs after Salmonella and Lm-FliC co-infection compared with mice infected with Salmonella alone, these cells were not sustained and were found only at levels below the limits of detection on day 21 after infection (Fig. 1). Hence, co-infection with Lm-FliC does not rescue the culling of adoptively transferred pathogen-specific CD4+ T cells dictated by Salmonella. Taken together, these results indicate that Salmonella triggers the active elimination of pathogen-specific CD4+ T cells after expansion during transition into the memory pool.
Culling of activated CD4+ T cells requires SPI-2 encoded virulence determinants
Given the efficiency whereby recombinant L. monocytogenes primes CD4+ T-cell expansion and the dominant impacts dictated by virulent Salmonella that result in the near complete elimination of adoptively transferred CD4+ T cells, related experiments sought to identify the Salmonella intrinsic features required for purging antigen-specific CD4+ T cells using co-infection with defined Salmonella mutant strains. We initially investigated proteins that form the structural framework of SPI-2 because the expression of these virulence factors is controlled by host cell Nramp1, and they participate in impeding dendritic cell antigen presentation and CD4+ T-cell priming.20,21,23 To determine the requirement for Salmonella SPI-2 in culling pathogen-specific CD4+ T cells, we compared the dynamics of adoptively transferred FliC-specific CD4+ T cells after co-infection with Lm-FliC and Salmonella containing targeted non-polar defects in ssaV, which encodes a structural component of the SPI-2 type III secretion system (strain HH109, STΔSPI-2), or an isogenic strain of wild-type Salmonella (strain SL12023).29,30 In striking contrast to the near complete elimination of FliC-specific CD4+ T cells after co-infection with wild-type Salmonella, FliC-specific CD45.1+ CD4+ T cells were maintained when STΔSPI-2 was used for co-infection instead (Fig. 2). Furthermore, the percent and total number of FliC-specific CD4+ T cells on day 21 after STΔSPI-2 and Lm-FliC co-infection, and the contraction rate compared with levels on day 5 were each indistinguishable from that when primed by Lm-FliC alone (Fig. 2). Hence, SPI-2-associated virulence determinants are essential for the elimination of adoptively transferred CD4+ T cells after Salmonella infection.
Figure 2.
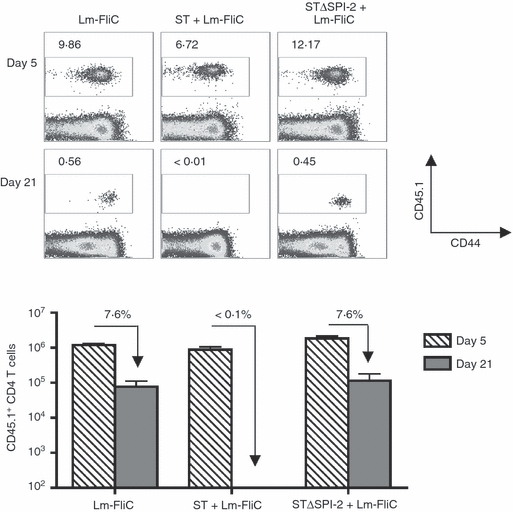
Elimination of FliC-specific CD4+ T cells during Salmonella infection is SPI-2 dependent. Percent (top) and total numbers (bottom) of FliC-specific CD45.1+ among CD4+ T cells at the indicated time-points after infection with recombinant Listeria monocytogenes expressing the FliC peptide (Lm-FliC), or co-infection with Lm-FliC and wild-type Salmonella (ST) or ΔSPI-2 Salmonella (STΔSPI-2). The percent FliC-specific CD45.1+ CD4+ T cells remaining on day 21 compared with day 5 are indicated for each infection condition. These results are representative of three independent experiments each with similar results containing six or seven mice per group. Bar, 1 SE.
CD4+ T-cell culling is not restricted to those with specificity for Salmonella-expressed antigens
Next, we examined if the culling of CD4+ T cells was restricted only to those with specificity for Salmonella-expressed antigens. These experiments used aflagellated Salmonella (ΔFliC, strain BC490) that do not stimulate the expansion of adoptively transferred FliC427–441-specific CD4+ T cells from SM1 TCR transgenic mice or isogenic wild-type Salmonella (strain SL1344) for co-infection with Lm-FliC (Fig. 3a).27,33 Compared with Lm-FliC infection alone, co-infection using either wild-type or ΔFliC Salmonella each with Lm-FliC primed similar levels of FliC-specific CD4+ T-cell expansion on day 5 after infection (Fig. 3a). Interestingly, however, these cells were not sustained and fell below the limits of detection by day 21 post-infection in mice co-infected with either ΔFliC or wild-type Salmonella (Fig. 3a). By contrast, FliC-specific CD4+ T cells were maintained at ∼ 5% of day 5 levels in mice infected with Lm-FliC alone without Salmonella. These results demonstrate that the elimination of CD4+ T cells during Salmonella infection is not restricted to those with specificity for Salmonella-expressed antigen, but extends to CD4+ T cells simultaneously stimulated by recombinant L monocytogenes.
Figure 3.
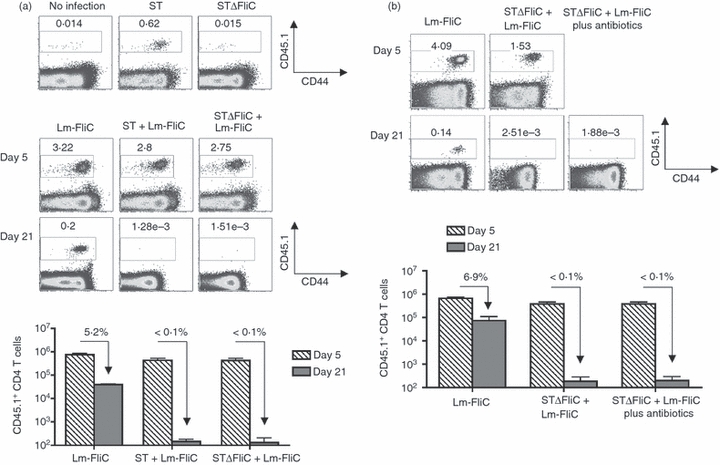
Elimination of CD4+ T cells during Salmonella infection is not restricted to those with specificity to Salmonella-expressed antigens. (a) Percent FliC-specific CD45.1+ among CD4+ T cells on day 5 after infection with wild-type Salmonella (ST) or aflagellated Salmonella (STΔFliC) (top). Percent and total numbers (bottom) of FliC-specific CD45.1+ among CD4+ T cells at the indicated time-points after infection with recombinant Listeria monocytogenes expressing the FliC peptide (Lm-FliC), or co-infection with Lm-FliC and wild-type (ST) or aflagellated Salmonella (STΔFliC). (b) Percent and total numbers of FliC-specific CD45.1+ among CD4+ T cells at the indicated time-points after infection with recombinant Lm expressing the FliC peptide (Lm-FliC), or co-infection with Lm-FliC plus aflagellated Salmonella (STΔFliC) in mice treated with enrofloxacin in the drinking water beginning day 5 after infection. The percent FliC-specific CD45.1+ CD4+ T cells remaining on day 21 compared with day 5 are indicated for each infection condition. These results are representative of three independent experiments each with similar results containing 6–10 mice per group. Bar, 1 SE.
Using a similar approach, these experiments were extended to investigate if progressively increasing Salmonella bacterial burden within the first 21 days after infection was required for the active elimination of antigen-specific CD4+ T cells primed by recombinant L monocytogenes. We found that treatment with antibiotics that efficiently eradicates Salmonella beginning on day 5 after infection31 did not prevent the near complete elimination of adoptively transferred FliC-specific CD4+ T cells in mice co-infected with Lm-FliC plus ΔFliC Salmonella (Fig. 3b). These results indicate that stimulation signals within the first few days after Salmonella infection dictate the later persistence of expanding antigen-specific CD4+ T cells.
Polyclonal CD4+ T cells among the endogenous repertoire escape Salmonella-induced culling
To further investigate if the culling of adoptively transferred FliC-specific CD4+ T cells during Salmonella infection is representative of the larger polyclonal CD4+ T-cell response, we measured the expansion and persistence of endogenous antigen-specific CD4+ T cells that are identified using I-Ab MHC class II tetramers. Given the large precursor frequency among endogenous CD4+ T cells in naive mice with specificity to the 2W1S52–68 I-Ab peptide,34 we enumerated the impacts of Salmonella co-infection on the dynamics of 2W1S-specific CD4+ T cells in mice primed by recombinant L monocytogenes engineered to express this peptide as an endogenous antigen (Lm-2W1S).32,34 Consistent with our previous findings after Lm-2W1S infection in C57BL/6 mice, Lm-2W1S infection in B6.129 F1 mice primed the robust expansion of 2W1S-specific CD4+ T cells that peak at day 7, and later contract to 10–20% of peak levels by day 21 (Fig. 4). Interestingly, and in sharp contrast to the near elimination of adoptively transferred FliC-specific CD4+ T cells primed by Salmonella and Lm-FliC co-infection that occurs regardless of Salmonella FliC expression (Fig. 3), co-infection with virulent Salmonella did not significantly impact the maintenance of 2W1S-specific CD4+ T cells primed by Lm-2W1S (Fig. 4). Specifically, the contraction of 2W1S-specific CD4+ T cells between days 7 and 21 after infection for mice infected with Lm-2W1S alone compared with those co-infected with Lm-2W1S and Salmonella was similar, each approaching ∼ 18%. These results suggest that the culling of activated CD4+ T cells dictated by virulent Salmonella does not extend to the endogenous repertoire of antigen-specific CD4+ T cells primed by recombinant L. monocytogenes.
Figure 4.
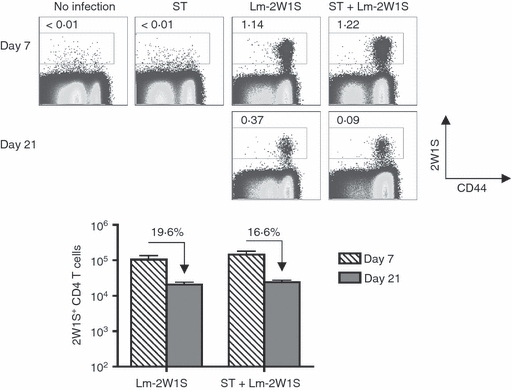
Endogenous antigen-specific CD4+ T cells identified with MHC class II tetramer escape elimination during Salmonella infection. Percent (top) and total numbers (bottom) of 2W1S tetramer positive among CD4+ T cells at the indicated time-points after infection with recombinant Listeria monocytogenes expressing the 2W1S peptide (Lm-2W1S), or co-infection with Lm-2W1S and wild-type Salmonella (ST). The percent 2W1S tetramer-positive CD4+ T cells remaining on day 21 compared with day 7 are indicated for each infection condition. These results are representative of three independent experiments each with similar results containing 6–10 mice per group. Bar, 1 SE.
Given the inherent differences related to tracking antigen-specific T cells among adoptively transferred cells based on expression of the CD45.1 congenic marker and endogenous cells based on MHC II tetramer staining, related experiments compared the dynamics of antigen-specific CD4+ T cells among the polyclonal repertoire after adoptive transfer. We transferred one spleen-equivalent of cells from naive CD45.1+ mice into recipient B6.129 (CD45.1−) F1 mice that were subsequently infected with Lm-2W1S alone or Lm-2W1S plus Salmonella, and enumerated the ensuing 2W1S-specific CD4+ T-cell response among CD45.1+ donor and CD45.1− endogenous cells. Compared with peak expansion levels, we found that 2W1S-specific CD4+ T cells were maintained to a similar degree among adoptively transferred CD45.1+ cells and endogenous CD45.1− cells at the later time-point after Lm-2W1S infection regardless of Salmonella co-infection (Fig. 5). Together, these results demonstrate that the sustained maintenance of antigen-specific CD4+ T cells among the polyclonal repertoire, unlike monoclonal cells from TCR transgenic mice, is not the result of inherent survival differences between adoptively transferred and endogenous cells.
Figure 5.
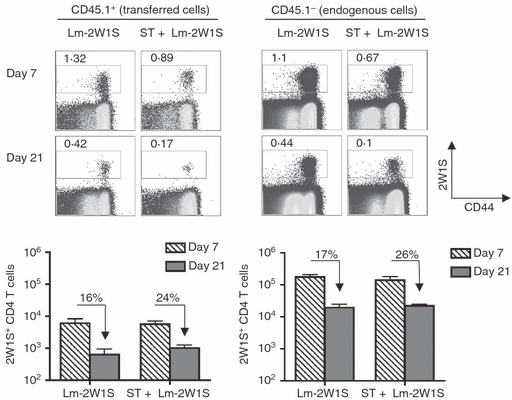
Survival of antigen-specific CD4+ T cells identified using MHC class II tetramer extends to adoptively transferred polyclonal cells. Percent (top) and total numbers (bottom) of 2W1S tetramer positive among adoptively transferred CD45.1+ or endogenous CD45.1− CD4+ T cells at the indicated time-points after infection with recombinant Listeria monocytogenes expressing the 2W1S peptide (Lm-2W1S), or co-infection with Lm-2W1S and wild-type Salmonella (ST). The percent 2W1S tetramer-positive CD4+ T cells remaining on day 21 compared with day 7 are indicated for each infection condition. These results are representative of three independent experiments each with similar results containing 6 to 8 mice per group. Bar, 1 SE.
Shifts in TCR variable segment usage among antigen-specific CD4+ T cells after Salmonella infection
The apparent discordance in susceptibility to elimination for monoclonal compared with polyclonal antigen-specific CD4+ T cells led us to investigate if Salmonella infection triggers the selective elimination of CD4+ T-cell subsets among the endogenous repertoire by measuring their relative usage of TCR variable β (Vβ) segments. Consistent with the shift in relative usage of specific Vβ segments among 2W1S tetramer-positive CD4+ T cells in unmanipulated C57BL/6 mice or after stimulation with cognate peptide plus lipopolysaccharide,34 we found 2W1S tetramer-positive cells expressing Vβ5.1/5.2 and Vβ11 were over-represented, while those expressing Vβ6, Vβ12 and Vβ14 were under-represented compared with bulk tetramer-negative CD4+ T cells early (day 7) after infection with either Lm-2W1S alone or co-infection with Lm-2W1S plus Salmonella (Fig. 6). However, and in sharp contrast to this early time-point, significant shifts in the relative usage of numerous specific TCR Vβ segments became apparent for 2W1S cells primed by Lm-2W1S plus Salmonella co-infection compared with Lm-2W1S infection alone by day 21 (Fig. 6). Specifically, the proportion of 2W1S tetramer-positive CD4+ T cells that express Vβ7, Vβ8.1/8.2, Vβ8.3 and Vβ10b were each significantly expanded (Vβ7, 6·3-fold increase, P<0·01; Vβ8.1/8.2, 1·7-fold increase, P<0·02; Vβ8.3, 2·0-fold increase, P<0·05; Vβ10b, 1·7-fold increase, P < 0·02) whereas those that express other undefined Vβ segments were reciprocally reduced (3·4-fold reduction, P<0·001) in Lm-2W1S mice co-infected with virulent Salmonella compared with those infected with Lm-2W1S alone (Fig. 6).
Figure 6.
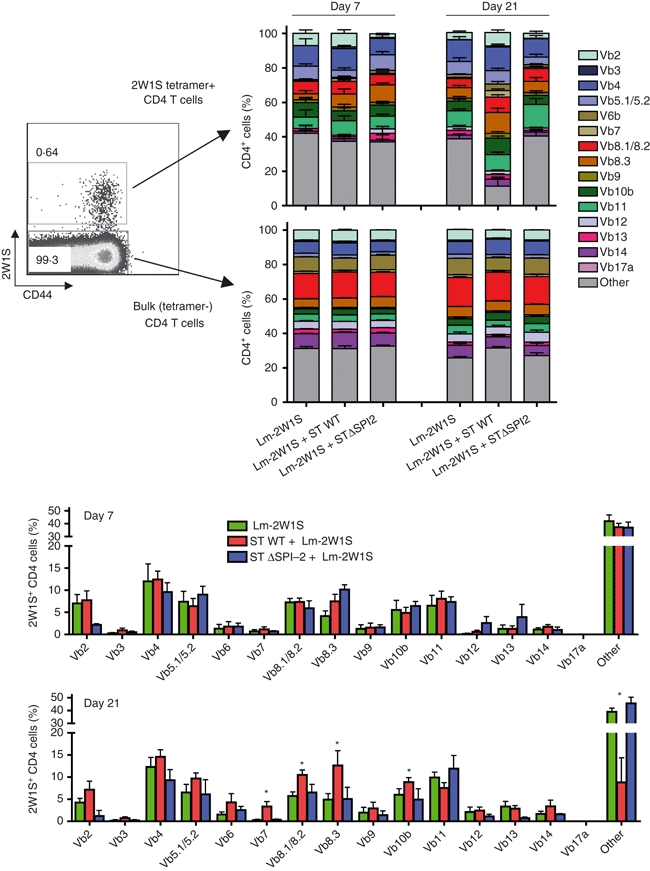
Shifts in T-cell receptor (TCR) Vβ repertoire usage among endogenous CD4+ T cells after Salmonella infection are SPI-2-dependent. Relative usage of TCR Vβ segments among 2W1S tetramer-positive or bulk CD4+ T cells in mice on days 7 or 21 after infection with recombinant Listeria monocytogenes expressing the 2W1S peptide (Lm-2W1S) alone, or after co-infection with Lm-2W1S plus either wild-type (ST) or SPI-2-deficient (STΔSPI-2) Salmonella. These results are representative of three independent experiments each with similar results containing 6–12 mice per group. Bar, 1 SE. *P<0·05.
Based on the requirement for Salmonella SPI-2 in culling adoptively transferred FliC-specific CD4+ T cells (Fig. 2), related experiments investigated if these shifts in TCR Vβ segment usage among endogenous antigen-specific CD4+ T cells after Salmonella infection were also SPI-2-dependent. We found that the relative usage of TCR Vβ segments did not change significantly for 2W1S-specific CD4+ T cells early after co-infection with Lm-2W1S and STΔSPI-2 (Fig. 6). Remarkably, however, at the later infection time-point (day 21), shifts in relative usage of Vβ segments for 2W1S tetramer cells primed by wild-type Salmonella were eliminated when the SPI-2-deficient strain was used for co-infection with Lm-2W1S (Fig. 6). Importantly, although these shifts occurred for CD4+ T cells without specificity for Salmonella-expressed antigen, they were restricted to cells stimulated by recombinant L. monocytogenes because neither infection with ΔSPI-2 nor wild-type Salmonella caused significant shifts in the relative usage of Vβ segments among bulk (tetramer-negative) CD4+ T cells at early or late infection time-points (Fig. 6). These results indicate that Salmonella shifts the clonal diversity of activated antigen-specific CD4+ T cells from early to late infection time-points through an SPI-2 dependent process.
Selective elimination of CD4+ T cells with high avidity for pathogen-associated antigens by Salmonella
Given the direct correlation between TCR avidity and the binding intensity of MHC II–peptide complexes,35,36 we also investigated if shifts in the TCR Vβ repertoire among activated antigen-specific CD4+ T cells by Salmonella represent a loss of high or low avidity clones by enumerating the relative intensity of tetramer staining among endogenous 2W1S-specific CD4+ T cells primed by Lm-2W1S with or without Salmonella co-infection. Early after infection (day 7), the intensity of 2W1S tetramer staining was not significantly different in cells recovered from Lm-2W1S-infected mice regardless of Salmonella co-infection (Fig. 7a). By contrast, at day 21 after infection, the intensity of 2W1S tetramer staining was sharply reduced (∼ 40%, P<0·01) for cells recovered from mice co-infected with wild-type, but not SPI-2-deficient, Salmonella each compared with those recovered from mice infected with Lm-2W1S alone (Fig. 7a). These results are each consistent with the non-significant changes in Vβ usage among 2W1S tetramer-positive cells early (day 7) after infection, and selective shifts among activated 2W1S-specific CD4+ T cells later (day 21) by wild-type Salmonella that require SPI-2 (Fig. 6). Together, these findings demonstrate that Salmonella use SPI-2-dependent virulence features to selectively purge high avidity antigen-specific CD4+ T cells, which re-shapes the repertoire of activated T cells that persist through later infection time-points.
Figure 7.
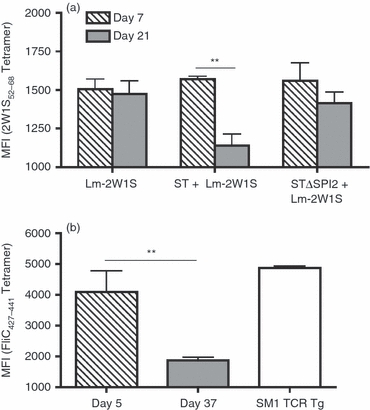
Selective elimination of CD4+ T cells that bind tetramer with high affinity by virulent Salmonella. (a) Mean fluorescence intensity (MFI) of 2W1S52–68 tetramer staining for antigen-specific CD4+ T cells recovered from mice on day 7 or 21 after Listeria monocytogenes (Lm) -2W1S infection alone, or after co-infection with Lm-2W1S plus either wild-type (ST) or SPI-2-deficient (STΔSPI-2) Salmonella. (b) Mean fluorescence intensity (MFI) of FliC427–441 tetramer staining for antigen-specific CD4+ T cells recovered from mice on day 5 or 37 after wild-type Salmonella infection compared with CD4+ T cell in SM1 TCR transgenic mice. These results are representative of two independent experiments containing 6 to 8 mice per group. Bar, 1 SE. **P<0·01.
To investigate if the selective loss of high avidity CD4+ T cells after Lm-2W1S and Salmonella co-infection explains the discordance between the progressive expansion of endogenous FliC-specific CD4+ T cells identified using MHC class II tetramer6 and the near complete elimination of adoptively transferred CD4+ T cells from TCR transgenic mice with the same specificity (Fig. 1), we compared the relative intensity of I-Ab FliC427–441 tetramer staining for endogenous polyclonal CD4+ T cells recovered from mice on days 5 and 37 after wild-type Salmonella infection with FliC-specific CD4+ T cells recovered from SM1 TCR transgenic mice. Consistent with the hypothesis that Salmonella selectively prevents the accumulation of activated CD4+ T cells with high avidity for pathogen-associated antigen, sharp reductions in the intensity of FliC427–441 tetramer staining for cells recovered on day 5 compared with day 37 after Salmonella infection were identified (Fig. 7b). In turn, FliC-specific CD4+ T cells from SM1 TCR transgenic mice bound FliC427–441 tetramer with the highest intensity, to levels even higher than that recovered from mice on post-infection day 5 (Fig. 7b). Hence, adoptively transferred FliC-specific CD4+ T cells from SM1 TCR transgenic mice like those endogenous cells that bind tetramer with the highest intensity early after infection are not sustained after expansion during Salmonella infection. Together with the elimination of FliC-specific CD4+ T cells by aflagellated Salmonella with Lm-FliC co-infection (Fig. 3) and shifts in intensity of tetramer staining for 2W1S-specific CD4+ T cells by wild-type Salmonella with Lm-2W1S co-infection (Fig. 7a), these results demonstrate that the culling of high avidity activated CD4+ T cells is not limited only to cells with specificity for Salmonella-expressed antigen, but extends to those activated by recombinant L. monocytogenes co-infection.
Discussion
Pathogens that cause persistent infection have developed intricate immune evasion strategies that facilitate their survival within susceptible hosts.3,4,37 For infection caused by intracellular bacteria within the Salmonella genus, host defence is primarily provided by CD4+ T cells,6–8,13,15,16 so circumventing protective CD4+ T-cell responses probably plays an important role for Salmonella in vivo survival and pathogenesis. In this report, we used virulent Salmonella to probe the dynamics of pathogen-specific CD4+ T cells at relatively early and later time-points during persistent infection. We find that adoptively transferred CD4+ T cells from TCR transgenic mice with specificity to the Salmonella FliC427–441 MHC class II peptide readily expand in response to virulent Salmonella infection, but are subsequently eliminated and not sustained. In contrast, adoptively transferred CD4+ T cells of the same specificity primed with recombinant Lm-FliC were sustained at 5–10% peak levels. Co-infection experiments with Lm-FliC and either wild-type or various Salmonella mutants demonstrate that the culling of activated CD4+ T cells is actively mediated by virulent Salmonella (Fig. 1), requires Salmonella SPI-2 encoded virulence determinants (Fig. 2), and is not limited only to CD4+ T cells with specificity for Salmonella-expressed antigens but extends to cells activated by L. monocytogenes co-infection (Fig. 3).
These initial series of experiments tracking the Salmonella-specific CD4+ T-cell response using adoptively transferred cells from TCR transgenic mice with fixed antigen specificity expand the significance of earlier observations in several unique aspects. First by using virulent Salmonella for infection in mice with the resistant allele of Nramp1, we demonstrate that the elimination of antigen-specific CD4+ T cells previously reported for attenuated Salmonella in mice homozygous for the susceptible Nramp1 allele extends to virulent Salmonella.22 Furthermore, the finding that Salmonella eliminates even adoptively transferred CD4+ T cells primed with recombinant L. monocytogenes, a potent inducer of long-lived T cells, indicates that virulent Salmonella actively overrides survival signals for pathogen-specific CD4+ T cells stimulated by L. monocytogenes. Second, these results establish an essential role for SPI-2-dependent virulence determinants in the culling of activated CD4+ T cells. In this regard, although a role for SPI-2-encoded virulence genes has been implicated in the loss of FliC-specific CD4+ T cells in Nramp1-susceptible mice, the virulence retained by SPI-2-deficient Salmonella precluded analysis of CD4+ T-cell survival beyond 6 days after infection.22 These limitations were bypassed in our studies by using isogenic wild-type and mutant Salmonella containing targeted defects in ssaV, which encodes an essential structural component of the SPI-2 type III secretion system for infection in Nramp1-resistant mice.29,30 Third, the sustained elimination of adoptively transferred FliC-specific CD4+ T cells in Lm-FliC and Salmonella co-infected mice initiated on antibiotic therapy within 5 days post-infection indicates that stimulation signals imprinted within the first few days after virulent Salmonella infection programme cells that expand but are non-sustained. Finally, the efficient and near complete elimination of FliC-specific CD4+ T cells even after co-infection with aflagellated Salmonella indicates that the culling of CD4+ T cells is not restricted to only those with specificity for Salmonella-expressed antigen, but also extends to activated cells primed using the unrelated intracellular bacterium L. monocytogenes. Hence, CD4+ T cells are primed for expansion after Salmonella infection in an antigen-specific manner, but eliminated in a non-antigen-specific fashion shortly after.
Despite the unambiguous nature of these results using adoptively transferred cells from TCR transgenic mice to track the CD4+ T-cell response after Salmonella infection, complementary experiments using MHC class II tetramers to identify antigen-specific cells among the polyclonal repertoire of endogenous cells revealed that the elimination of activated CD4+ T cells after Salmonella infection was incomplete (Fig. 4). This discordance was not the result of the continuous refilling of the endogenous cell compartment from thymic emigrants or other inherent survival differences for adoptively transferred compared with endogenous cells because the persistence of antigen-specific CD4+ T cells identified using MHC class II tetramers with Salmonella co-infection occurred to a similar extent for polyclonal adoptively transferred compared with endogenous CD4+ T cells (Fig. 5). Instead, by examining the relative intensity of tetramer staining and TCR Vβ segment usage among tetramer-positive cells, we find the selective loss of activated CD4+ T cells that bind tetramer with the highest intensity that is associated with dramatic shifts in relative usage of TCR Vβ segments from early to later time-points after co-infection with wild-type, but not SPI-2-deficient Salmonella (Fig. 6). Hence, Salmonella through an SPI-2-dependent process modifies the repertoire and clonal diversity of activated CD4+ T cells that persist later after infection by selectively eliminating those with highest avidity. These results reconcile the apparent discordance between the near complete elimination of adoptively transferred monoclonal CD4+ T cells from TCR transgenic mice selected based on high avidity to cognate antigen,27 and the maintenance of pathogen-specific CD4+ T cells among polyclonal endogenous cells at later infection time-points,6 and suggest that only CD4+ T-cell subsets with reduced or intermediate avidity for pathogen-associated antigens are retained. In this regard, although the percentage of activated CD44hi, CD62Llo, CD69hi and interferon-γ-producing CD4+ T cells each progressively increases beginning on day 5 through to day 31 during persistent Salmonella infection,6 the specificity of these cells remaining at later infection time-points are probably skewed to those with reduced avidity for Salmonella antigens. The identification of other Salmonella-specific antigens, and the development of tools to track the CD4+ T-cell response to these antigens are required to more comprehensively dissect differences in affinity for Salmonella-specific CD4+ T cells at early and late infection time-points.
The discordance in survival between adoptively transferred CD4+ T cells from TCR transgenic mice with fixed specificity and the endogenous polyclonal repertoire of antigen-specific CD4+ T cells identified using MHC class II tetramers during Salmonella infection are congruent with the elimination of relatively low avidity lymphocytic choriomeningitis virus GP61–80-specific CD4+ T cells from SMARTA TCR transgenic mice and maintenance of CD4+ T cells with higher avidity for the same antigen among the endogenous repertoire after recombinant L. monocytogenes infection.38 Although these differences in survival for GP61–80-specific CD4+ T cells after recombinant L. monocytogenes or lymphocytic choriomeningitis virus infection could be attributed to TCR avidity for antigens expressed by each respective pathogen, other factors dictate the culling of CD4+ T cells after Salmonella infection because the elimination of FliC-specific CD4+ T cells occurs even after infection with aflagellated Salmonella, which does not stimulate the expansion of these cells (Fig. 3). Similarly, virulent Salmonella that does not prime the expansion of 2W1S-specific CD4+ T cells nevertheless triggers dramatic shifts in the relative usage of TCR Vβ segments among 2W1S tetramer-positive cells from early to late infection time-points that is associated with the selective loss of high avidity 2W1S-specific CD4+ T-cell clones. Accordingly, these findings demonstrate that Salmonella actively curtails the survival of high avidity activated T cells in a non-antigen-specific fashion. Taken together, these results indicate that the culling of antigen-specific CD4+ T cells based on TCR avidity is a more generalizable phenomenon that occurs after infection with pathogens that cause acute and persistent infection. For persistent Salmonella infection in particular, identifying the specific SPI-2-dependent virulence factors and establishing if the loss of high avidity activated T cells is restricted only to the CD4+ subset or more broadly applicable to CD8+ T cells represents important areas for further investigation.
Acknowledgments
We thank Drs Marc Jenkins and Stephen McSorley for providing I-Ab 2W1S52–68 and FliC427–441 tetramers. This research was supported by grants R01AI087830 (NIAID to SSW), F30DK084674 (NIDDK to JHR), F31AI091298 (NIAID to MRN), and University of Minnesota Grant-in-Aid.
Disclosures
The authors have no conflicts of interest, or financial conflicts to disclose.
References
- 1.Parry CM, Hien TT, Dougan G, White NJ, Farrar JJ. Typhoid fever. N Engl J Med. 2002;347:1770–82. doi: 10.1056/NEJMra020201. [DOI] [PubMed] [Google Scholar]
- 2.Crump JA, Mintz ED. Global trends in typhoid and paratyphoid fever. Clin Infect Dis. 2010;50:241–6. doi: 10.1086/649541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monack DM, Mueller A, Falkow S. Persistent bacterial infections: the interface of the pathogen and the host immune system. Nat Rev Microbiol. 2004;2:747–65. doi: 10.1038/nrmicro955. [DOI] [PubMed] [Google Scholar]
- 4.Tischler AD, McKinney JD. Contrasting persistence strategies in Salmonella and Mycobacterium. Curr Opin microbiol. 2010;13:93–9. doi: 10.1016/j.mib.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monack DM, Bouley DM, Falkow S. Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNgamma neutralization. J Exp Med. 2004;199:231–41. doi: 10.1084/jem.20031319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johanns TM, Ertelt JM, Rowe JH, Way SS. Regulatory T cell suppressive potency dictates the balance between bacterial proliferation and clearance during persistent Salmonella infection. PLoS pathog. 2010;6:e1001043. doi: 10.1371/journal.ppat.1001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaufmann SH. Immunity to intracellular bacteria. Annu Rev Immunol. 1993;11:129–63. doi: 10.1146/annurev.iy.11.040193.001021. [DOI] [PubMed] [Google Scholar]
- 8.Mittrucker HW, Kaufmann SH. Immune response to infection with Salmonella typhimurium in mice. J Leukoc Biol. 2000;67:457–63. doi: 10.1002/jlb.67.4.457. [DOI] [PubMed] [Google Scholar]
- 9.Guilloteau L, Buzoni-Gatel D, Bernard F, Lantier I, Lantier F. Salmonella abortusovis infection in susceptible BALB/c by mice: importance of Lyt-2+ and L3T4+ T cells in acquired immunity and granuloma formation. Microb Pathog. 1993;14:45–55. doi: 10.1006/mpat.1993.1005. [DOI] [PubMed] [Google Scholar]
- 10.Nelson MR, Shanson DC, Hawkins DA, Gazzard BG. Salmonella, Campylobacter and Shigella in HIV-seropositive patients. AIDS (London, England) 1992;6:1495–8. doi: 10.1097/00002030-199212000-00012. [DOI] [PubMed] [Google Scholar]
- 11.Rajagopalan P, Kumar R, Malaviya AN. Immunological studies in typhoid fever. II. Cell-mediated immune responses and lymphocyte subpopulations in patients with typhoid fever. Clin Exp Immunol. 1982;47:269–74. [PMC free article] [PubMed] [Google Scholar]
- 12.Hess J, Ladel C, Miko D, Kaufmann SH. Salmonella typhimurium aroA– infection in gene-targeted immunodeficient mice: major role of CD4+ TCR-alpha beta cells and IFN-gamma in bacterial clearance independent of intracellular location. J Immunol. 1996;156:3321–6. [PubMed] [Google Scholar]
- 13.Nauciel C. Role of CD4+ T cells and T-independent mechanisms in acquired resistance to Salmonella typhimurium infection. J Immunol. 1990;145:1265–9. [PubMed] [Google Scholar]
- 14.Sinha K, Mastroeni P, Harrison J, de Hormaeche RD, Hormaeche CE. Salmonella typhimurium aroA, htrA, and aroD htrA mutants cause progressive infections in athymic (nu/nu) BALB/c mice. Infect Immun. 1997;65:1566–9. doi: 10.1128/iai.65.4.1566-1569.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pie S, Truffa-Bachi P, Pla M, Nauciel C. Th1 response in Salmonella typhimurium-infected mice with a high or low rate of bacterial clearance. Infect Immun. 1997;65:4509–14. doi: 10.1128/iai.65.11.4509-4514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mastroeni P, Villarreal-Ramos B, Hormaeche CE. Role of T cells, TNF alpha and IFN gamma in recall of immunity to oral challenge with virulent salmonellae in mice vaccinated with live attenuated aro–Salmonella vaccines. Microb Pathog. 1992;13:477–91. doi: 10.1016/0882-4010(92)90014-f. [DOI] [PubMed] [Google Scholar]
- 17.Matsui K, Arai T. Salmonella infection-induced non-responsiveness of murine splenic T-lymphocytes to interleukin-2 (IL-2) involves inhibition of IL-2 receptor gamma chain expression. FEMS Immunol Med Microbiol. 1998;20:175–80. doi: 10.1111/j.1574-695X.1998.tb01125.x. [DOI] [PubMed] [Google Scholar]
- 18.Srinivasan A, McSorley SJ. Pivotal advance: exposure to LPS suppresses CD4+ T cell cytokine production in Salmonella-infected mice and exacerbates murine typhoid. J Leukoc Biol. 2007;81:403–11. doi: 10.1189/jlb.0306194. [DOI] [PubMed] [Google Scholar]
- 19.Mittrucker HW, Kohler A, Kaufmann SH. Characterization of the murine T-lymphocyte response to Salmonella enterica serovar Typhimurium infection. Infect Immun. 2002;70:199–203. doi: 10.1128/IAI.70.1.199-203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halici S, Zenk SF, Jantsch J, Hensel M. Functional analysis of the Salmonella pathogenicity island 2-mediated inhibition of antigen presentation in dendritic cells. Infect Immun. 2008;76:4924–33. doi: 10.1128/IAI.00531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tobar JA, Carreno LJ, Bueno SM, Gonzalez PA, Mora JE, Quezada SA, Kalergis AM. Virulent Salmonella enterica serovar typhimurium evades adaptive immunity by preventing dendritic cells from activating T cells. Infect Immun. 2006;74:6438–48. doi: 10.1128/IAI.00063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srinivasan A, Nanton M, Griffin A, McSorley SJ. Culling of activated CD4 T cells during typhoid is driven by Salmonella virulence genes. J Immunol. 2009;182:7838–45. doi: 10.4049/jimmunol.0900382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zaharik ML, Vallance BA, Puente JL, Gros P, Finlay BB. Host–pathogen interactions: host resistance factor Nramp1 up-regulates the expression of Salmonella pathogenicity island-2 virulence genes. Proc Natl Acad Sci USA. 2002;99:15705–10. doi: 10.1073/pnas.252415599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malo D, Vogan K, Vidal S, et al. Haplotype mapping and sequence analysis of the mouse Nramp gene predict susceptibility to infection with intracellular parasites. Genomics. 1994;23:51–61. doi: 10.1006/geno.1994.1458. [DOI] [PubMed] [Google Scholar]
- 25.Gruenheid S, Gros P. Genetic susceptibility to intracellular infections: Nramp1, macrophage function and divalent cations transport. Curr Opin microbiol. 2000;3:43–8. doi: 10.1016/s1369-5274(99)00049-1. [DOI] [PubMed] [Google Scholar]
- 26.Luu RA, Gurnani K, Dudani R, Kammara R, van Faassen H, Sirard JC, Krishnan L, Sad S. Delayed expansion and contraction of CD8+ T cell response during infection with virulent Salmonella typhimurium. J Immunol. 2006;177:1516–25. doi: 10.4049/jimmunol.177.3.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McSorley SJ, Asch S, Costalonga M, Reinhardt RL, Jenkins MK. Tracking salmonella-specific CD4 T cells in vivo reveals a local mucosal response to a disseminated infection. Immunity. 2002;16:365–77. doi: 10.1016/s1074-7613(02)00289-3. [DOI] [PubMed] [Google Scholar]
- 28.McSorley SJ, Cookson BT, Jenkins MK. Characterization of CD4+ T cell responses during natural infection with Salmonella typhimurium. J Immunol. 2000;164:986–93. doi: 10.4049/jimmunol.164.2.986. [DOI] [PubMed] [Google Scholar]
- 29.Shea JE, Beuzon CR, Gleeson C, Mundy R, Holden DW. Influence of the Salmonella typhimurium pathogenicity island 2 type III secretion system on bacterial growth in the mouse. Infect Immun. 1999;67:213–9. doi: 10.1128/iai.67.1.213-219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deiwick J, Nikolaus T, Shea JE, Gleeson C, Holden DW, Hensel M. Mutations in Salmonella pathogenicity island 2 (SPI2) genes affecting transcription of SPI1 genes and resistance to antimicrobial agents. J Bacteriol. 1998;180:4775–80. doi: 10.1128/jb.180.18.4775-4780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johanns TM, Law CY, Kalekar LA, O'Donnell H, Ertelt JM, Rowe JH, Way SS. Early eradication of persistent Salmonella infection primes antibody-mediated protective immunity to recurrent infection. Microbes infect. 2011;13:322–30. doi: 10.1016/j.micinf.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ertelt JM, Rowe JH, Johanns TM, Lai JC, McLachlan JB, Way SS. Selective priming and expansion of antigen-specific Foxp3− CD4+ T cells during Listeria monocytogenes infection. J Immunol. 2009;182:3032–8. doi: 10.4049/jimmunol.0803402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johanns TM, Ertelt JM, Lai JC, Rowe JH, Avant RA, Way SS. Naturally occurring altered peptide ligands control Salmonella-specific CD4+ T cell proliferation, IFN-gamma production, and protective potency. J Immunol. 2010;184:869–76. doi: 10.4049/jimmunol.0901804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, Jenkins MK. Naive CD4+ T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–13. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crawford F, Kozono H, White J, Marrack P, Kappler J. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity. 1998;8:675–82. doi: 10.1016/s1074-7613(00)80572-5. [DOI] [PubMed] [Google Scholar]
- 36.Moon JJ, Dash P, Oguin TH, III, McClaren JL, Chu HH, Thomas PG, Jenkins MK. Quantitative impact of thymic selection on Foxp3+ and Foxp3− subsets of self-peptide/MHC class II-specific CD4+ T cells. Proc Natl Acad Sci USA. 2011;108:14602–7. doi: 10.1073/pnas.1109806108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young D, Hussell T, Dougan G. Chronic bacterial infections: living with unwanted guests. Nat Immunol. 2002;3:1026–32. doi: 10.1038/ni1102-1026. [DOI] [PubMed] [Google Scholar]
- 38.Williams MA, Ravkov EV, Bevan MJ. Rapid culling of the CD4+ T cell repertoire in the transition from effector to memory. Immunity. 2008;28:533–45. doi: 10.1016/j.immuni.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]


