Abstract
The complete genome of the Orgyia leucostigma nucleopolyhedrovirus (OrleNPV) isolated from the whitemarked tussock moth (Orgyia leucostigma, Lymantridae: Lepidoptera) was sequenced, analyzed, and compared to other baculovirus genomes. The size of the OrleNPV genome was 156,179 base pairs (bp) and had a G+C content of 39%. The genome encoded 135 putative open reading frames (ORFs), which occupied 79% of the entire genome sequence. Three inhibitor of apoptosis (ORFs 16, 43 and 63), and five baculovirus repeated ORFs (bro-a through bro-e) were interspersed in the OrleNPV genome. In addition to six direct repeat (drs), a common feature shared among most baculoviruses, OrleNPV genome contained three homologous regions (hrs) that are located in the latter half of the genome. The presence of an F-protein homologue and the results from phylogenetic analyses placed OrleNPV in the genus Alphabaculovirus, group II. Overall, OrleNPV appears to be most closely related to group II alphabaculoviruses Ectropis obliqua (EcobNPV), Apocheima cinerarium (ApciNPV), Euproctis pseudoconspersa (EupsNPV), and Clanis bilineata (ClbiNPV).
Keywords: alphabaculovirus, baculovirus, white-marked tussock moth
1. Introduction
The family Baculoviridae consists of rod-shaped, enveloped, and occluded viruses mainly pathogenic to insects in the orders Lepidoptera, Diptera and Hymenoptera [1]. In recent years, there has been continued interest in baculovirus genome studies mainly due to their application as biopesticides of major insect pests in forests and agriculture [2–4], and also as vectors for protein expression and gene therapy [5–6].
Baculoviruses genomes are characterized by their large, covalently-closed, circular, double-stranded DNA molecules containing numerous open reading frames (ORFs) which are distributed on both strands. Baculoviruses are classified into four genera based on host specificities and phylogenetic analysis; Alphabaculovirus (lepidopteran-specific nucleopolyhedroviruses or NPVs), Betabaculovirus (lepidopteran-specific granuloviruses or GVs), Gammabaculovirus (hymenopteran-specific NPVs), and Deltabaculovirus (dipteran-specific NPVs) [7]. Alphabaculoviruses are subdivided further, based, in part, on the presence of genes encoding different glycoproteins that serve similar functions associated with budded virions (BV), gp64 in group I and f-protein in group II [8–10]. Baculovirus genomes range in size from 81,755 bp for Neodiprion lecontei NPV (NeleNPV) to 178,733 bp for Xestia c-nigrum GV (XecnGV) [11,12]. To date, 58 baculovirus genomes including 41 Alphabaculovirus, 13 Betabaculovirus, three Gammabaculovirus and one Deltabaculovirus have been sequenced and are available in the National Centre for Biotechnology Information (NCBI) database (Table 1).
Table 1.
Characteristics of baculovirus genomes. The 58 currently sequenced baculovirus genomes are summarized according to their current classification scheme and alphabetical order.
| Genus (Group) | Virus name | Abbreviation | Length (bp) | G+C (%) | No. of ORFs | 1 No. of hrs | 2 No. of bro | GenBank accession | 3 Reference |
|---|---|---|---|---|---|---|---|---|---|
| Alphabaculovirus (Group I) | Antheraea pernyi NPV | AnpeNPV | 126,629 | 53 | 147 | 3 | 2 | DQ486030 | [69] |
| Antheraea pernyi MNPV-L2 | AnpeMNPV-L2 | 126,246 | 53 | 145 | 6 | 2 | EF207986 | [70] | |
| Anticarsia gemmatalis MNPV-2D | AgMNPV-2D | 132,239 | 44 | 152 | 9 | 8 | DQ813662 | [71] | |
| Autographa californica MNPV-C6 | AcMNPV-C6 | 133,894 | 40 | 156 | 9 | 1 | L22858 | [17] | |
| Bombyx mori NPV-T3 | BmNPV-T3 | 128,413 | 40 | 143 | 7 | 5 | L33180 | [72] | |
| Bombyx mandarina NPV-S1 | BomaNPV-S1 | 126,770 | 40 | 141 | 7 | 3 | FJ882854 | [73] | |
| Choristoneura fumiferana MNPV | CfMNPV | 129,593 | 50 | 146 | 5 | 1 | AF512031 | [30] | |
| Choristoneura fumiferana DEF MNPV | CfDEFMNPV | 131,160 | 45 | 149 | 13 | 2 | AY327402 | [74] | |
| Epiphyas postivittana NPV | EppoNPV | 118,584 | 40 | 136 | 5 | 1 | AY043265 | [75] | |
| Hyphantria cunea NPV | HycuNPV | 132,959 | 45 | 148 | 6 | 5 | AP009046 | [76] | |
| Maruca vitrata NPV | MaviNPV | 111,953 | 39 | 126 | 5 | 0 | EF125867 | [77] | |
| Orgyia pseudotsugata MNPV | OpMNPV | 131,990 | 55 | 152 | 5 | 3 | U75930 | [29] | |
| Plutella xylostella MNPV (CL3) | PlxyMNPV | 134,417 | 40 | 153 | 9 | 2 | DQ457003 | [78] | |
| Rachiplusia ou MNPV-R1 | RoMNPV-R1 | 131,526 | 39 | 149 | 9 | 0 | AY145471 | [79] | |
| Alphabaculovirus (Group II) | Adoxophyes honmai NPV | AdhoNPV | 113,220 | 35 | 125 | 4 | 4 | AP006270 | [80] |
| Adoxophyes orana NPV | AdorNPV | 111,724 | 35 | 121 | 4 | 3 | EU591746 | [81] | |
| Agrotis ipsilon MNPV | AgipMNPV | 155,122 | 48 | 163 | 7 | 5 | EU839994 | [82] | |
| Agrotis segetum NPV-A | AgseNPV-A | 147,544 | 45 | 153 | 5 | 4 | DQ123841 | [83] | |
| Apocheima cinerarium NPV | ApciNPV | 123,876 | 45 | 118 | 4 | 0 | FJ914221 | NP | |
| Chrysodeixis chalcites NPV | ChchNPV | 149,622 | 39 | 151 | 0 | 4 | AY864330 | [57] | |
| Clanis bilineata NPV | ClbiNPV | 135,454 | 37 | 139 | 0 | 3 | DQ504428 | [21] | |
| Ectropis obliqua NPV | EcobNPV | 131,204 | 37 | 126 | 3 | 2 | DQ837165 | [28] | |
| Euproctis pseudoconspersa NPV | EupsNPV | 141,291 | 40 | 139 | 4 | 2 | FJ227128 | [84] | |
| Helicoverpa armigera SNPV-G4 | HearSNPV-G4 | 131,405 | 39 | 135 | 5 | 3 | AF271059 | [85] | |
| Helicoverpa armigera SNPV-C1 | HearSNPV-C1 | 130,759 | 38 | 137 | 5 | 3 | AF30304 | [86] | |
| Helicoverpa armigera MNPV | HearMNPV | 154,196 | 40 | 162 | 4 | 6 | EU730893 | NP | |
| Helicoverpa armigera SNPV NNg1 | HearSNPV-NNg1 | 132,425 | 39 | 143 | 5 | 4 | AP010907 | [87] | |
| Helicoverpa zea SNPV | HzSNPV | 130,869 | 39 | 139 | 5 | 2 | AF334030 | [88] | |
| Leucania separata NPV | LeseNPV | 168,041 | 48 | 169 | 8 | 10 | AY394490 | [89] | |
| Lymantria dispar MNPV | LdMNPV | 161,046 | 57 | 164 | 13 | 16 | AF081810 | [43] | |
| Lymantria xylina MNPV | LyxyMNPV | 156,344 | 53 | 157 | 13 | 14 | GQ202541 | [48] | |
| Mamestra configurata NPV-A (90/2) | MacoNPV-A (90/2) | 155,060 | 41 | 169 | 4 | 8 | U59461 | [46] | |
| Mamestra configurata NPV-A (90/4) | MacoNPV-A (90/4) | 153,656 | 168 | 4 | 7 | AF539999 | [90] | ||
| Mamestra configurata NPV-96B | MacoNPV-B | 158,482 | 40 | 169 | 4 | 7 | AYI26275 | [47] | |
| Orgyia leucostigma NPV | OrleNPV | 156,179 | 39 | 135 | 3 | 5 | EU309041 | TP | |
| Spodoptera exigua MNPV | SeMNPV | 135,611 | 43 | 139 | 6 | 0 | AF169823 | [45] | |
| Spodoptera frugiperda MNPV-3AP2 | SfMNPV-3AP2 | 131,330 | 40 | 143 | 8 | 1 | EF035042 | [91] | |
| Spodoptera frugiperda MNPV-19 | SfMNPV-19 | 132,565 | 40 | 141 | 8 | 1 | EU258200 | [92] | |
| Spodoptera litura MNPV-G2 | SpltMNPV-G2 | 139,342 | 42 | 141 | 17 | 2 | AF325155 | [22] | |
| Spodoptera litura MNPV- II | SpltMNPV- II | 148,634 | 44 | 147 | 7 | 2 | EU780426 | NP | |
| Trichoplusia ni SNPV | TnSNPV | 134,394 | 39 | 144 | 0 | 2 | DQ017380 | [93] | |
| Betabaculovirus | Adoxophyles orona GV | AdorGV | 99,657 | 34 | 119 | 0 | 0 | AF547984 | [94] |
| Agrotis segetum GV | AgseGV | 131,680 | 37 | 132 | 0 | 0 | AY522332 | NP | |
| Choristoneura occidentalis GV | ChocGV | 104,710 | 32 | 116 | 5 | 0 | DQ333351 | [61] | |
| Clostera anachoreta GV | ClanGV | 101,487 | 44 | 123 | 4 | 0 | HQ116624 | [95] | |
| Cryptophlebia leucotreta GV | CrleGV | 110,907 | 32 | 128 | 3 | 0 | AY229987 | [96] | |
| Cydia pomonella GV | CpGV | 123,500 | 45 | 143 | 0 | 1 | U53466 | [97] | |
| Helicoverpa armigera GV | HearGV | 169,794 | 40 | 179 | 9 | 10 | EU255577 | [98] | |
| Phthorimaea operculella GV | PhoGV | 119,217 | 35 | 130 | 12 | 1 | AF499596 | NP | |
| Pieris rapae GV | PrGV | 108,592 | 33 | 120 | 0 | 0 | GQ884143 | [99] | |
| Plutella xylostella GV | PxGV | 100,999 | 40 | 120 | 4 | 0 | AF270937 | [100] | |
| Pseudaletia unipuncta GV | PsunGV | 176,677 | 39 | 183 | 9 | 7 | EU678671 | NP | |
| Spodoptera litura GV | SpltGV | 124,121 | 38 | 136 | 0 | 6 | DQ288858 | [101] | |
| Xestia c-nigrum GV | XecnGV | 178,733 | 40 | 181 | 9 | 7 | AF162221 | [15] | |
| Gammabaculovirus | Neodiprion abietis NPV | NeabNPV | 84,264 | 33 | 93 | 5 | 0 | DQ317692 | [102] |
| Neodiprion lecontei NPV | NeleNPV | 81,755 | 33 | 89 | 0 | 0 | AY349019 | [12] | |
| Neodiprion sertifer NPV | NeseNPV | 86,462 | 33 | 90 | 6 | 0 | AY430810 | [11] | |
| Deltabaculovirus | Culex nigripalpus NPV | CuniNPV | 108,252 | 109 | 50 | 4 | 6 | AF403738 | [56] |
hrs and bro refers to homologous regions and baculovirus repeated ORFs, respectively.
(TP: this publication; NP: not published, please reference GenBank accession number.)
Baculoviruses exhibit two phenotypically distinct phenotypes: (i) budded virions (BV), and (ii) occlusion derived virions (ODV) that are produced during the early and late phases of virus replication, respectively. The ODV phenotypes for granuloviruses (GV) (Betabaculovirus) are embedded in a small granulin protein matrix unlike those of nucleopolyhedroviruses (NPV) that are contained in a large polyhedrin matrix. The OBs of GVs contain single virions while NPVs contain multiple virions within the polyhedrin matrix.
The ODVs of the alphabaculovirus infecting the whitemarked tussock moth (WMTM) Orgyia leucostigma (L.) have single nucleocapsids per virion. The WMTM is a common pest of balsam fir trees (Abies balsamea (L.) Miller) in Canada but will feed on a number of conifer, decidiuous trees, and agricultural crops such as blueberries (Vaccinium spp.). OrleNPV has been documented to contribute to the collapse of WMTM populations in Atlantic Canada [13]. Experimental applications of OrleNPV for the control of WMTM infestations have been made in Nova Scotia [14]. To better understand the molecular basis of OrleNPV pathogenicity, viral DNA from a Nova Scotia isolate was purified, sequenced and compared with other baculoviruses using various phylogenetic tools. The OrleNPV genome was found to be 156,179 bp in size and was confirmed to belong in the genus Alphabaculovirus, group II.
2. Results and Discussion
2.1. Nucleotide Sequence Analysis
The circular OrleNPV genome was 156,179 bp in size, making it the eighth largest baculovirus genome sequenced to date, with others ranging from 81,755 bp for Neodiprion lecontei NPV (NeleNPV) [12] to 178,733 bp for Xestia c-nigrum GV (XecnGV) [15]. The OrleNPV genome is AT-rich having only 39% G+Cs, lower than the average GC content of group I (44.9 %) and group II (41.6%) alphabaculoviruses [16]. As originally defined, [17], the adenine of methionine start codon of the polyhedrin gene represented the zero point on the OrleNPV physical map (Figure 1), and was designated as ORF 1 (Table 2). Overall, 135 putative ORFs, with a minimal size of 50 amino acids and with promoter motifs corresponding to various transcriptional profiles, were detected and encoded for 79% of the total genome (Tables 1 and 2). Fifty-eight (42.9%) of the ORFs were forward oriented, whereas 77 (57.1%) were reverse oriented (Figure 1). Interestingly, OrleNPV encodes for relatively fewer genes (135) compared to other baculoviruses where the average number of ORFs is 140.5. This could be attributed to the presence of several copies of hrs, drs, and large intergenic regions, which collectively account for 21 % on the entire genome. Multiple copies of hrs and drs may be a replication strategy for the virus to initiate DNA replication from several regions leading to rapid rate of DNA replication in the host system.
Figure 1.
Linear map of Orgyia leucostigma nucleopolyhedrovirus (OrleNPV) genome. The arrows and arrow heads indicate the orientation of the predicted open reading frames (ORFs). The rectangles indicate homologous regions (hrs). The number above the horizontal lines denotes the nucleotide position (bp) relative to the start codon of polyhedrin ORF. Thirty-one baculovirus core genes are shown in bold letters.
Table 2.
OrleNPV genome annotation.
| ORF Name 1 | Position 2 | Intergenic Length Prom | Homologues (% aa identity) 4 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Left | Dir | Right | (bp) | (aa) | motifs 3 | EcobNPV | ChchNPV | OpMNPV | AcMNPV | ||
| 1 | polyhedrin | 1 | > | 738 | 51 | 246 | L | 1(95) | 1(94) | 3(86) | 8(89) |
| 2 | orf1629 | 789 | < | 2531 | −7 | 581 | ? | 2(26) | 2(28) | - | 9(25) |
| 3 | pk1 | 2524 | > | 3327 | 293 | 268 | L | 3(61) | 3(49) | 1(34) | 10(37) |
| 4 | hoar | 3620 | < | 5245 | 523 | 542 | E | 4(28) | 4(23) | - | - |
| rep1 | 4273 | 4348 | |||||||||
| 5 | orle001 | 5768 | > | 6715 | 232 | 316 | ? | - | - | - | - |
| rep2 | 6725 | 6809 | |||||||||
| 6 | odv-e56 | 6947 | > | 8080 | 124 | 378 | L | 5(71) | 7(39) | 146(59) | 148(59) |
| 7 | ie-1 | 8204 | < | 10120 | 70 | 639 | E | 6(52) | 16(31) | 145(30) | 147(33) |
| 8 | ep23 | 10190 | > | 10792 | 118 | 201 | ? | 7(36) | 15(36) | 144(38) | 146(30) |
| 9 | chtB1 | 10910 | < | 11185 | 17 | 92 | L | 8(64) | 14(53) | 142(44) | 145(52) |
| 10 | odv-e27 | 11202 | < | 12062 | 89 | 287 | L | 9(64) | 13(62) | 141(47) | 144(51) |
| 11 | odv-e18 | 12151 | < | 12420 | 14 | 90 | L | 10(42) | - | - | - |
| 12 | p49 | 12434 | < | 13921 | 22 | 496 | L | 11(57) | 11(57) | 139(44) | 142(47) |
| 13 | ie-0 | 13943 | < | 14794 | 665 | 284 | L | 12(49) | 10(39) | 138(27) | 141(31) |
| rep3 | 15148 | 15218 | |||||||||
| 14 | me53 | 15459 | > | 16604 | 90 | 382 | E,L | 13(43) | 8(34) | 137(23) | 139(23) |
| 15 | p74 | 16694 | > | 18649 | 119 | 652 | L | 14(71) | 17(57) | 134(59) | 138(58) |
| 16 | iap-3 | 18768 | < | 19052 | 208 | 95 | E,L | - | - | 35(52) | - |
| 17 | dbp-1 | 19260 | > | 20042 | 28 | 261 | E | 27(23) | 22(20) | 43(28) | 25(27) |
| 18 | lef6 | 20070 | > | 20666 | 253 | 199 | L | 16(23) | 21(34) | 40(35) | 28(38) |
| 19 | unknown | 20919 | < | 21179 | 212 | 87 | ? | 17(39)- | 20(40)- | 39(34)- | 29(28)- |
| 20 | p26-a | 21391 | > | 22224 | 45 | 278 | L | 18(45) | 19(44) | 132(31) | 136(31) |
| 21 | p10 | 22269 | > | 22544 | 233 | 92 | L | 19(83) | 18(68) | - | - |
| 22 | cathepsin | 22777 | < | 23760 | 2298 | 328 | E,L | 51(79) | 64(56) | 125(64) | 127(70) |
| rep4 | 25452 | 25626 | |||||||||
| 23 | bro-a | 26058 | < | 26651 | 345 | 198 | ? | - | 114(27) | - | - |
| 24 | chitinase | 26996 | > | 28690 | 130 | 565 | L | 50(72) | 65(68) | 124(70) | 126(69) |
| 25 | unknown | 28820 | < | 29464 | 1924 | 215 | ? | - | - | - | - |
| 26 | unknown | 31388 | < | 31858 | 1052 | 157 | ? | - | - | - | - |
| 27 | ctl-1 | 32910 | < | 33068 | 123 | 53 | E, L | 47(62) | 74(86) | 136(63) | 3(41) |
| 28 | dna-ligase | 33191 | > | 35014 | 1335 | 608 | ? | - | - | - | - |
| rep5 | 35314 | 36297 | |||||||||
| 29 | gp37 | 36349 | < | 37152 | 358 | 268 | L | 49(57) | 67(64) | 69(57) | 64(58) |
| 30 | bro-b | 37510 | < | 38265 | 213 | 252 | ? | - | - | - | - |
| 31 | lef9 | 38478 | < | 39959 | 103 | 494 | ? | 46(78) | 52(74) | 65(63) | 62(68) |
| 32 | fp25 | 40062 | > | 40685 | 506 | 208 | E, L | 45(70) | 51(68) | 64(55) | 61(54) |
| 33 | chaB2 | 41191 | > | 41502 | 308 | 104 | ? | 44(46) | 50(48) | 63(48) | 60(46) |
| 34 | chaB1 | 41810 | > | 42331 | 72 | 174 | ? | 43(46) | 49(64) | 62(49) | 59(54) |
| 35 | unknown | 42403 | < | 42891 | 365 | 163 | ? | 42(58) | 48(43) | 61(35) | 57(40) |
| 36 | unknown | 43256 | < | 43543 | −55 | 96 | ? | 41(38) | 47(51) | - | - |
| 37 | unknown | 43488 | < | 43724 | 354 | 79 | ? | 40(46) | - | - | - |
| 38 | vp1054 | 44078 | < | 45112 | 18 | 345 | E | 39(50) | 45(51) | 58(35) | 54(38) |
| 39 | unknown | 45130 | > | 45396 | 6 | 89 | ? | - | 43(42) | - | - |
| 40 | unknown | 45402 | > | 46421 | 361 | 340 | ? | 36(24) | 42(31) | - | - |
| 41 | unknown | 46782 | < | 47198 | 54 | 139 | ? | 35(63) | 41(55) | 56(47) | 53(50) |
| 42 | unknown | 47252 | > | 47863 | 392 | 204 | ? | 34(26) | 40(32) | - | - |
| 43 | iap-2 | 48255 | > | 48728 | 43 | 158 | E | 54(29) | 62(38) | 74(29) | 71(28) |
| 44 | djbd | 48771 | < | 49754 | −42 | 328 | E,L | 33(27) | 38(23) | - | - |
| 45 | lef8 | 49712 | > | 52450 | −563 | 913 | E | 32(72) | 37(62) | 54(59) | 50(63) |
| 46 | unknown | 51887 | < | 52279 | 491 | 131 | ? | - | - | - | 17(60) |
| 47 | unknown | 52770 | > | 53852 | 219 | 361 | ? | - | - | 11(26) | 11(25) |
| 48 | unknown | 54071 | < | 54250 | 317 | 60 | ? | 30(46) | 35(38) | - | - |
| 49 | bro-c | 54567 | > | 55595 | 103 | 343 | ? | 76(56) | 69(45) | - | 2(21) |
| 50 | lef12 | 55698 | < | 56429 | 42 | 244 | ? | - | - | 46(38) | 41(48) |
| 51 | odv-e66a | 56471 | < | 58537 | 462 | 689 | ? | 31(29) | 101(39) | 50(29) | 46(30) |
| 52 | p47 | 58999 | > | 60183 | 290 | 395 | E | 29(63) | 33(61) | 45(49) | 40(52) |
| 53 | dbp-2 | 60473 | < | 61420 | 386 | 316 | E | 15(24) | 22(32) | 43(24) | 25(28) |
| 54 | bv-e31 | 61806 | > | 62489 | −66 | 228 | ? | 26(63) | 30(51) | 22(57) | 38(57) |
| 55 | lef11 | 62423 | > | 62839 | −76 | 139 | ? | 25(58) | 29(51) | 23(34) | 37(37) |
| 56 | 39K/pp31 | 62763 | > | 63659 | 277 | 299 | E | 24(51) | 28(33) | 24(34) | 36(34) |
| 57 | unknown | 63936 | < | 64151 | 47 | 72 | ? | 22(45) | - | - | - |
| 58 | ubiquitin | 64195 | < | 64443 | −203 | 83 | L | 21(83) | - | - | - |
| 59 | unknown | 64240 | > | 64479 | 13 | 80 | ? | - | - | - | - |
| 60 | unknown | 64492 | > | 65079 | 144 | 196 | ? | 20(50) | 25(31) | 26(37) | 34(30) |
| 61 | bro-d | 65223 | < | 66503 | 419 | 427 | E | 89(25) | - | - | 2(28) |
| 62 | p26-b | 66922 | < | 67611 | 124 | 230 | ? | 18(28) | 19(30) | - | 136(27) |
| 63 | iap-2 | 67735 | < | 68691 | 150 | 319 | E | 54(33) | 62(32) | - | 71(27) |
| 64 | unknown | 68841 | < | 69230 | −1 | 130 | ? | 55(65) | 61(52) | 73(41) | 68(43) |
| 65 | lef3 | 69229 | > | 70461 | 278 | 411 | L | 56(41) | 60(32) | 72(28) | 67(26) |
| 66 | desmoplakin | 70739 | < | 73192 | −1 | 818 | L | 57(40) | 59(26) | 66(20) | |
| 67 | DNA-poly | 73191 | > | 76406 | 68 | 1072 | E | 58(57) | 58(55) | 70(41) | 65(42) |
| 68 | unknown | 76474 | < | 76863 | 7 | 130 | ? | 59(57) | 57(42) | 78(26) | - |
| 69 | unknown | 76870 | < | 77124 | 114 | 85 | ? | 60(88) | 56(83) | 79(40) | 76(41) |
| 70 | vlf1 | 77238 | < | 78404 | 153 | 389 | L | 61(84) | 76(75) | 80(70) | 77(74) |
| 71 | unknown | 78557 | < | 78901 | 41 | 115 | ? | 62(38) | - | - | - |
| 72 | gp41 | 78942 | < | 80156 | 5 | 405 | L | 63(68) | 78(49) | 83(45) | 80(55) |
| 73 | unknown | 80161 | < | 80754 | −67 | 198 | ? | 64(65) | 79(69) | 84(50) | 81(52) |
| 74 | tlp-20 | 80687 | < | 81430 | −31 | 248 | L | 65(42) | 80(47) | - | 82(34) |
| 75 | vp91 | 81399 | > | 83870 | 167 | 824 | L | 66(45) | 81(42) | 86(39) | 83(39) |
| 76 | cg30 | 84037 | < | 84801 | 129 | 255 | ? | 67(26) | - | 89(25) | 88(21) |
| 77 | vp39 | 84930 | < | 85961 | −1 | 344 | L | 68(51) | 82(47) | 90(46) | 89(41) |
| 78 | lef4 | 85960 | > | 87393 | 162 | 478 | E | 69(53) | 83(51) | 91(42) | 90(46) |
| 79 | unknown | 87555 | < | 87962 | 196 | 136 | ? | 70(49) | - | - | - |
| 80 | unknown | 88158 | < | 88913 | −1 | 252 | ? | 71(57) | 84(55) | 93(38) | 92(42) |
| 81 | unknown | 88912 | > | 89382 | 0 | 157 | ? | 72(69) | 85(63) | 94(40) | 93(46) |
| 82 | odv-e25 | 89382 | > | 90047 | 152 | 222 | E,L | 73(68) | 86(64) | 95(39) | 94(43) |
| 83 | helicase | 90199 | < | 93912 | −43 | 1238 | E,L | 74(61) | 87(52) | 96(35) | 95(43) |
| 84 | odv-e28 | 93869 | > | 94384 | 93 | 172 | ? | 75(69) | 88(65) | 97(50) | 96(54) |
| 85 | 38K | 94477 | < | 95424 | −140 | 316 | L | 77(60) | 91(55) | 99(43) | 98(44) |
| 86 | lef5 | 95284 | > | 96198 | 26 | 305 | ? | 78(61) | 92(59) | 100(44) | 99(46) |
| 87 | p6.9 | 96224 | < | 96499 | 68 | 92 | L | 79(57) | 93(69) | 101(62) | 100(60) |
| rep6 | 96350 | 96431 | |||||||||
| 88 | p40 | 96567 | < | 97697 | 55 | 377 | E,L | 80(59) | 94(50) | 102(41) | 101(38) |
| 89 | p12 | 97752 | < | 98105 | −4 | 118 | E,L | 81(56) | 95(34) | 103(30) | 102(31) |
| 90 | p45 | 98101 | < | 99282 | 164 | 394 | E, L | 82(67) | 96(54) | 104(39) | 103(41) |
| 91 | vp80 | 99446 | > | 101776 | 107 | 777 | ? | 83(67) | 97(32) | 105(27) | 104(26) |
| 92 | unknown | 101883 | > | 102047 | 41 | 55 | ? | 84(69) | - | - | - |
| 93 | odv-ec43 | 102088 | > | 103161 | 38 | 358 | L | 85(69) | 99(53) | 109(46) | 109(47) |
| 94 | unknown | 103199 | > | 103453 | 527 | 85 | ? | 86(54) | 100(39) | - | - |
| hr1 | 103528 | 103833 | |||||||||
| 95 | dUTPase | 103980 | > | 104399 | 110 | 140 | E | - | 119(49) | - | - |
| 96 | unknown | 104509 | < | 105501 | 118 | 331 | ? | 90(40) | - | - | 112(45) |
| 97 | endonuclease | 105619 | < | 106029 | 216 | 137 | ? | 88(40) | - | - | - |
| 98 | nrk1 | 106245 | > | 107324 | 101 | 360 | ? | 114(42) | 106(33) | 31(28) | 33(30) |
| 99 | unknown | 107425 | < | 108075 | 93 | 217 | ? | 116(64) | 107(62) | 107(58) | 106(55) |
| 100 | bro-e | 108168 | < | 109211 | 129 | 348 | ? | 76(59) | 69(47) | - | - |
| 101 | pagr | 109340 | < | 111118 | 307 | 593 | ? | 113(23) | - | - | - |
| 102 | pif3 | 111425 | < | 112030 | 66 | 202 | ? | 111(23) | 110(50) | 115(52) | 115(54) |
| 103 | unknown | 112096 | < | 112998 | 302 | 301 | ? | 110(28) | - | - | - |
| 104 | unknown | 113300 | < | 115057 | 193 | 586 | ? | - | 113(27) | - | - |
| 105 | sod | 115250 | > | 115720 | 95 | 157 | L | 109(61) | 115(71) | 29(47) | 31(75) |
| 106 | ctl-2 | 115815 | < | 115973 | 311 | 53 | ? | 47(47) | 74(50) | 30(50) | 3(77) |
| 107 | helicase-2 | 116284 | < | 117591 | 192 | 436 | ? | - | - | - | - |
| 108 | unknown | 117783 | > | 118103 | 205 | 107 | ? | - | - | - | - |
| 109 | calyx/pep | 118308 | < | 119270 | 545 | 321 | L | 108(63) | 121(50) | 129(31) | 131(30) |
| 110 | orle002 | 119815 | > | 120387 | 202 | 191 | ? | - | - | - | - |
| 111 | unknown | 120589 | > | 121710 | 135 | 374 | ? | 107(52) | 124(44) | 113(62) | - |
| 112 | pif2 | 121845 | < | 122999 | 977 | 385 | L | 106(81) | 148(68) | 20(63) | 22(66) |
| 113 | pkip | 123976 | < | 124533 | 113 | 186 | L | 104(40) | 146(29) | - | - |
| 114 | lef2 | 124646 | < | 125275 | −43 | 210 | L | 103(50) | 136(46) | 6(38) | 6(40) |
| 115 | unknown | 125232 | < | 125630 | 232 | 133 | ? | - | - | - | - |
| 116 | p24 | 125862 | > | 126572 | 21 | 237 | L | 101(57) | 134(55) | 127(36) | 129(37) |
| 117 | unknown | 126593 | < | 127081 | 75 | 163 | ? | 100(38) | - | - | - |
| 118 | gp16 | 127156 | > | 127458 | 2813 | 101 | L | 99(48) | 133(49) | 128(36) | 130(40) |
| hr2 | 127459 | 130270 | |||||||||
| 119 | odv-e66b | 130271 | > | 132241 | 250 | 657 | E, L | 31(41) | 101(62) | 50(44) | 46(30) |
| 120 | rr2 | 132491 | < | 133507 | 189 | 339 | E | 117(76) | 122(61) | - | - |
| 121 | unknown | 133696 | > | 134166 | 244 | 157 | ? | - | - | - | - |
| 122 | unknown | 134410 | < | 134859 | 9 | 150 | ? | 91(76) | 126(28) | - | - |
| 123 | unknown | 134868 | > | 136097 | 90 | 410 | ? | 92(34) | 125(29) | - | - |
| 124 | alk-exo | 136187 | > | 137380 | 1145 | 398 | E | 93(48) | 127(44) | 131(37) | 133(39) |
| 125 | fgf | 138525 | > | 139562 | 432 | 346 | L | 95(36) | 130(31) | 27(27) | 32(30) |
| 126 | pif1 | 139994 | < | 141586 | 600 | 531 | L | 98(66) | 131(54) | 119(53) | 119(54) |
| hr3 | 141587 | 142185 | |||||||||
| 127 | f-protein | 142186 | < | 144276 | 368 | 697 | E,L | 118(55) | 150(42) | 21(23) | 23(21) |
| 128 | unknown | 144644 | > | 147478 | 308 | 945 | ? | 119(40) | 143(28) | - | - |
| 129 | unknown | 147786 | < | 148379 | 241 | 198 | ? | 121(40) | 142(28) | - | - |
| 130 | egt | 148620 | < | 150149 | 250 | 510 | E | 122(61) | 141(63) | 14(45) | 15(46) |
| 131 | unknown | 150399 | < | 150758 | 61 | 120 | ? | 123(44) | 139(49) | - | - |
| 132 | lef1 | 150819 | > | 151508 | 28 | 230 | E | 124(53) | 138(50) | 13(43) | 14(46) |
| 133 | 38.7K | 151536 | > | 152678 | 104 | 381 | ? | 125(38) | 137(29) | 12(24) | 13(25) |
| 134 | unknown | 152782 | < | 153459 | 182 | 226 | ? | - | - | - | - |
| 135 | rr1 | 153641 | < | 155923 | 257 | 761 | E | 126(66) | - | - | - |
ORFs were named starting from the polyhedrin gene (ORF1) to ribonuclease reductase 1 (ORF135).
Non-coding sequences including direct repeat region and the hrs are printed in bold. The direction of each gene is indicated by arrow heads.
Promoter motif for each gene is designated as Early (E) or Late (L) based on the consensus elements.
Respective homologues for EcobNPV, ChchNPV, OpMNPV, and AcMNPV ORFs are shown followed by the percentage amino acid identities in the brackets.
2.2. Gene Homology
The OrleNPV genome was compared to Autographa californica MNPV (AcMNPV), Chrysodeixis chalcites NPV (ChchNPV), Ectropis obliqua NPV (EcobNPV), and Orgyia pseudotsugata MNPV (OpMNPV) genomes (Table 2). All 62 conserved alphabaculovirus genes, including 31 core genes found in all baculoviruses [12,16,18], were clearly identified in OrleNPV. Of the 135 OrleNPV putative ORFs, 134 were homologous to other baculovirus ORFs and one ORF (ORF5, Orle001) was unique to OrleNPV (Table 3). OrleNPV genome shared the largest number of homologues with group II alphabaculoviruses: 114 (84%) with EcobNPV and 103 (76%) with ChchNPV. The number of homologues in common between OrleNPV and group I alphabaculoviruses, AcMNPV and OpMNPV were 87 (64%) and 81 (60%), respectively. The mean amino acid identity between OrleNPV and group II alphabaculoviruses was EcobNPV 52.4% ± 16.6 standard deviation, and ChchNPV 48.2% ± 15.1. The highest mean amino acid identity between OrleNPV and a group I alphabaculovirus was with AcMNPV at 43.1% ± 14.7. These results support OrleNPV as a group II alphabaculovirus with further evidence provided by the presence of the group II specific f-protein gene and absence of the group I specific gp64 [9,10,18,19]. Polyhedrin was the most conserved ORF between OrleNPV and other alphabaculoviruses with highest shared putative amino acid identities to EcobNPV (96%), ChchNPV (95%), AcMNPV (89%), and OpMNPV (86%) (Table 2). Very late expression factor 1 (vlf-1, ORF70) was the second most conserved ORFs between OrleNPV and other alphabaculoviruses; EcobNPV (84%), ChchNPV (75%), AcMNPV (74%), and OpMNPV (70%).
Table 3.
OrleNPV unknown and unique ORFs. This table contains some unknown and unique ORFs and their respective genomic positions. ORF 5 (orle001) is unique to OrleNPV.
| ORF | Name | Position | Homologue (% aa identity) 1 | Virus |
|---|---|---|---|---|
| 5 | orle001 | 5768–6715 | unique | OrleNPV |
| 25 | unknown | 28820–29464 | APNV_p040 (25%) | AnpeNPV |
| 26 | unknown | 31388–31858 | McAVgp043 (31%) | MacoNPV-A |
| 28 | dna ligase | 33191–35014 | LyxMNPV_gp020 (32%) | LyxMNPV |
| 59 | unknown | 64240–64479 | APNV-p121 (43%) | AnpeNPV |
| 107 | helicase-2 | 116284–117591 | Ld-helicase-2 (60%) | LdMNPV |
| 108 | unknown | 117783–118103 | EupsNPV_gp105 (48%) | EupsNPV |
| 110 | orle002 | 119815–120387 | Agip90 (30%) | AgipMNPV |
| 115 | unknown | 125232–125630 | ORF88_ ApciNPV (41%) | ApciNPV |
| 121 | unknown | 133696–134166 | ORF84_AgseGV (55%) | AgseGV |
| 134 | unknown | 152782–153459 | LdOrf-24_LdMNPV (23%) | LdMNPV |
Baculovirus homologues with E-value better than the threshold are shown with corresponding amino acid identities in brackets. ORF 110 (orle002), a C3HC4 type Zinc finger protein, is the only ORF with E-value worse than threshold. E-values were generated following the previous method [11].
2.3. Gene Organization
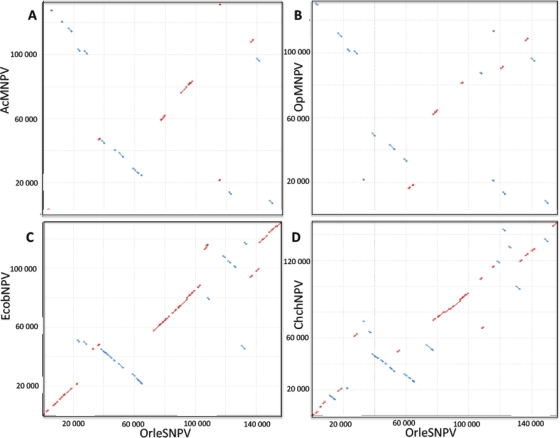
Gene parity plots were used to reveal the gene order between two baculoviruses, where closely related baculoviruses show colinear arrangement of genes and, conversely, colinearity decreases with increased divergence between baculoviruses [12,20]. Comparisons of relative gene order between OrleNPV and EcobNPV, ChchNPV, AcMNPV or OpMNPV homologues revealed five gene clusters that were conserved in these six alphabaculoviruses (Figure 2). The gene clusters consisted of 57 genes where 22 (76%) core baculovirus genes and 23 (72%) lepidopteran specific genes were identified. Gene cluster 5 showed the largest collinear segment which contained a core cluster of four genes, helicase, ac96 (odv-e28), 38K and lef5, that are thought to be present in all baculoviruses [18]. The gene arrangements of these five gene clusters indicate further that OrleNPV is more closely related to group II than group I alphabaculoviruses. Inversions in gene clusters 3 and 4 were identified in all six of these group I alphabaculovirues (Figure 2) and ChchNPV, a group II alphabaculovirus, showed inversion orientations in gene clusters 2 and 5 similar to group I alphabaculoviruses, AcMNPV and OpMNPV. These results are in agreement with a recent study, which showed similar gene arrangements between ClbiNPV and OrleNPV [21].
Figure 2.
Gene-parity plot analysis. Gene parity comparison of gene order of OrleNPV with (A) Autographa californica MNPV (AcMNPV), (B) Orgyia pseudotsugata MNPV (OpMNPV), (C) Ectropis obliqua (EcobNPV), and (D) Chrysodeixis chalcites NPV (ChchNPV). Blue dots indicate inversions in relation to the OrleNPV gene order. Numbers on the x and y axes are nucleotide position (bp) relative to the polyhedrin start codon.
2.4. Homologous Regions and Direct Repeats
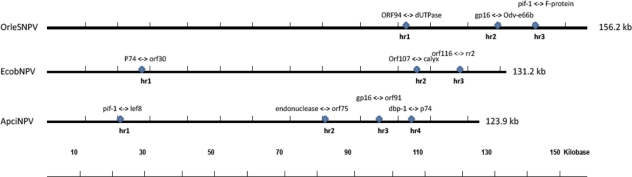
Homologous regions (hrs) are cis-acting elements present in many baculovirus genomes and most are characterized by the presence of several copies of direct repeats and imperfect palindromes. Comparisons of 57 sequenced baculovirus genomes showed that 47 contain hrs and eight, including Trichoplusia ni SNPV (TnSNPV), Adoxophyles orona GV (AdorGV), Agrotis segetum GV (AgseGV), Cydia pomonella GV (CpGV), Crytophlebia leucotreta GV (CrleGV), Pieris rapae GV (PrGV), SlGV, and NeleNPV have no hrs (Table 1). Spodoptera litura SMNPV genome (SpltMNPV-G2), contains 17 hrs, which is the highest number of hrs reported to date [22]. Although the in vivo origin(s) of baculovirus DNA replication (oris) remain unclear, hrs have been implicated as putative oris, and as enhancers of early gene expression using transient replication assays [23–25]. In addition, the interpalindromic sequences of baculovirus hrs contain certain cAMP response elements (CRE), which could bind host transcriptions factors of the bZIP family and possibly provide a synergy for the binding of IE-1 to the hrs [26,27]. In other baculovirus genera, particularly some betabaculoviruses, hrs exhibit both spatial and sequence heterogeneity and are devoid of the consensus palidromic sequences such as those found in the type baculovirus AcMNPV. For example, of the 9 hrs of XecnGV, hrs 1–8 contains about 120 bp long imperfect 3–6 direct repeats with no obvious palindromes and are interspersed within AT-rich regions, but the other hr is located within an ORF [15]. The OrleNPV genome contained three hrs that were located in the latter half of the genome (Table 2). Like other alphabaculoviruses, OrleNPV hrs were characterized by the presence of a single tandemly repeated imperfect palindrome sequence within a direct repeat of two to three copies. In AcMNPV genome, however, hrs have common repetitive elements with an imperfect palindrome of about 30 bp and are interspersed at multiple loci in the genome [27]. The size of the three OrleNPV hrs was variable (hr1 305 bp, hr2 219 bp and hr3 324 bp, (Table 2). Similar variations were observed in EcobNPV, which appears to be closely related to OrleNPV [28]. However, given the different numbers of hrs, the combined size of the OrleNPV hrs is relatively smaller (848 bp or 0.5% of the genome) compared to the combined size of the EcobNPV hrs (3541 bp or 2.7% of the genome). Nevertheless, the similarity in number and arrangement of the hrs (Figure 3), coupled with the phylogenetic analysis (Figure 4) strongly support the possibility of the two viruses sharing a common ancestor. In both OrleNPV and EcobNPV, there are indications of genome variability and arrangements around hr3. Here, two copies of OrleNPV ribonucleotide reductase genes (rr1 and rr2) are located on either side of the hr3 and in the same orientation. In EcobNPV genome, however, both rr1 and rr2 genes appear on the right side of hr3 and in opposite orientations. Moreover, EcobNPV pif1 gene does not flank hr3 as is the case of OrleNPV genome. Together, these findings suggest that, although both viruses appear to share a common ancestor (Figure 4), gene rearrangements around hr3 could have occurred through homologous recombination as the two viruses adapted in their specific hosts. Similar observations have been reported in other alphabaculoviruses although at different hr loci [29–31].
Figure 3.
Comparisons of hrs distribution. Linear map showing relative genomic locations of hrs (blue diamond shapes) of closely related baculoviruses; OrleNPV, EcobNPV and ApciNPV. ORFs flanking each of the hrs are indicated above each hr.
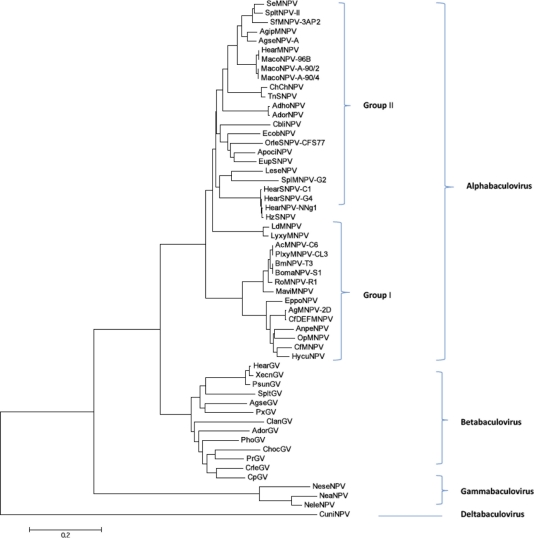
Figure 4.
Baculovirus Phylogeny. The phylogenetic tree was generated from concatenated amino acid sequences alignment of lef-8 and pif-2 genes of 56 of the 58 currently sequenced genomes. Evolutionary analyses were conducted in using MEGA5 software.
In addition to hrs, the OrleNPV genome contained six direct AT-rich repeat regions (drs) (except rep 5) containing 2 to 10 copies of direct tandem repeat sequences, ranging in size from 31 to 97 bp (Table 2). Common in other baculoviruses, drs have been implicated as origins of DNA replication [11,32,33]. They have also been shown to be more complex in structure compared to hrs and have similar sequence organization to those of eukaryotic oris [34–37]. Interestingly, the six drs located in the first half of the OrleNPV genome are absent in EcobNPV, ApciNPV, and Ectropis pseudoconspersa NPV (EupsNPV) in spite of their evolutionary relatedness. This observation suggests that these viruses may utilize different in vivo origins of DNA replication. This notion is supported by recent findings, which showed that none of the hrs is essential for baculovirus DNA replication [38]. Although structurally distinct to those of OrleNPV, non-hr repeats, have been reported in OpMNPV [29]. Since non-hrs mimics eukaryotic oris, and that OrleNPV and OpMNPV infect the same hosts, it is likely that the non-hrs were acquired from the host and may play an important role in viral DNA replication. It is worth noting that the only gene unique to OrleNPV (orle001) is flanked by rep1 and rep2 (Table 2). This gene may have been acquired from insect host and thus the drs may play a role in the horizontal transfer of genes into viral genomes. Although the molecular function of orle001 is unknown, based on its close proximity to these non-hr oris, it is also possible that orle001 product interacts with other replication machinery at the drs cis-acting elements during viral gene expression or DNA replication.
2.5. Baculovirus-Repeated ORFs
Bro genes are common in many baculovirus genomes and constitute a family of repetitive genes which vary in number and distribution. Among the current, sequenced baculovirus genomes, LdMNPV has the highest number of bro genes (16 copies), but they are absent in 13 baculovirus genomes including Maruca vitrata NPV (MaviNPV), Rachiplusia ou MNPV (RoMNPV), ApciNPV, Spodoptera exigua MNPV (SeMNPV), AdorGV, AgseGV, (ChocGV), CrleGV, PrGV, Plutella xylostella GV (PxGV), Neodiprion abietis NPV (NeabNPV), NeleNPV, and Neodiprion sertifer NPV (NeseNPV) (Table 1). In addition to baculoviruses, homologues of bro genes have been reported in ascoviruses, iridoviruses, entomopoxviruses, and class II transposons of prokaryotes [39]. Although their role during baculovirus replication is not clear, bro gene products have been implicated as potential DNA binding protein involved in host transcriptional regulation and DNA replication [40,41]. Other putative functions of bro genes, which may depend on the target insect host, include enhancement of late phase of virus replication [39] and as CRM1-dependent nuclear export shuttle proteins [42]. Five bro genes were identified in OrleNPV (ORFs 23, 30, 49, 61 and 100) and were designated as bro-a, bro-b, bro-c, bro-d, and bro-e based on their order of appearance in the genome (Table 2). Bro-a shares 27% and 38% identity with its respective homologues in ChchNPV and LdMNPV. Bro-b shares 25% identity with its LdMNPV homologue. Bro-c shares between 21% to 56% identity with its homologues in AcMNPV, EcobNPV, ChchNPV, and LdMNPV. Bro-d shares between 25% to 28% identity with its homologues in AcMNPV, EcobNPV, and LdMNPV. Bro-e shares between 47% to 64% identity with its homologues in EcobNPV, ChchNPV and LdMNPV. Based on LdMNPV Bro protein classification, bro-d belongs to group I, bro-b, bro-c, and bro-e (where bro-c and bro-e are more similar to each other than bro-b) belong to group II, and bro-a belongs to group III [43]. As shown in the phylogenic tree (Figure 4), ApciNPV is phylogenetically related to OrleNPV but lacks bro genes suggesting that the former may have lost bro genes by recombination as the viruses specialized to their hosts. Phylogenetic analysis of a multigene family of bro-like genes of other invertebrate DNA viruses and bacteria suggested that bro genes resulted from recombination events leading to loss, duplication, and acquisition of genes by horizontal gene transfer [39].
2.6. Genes with Two Homologues
Three genes, odv-e66, p26, and dbp, have two copies interspersed within the OrleNPV genome. A homologue of odv-e66 (ac46) has been shown to contain an inner nuclear membrane sorting motif at the N-terminal 33 aa, which promotes selective intracellular trafficking of proteins to the inner nuclear membrane [44]. Two copies of odv-e66 are also present in four other alphabaculoviruses including; EcobNPV (Ecob31 and Ecob48), SeMNPV (Se57 and Se114), Mamestra configurata NPV-A (MacoNPV-A) (ORFs 78 and 114), and MacoNPV-B (ORFs 77 and 143) [28,45–47]. Like OrleNPV, two copies of auxiliary gene p26 [ORFs 20 (p26a) and 62 (p26b)] can be found in other group II alphabaculoviruses including SeMNPV, AgseNPV-A, MacoNPV-B, ChchNPV, and TnSNPV and it has also been shown to be present in the group I alphabaculovirus, CfMNPV (Cf7 and Cf128) [30]. The placement of p26a (ORF 20) between Ac29 (ORF19) and p10 gene regions in OrleNPV was conserved as its homologues in EcobNPV (Ecob18), SeMNPV (Se129), AgseNPV-A (Asnv142), MacoNPV-B (MacoB157), ChchNPV (Chch19) and TnSNPV (Tn19) are also located between these two ORFs. However, the location of p26b (ORF62) homologues was more divergent in these same baculoviruses. OrleNPV is the fifth baculovirus identified to contain two copies of DNA-binding protein [dbp-1 (ORF 17), dbp-2 (ORF53)], the others being EcobNPV [28], LdMNPV [43], ClbiNPV [21], and Lymantria xylina MNPV (LyxyMNPV) [48]. A homologue of this gene in AcMNPV (ac25) has been implicated as encoding a single stranded DNA binding protein required for synthesis of normal levels of viral DNA, formation of virogenic stroma, and processing of replication intermediates by binding nascent DNA molecules [49,50]. OrleNPV dbp-1 shares low identities with Ecob27 (23 %) and Ld37 (25%) dbp-1 and OrleNPV dbp-2 shares slight higher identities with Ecob15 (27%) (Table 2). DBP-2 (ORF53) shared high aa identity with EupsNPV GP044 (47 %). Although the significance of having two copies of the above mentioned three genes in OrleNPV and other alphabaculoviruses is unclear, it is possible that one copy may act as a genetic backup to compensate the function in case either of the copy becomes dysfunctional due to mutations. Alternatively, gene duplication may contribute to unconstrained evolution, thus, possible adaptation to new environments (hosts).
2.7. Inhibitors of Apoptosis (iap)
Baculovirus inhibitors of apoptosis (iap) genes help to circumvent insect defense mechanisms involving programmed death of virus-infected cells and may act as host range factors [51]. Some alphabaculoviruses such as AcMNPV, RoMNPV, Bombyx mori NPV (BmNPV), Maruca vitrata NPV (MaviNPV), and SpltMNPV contain the p35 family of anti-apoptosis genes in addition to the iap family of genes. Both families of anti-apoptotic genes have been shown to have complementary functions, for example, replication of AcMNPV p35 mutant was rescued by iap genes from other baculoviruses [52,53]. OrleNPV genome contained three iap genes but lacks p35. ORF 16 (iap3) showed highest identity with group I alphabaculovirus, Choristoneura fumiferana MNPV (CfMNPV) iap3 (61%) and with group II, TnSNPV iap1 (54%). ORF 43 (iap2), however, showed highest identity to EupsNPV iap1 (35%) and Anticarsia gemmatalis MNPV (AgMNPV-2D) iap-3 (30%) in respective group II and I alphabaculoviruses. The other copy of OrleNPV iap2 (ORF 63) showed highest identities with iap2 of group II alphabaculoviruses, EcobNPV (33%) and ChchNPV (32%) (Table 2). All three proteins contained one copy of baculovirus inhibitor of apoptosis repeats (BIRs). BIRs contain a zinger-finger domain and a RING domain at the C-terminus and are involved in binding apoptosis-inducing proteins [54,55]. ORF 43 and 63 contained a zinc finger at the C-terminus, whereas ORF16 lacked a zinc finger domain which suggests that they may be involved in binding different forms of host apoptosis proteins.
2.8. Auxiliary Genes
Baculovirus auxillary genes are not essential for viral gene expression, DNA replication or morphogenesis, but provide a selective advantage for virus survival [19]. OrleNPV genome contained 11 auxiliary genes including; alkaline exonuclease (alk-exo), chitinase, cathepsin L-like proteinase (v-cath), ecdysteroid UDP glycosyltransferase (egt), fibroblast growth factor (fgf), p10, protein kinase-1 (pk-1), superoxide dismutase (sod), ubiquitin (ubi), conotoxin-like protein 1 and 2 (ctl-1 and ctl-2). Most of these genes exhibited 45–83% putative amino acid identities to their respective homologues in EcobNPV [28] (Table 2). OrleNPV genome encodes two copies of ctl genes as do Hyphantria cunea NPV (HycuNPV), OpMNPV and LdMNPV genomes. OrleNPV ctl-1 shares 86% putative amino acid identity with its ChchNPV homologue [57] (Table 2). Auxiliary genes not found in OrleNPV were actin rearrangement-inducing factor-1 (arif-1), proliferating nuclear antigen (pcna), viral enhancing factor-1 (vef-1) and ptp-1.
2.9. Phylogenetic Analysis
To date, 31 core genes have been used to elucidate the evolutionary relationships of various baculovirus species [16]. In this study, the phylogenetic tree was generated based on the concatemers of ac22 homologue (pif-2) and lef-8 genes. These two genes were previously shown to generate robust tree that is comparable to that of baculovirus core genes [58] from the current baculovirus genomes available in the NCBI database. The tree showed a clear separation of baculoviruses into their current scheme of classification (Figure 4) [7]. In addition, group I alphabaculoviruses and betabaculoviruses showed clear separation into two distinct clades (a and b), which have been reported in previous studies using the analysis of all baculovirus core genes [7,16,58]. This information further highlights the significance of these two conserved genes, albeit non-essential for replication, in elucidating the evolution of baculoviruses in their respective hosts [58]. Our results indicate that OrleNPV is most closely related to EcobNPV, ApciNPV, EupsNPV, and ClbiNPV (Figure 4).
3. Experimental Section
3.1. Virus Propagation, DNA Extraction and Purification
Third-instar O. leucostigma larvae were fed OrleNPV at a rate of 5 × 104 OBs per larva. Infected larvae were reared on artificial diet and were monitored for viral pathogenesis at various days post infection (dpi). Occlusion bodies were isolated from infected larvae as previously described with minor modifications [59,60]. In brief, 20 larvae were collected 10 dpi, homogenized with a hand blender, and stirred for 2 h in 0.5% SDS. The homogenate was filtered through cheese cloth and centrifuged at 7000 × g for 15 min at 15 °C. The pellet containing the OBs and insect debris was washed three times in deionized water by centrifugation as describe above. The final pellet of purified OBs was resuspended in 5 mL of sterile distilled water. OBs were dissolved in an alkaline solution (1.0 M sodium carbonate and 0.4 M sodium thioglycolate) to release the occlusion derived virus (ODV). The solution was centrifuged at 1000 × g to remove debris and undissolved OBs. ODV was purified in a continuous 10–45% sucrose gradient as previously described [61]. The band containing the ODV was collected, diluted with TE (10mM Tris and 1.0 mM EDTA, pH 8.0), and centrifuged at 22000 rpm (SW28 rotor Beckman ultracentrifuge) for 2 h at 4 °C. The pellet was suspended in 500 μL of TE and digested with proteinase K (25 μL, 20 mg/mL) along with 1% N-lauryl-sarcosine (final concentration) for 2 h at 37 °C. Viral DNA was further purified by a CsCl gradient approach as previously described [60] followed by dialysis of viral DNA against several changes of TE for 72 h at 4 °C. Purified DNA was resuspended in TE, and quantified spectrophometrically to ascertain DNA yield and purity.
3.2. DNA Sequencing and Analysis
OrleNPV genome was determined using the shotgun sequence approach as previously described [12]. In brief, total genomic DNA was sheared into small random fragments by nebulization and cloned to generate an overlapping genomic library. Purified template DNA from the genomic library was sequenced in an ABI Prisms® 3700 analyzer using the BigDye® terminator chemistry (v.3.0/3) as per Agencourt Bioscience Corporation (Beverly, MA, USA) sequencing specifications. The overall sequence obtained accounted for a 12-fold genomic coverage. The OrleNPV genome was assembled by Agencourt BioScience. Lasergene DNAStar SeqManPro (version 7.2) was used to manually edit and verify the contiguous sequence data. Genomic data was remotely submitted to Emboss software suite and start-to-stop translational searching was performed using getOrf program [62]. Open reading frames (ORFs) encoding 50 or more amino acids with minimal overlap were accepted as putative genes based on the established criteria [17]. NCBI web blast perl script was used to submit relevant ORFs to the GenBank. Homologue identification was done using standard protein-protein BLAST (blastp) with default settings [63].
Sequences 160 bp upstream of the predicted ORF start codons were analyzed for potential promoter motifs using WebLogo [64]. Upstream sequences were also scanned for exact matches using regular expression notation native to perl for commonly known baculovirus promoter elements, including the TATA sequence (TATA/TATAW), CAKT and DTAAG, within 160 bp or 40 to 20 bp regions. Further motif optimizations were preformed on BioProspector and AlignACE results using BioOptimizer v 3.0 [65]. Repeat regions including hrs and direct repeat regions were identified using Tandem repeat finder [66], Emboss Palindrome [62], and Reputer [67].
3.3. Gene homology and Phylogenetic Analysis
Predicted ORFs were compared with homologues in four alphabaculoviruses, which included AcMNPV; NCBI reference (NC_001623) MNPV OpMNPV (NC_001875), ChchNPV (NC_007151), EcobNPV (NC_008586). Gene parity plots were used to analyze the gene order of OrleNPV relative to those representatives of group I and group II alphabaculoviruses mentioned above. Phylogenetic tree was generated using concatenated amino acid sequences derived from lef-8 and pif-2 genes for the 56 complete baculovirus genomes that were available in the NCBI database at the time of analysis. The tree was inferred using neighbour-joining method using the MEGA5 program [68].
4. Conclusions
OrleNPV genome is the eighth largest baculovirus genome sequenced to date (157, 179 bp) and encodes for a total of 135 ORFs with one being unique to the virus. Interspersed within the genome are three hrs, five bros, and three iap genes, which are common features in most baculovirus genomes. Duplicate copies of dbp, odv-e66, and p26 were present in OrleNPV genome. Based on phylogenetic analysis and gene arrangements, OrleNPV appears to be most closely related to the group II alphabaculoviruses EcobNPV, ApciNPV, EupsNPV, and ClbiNPV. Together, the OrleNPV genomic data would be helpful components for future analysis of emerging baculovirus genomes and also in providing a deeper understanding of the molecular basis of OrleNPV pathogenicity.
Acknowledgments
This research was funded through grants awarded to CJL by the Canadian Forest Service research programs, the Canadian Biotechnology Strategy and the Canadian Regulatory System for Biotechnology. The authors gratefully acknowledge the thoughtful reviews of the manuscript provided by Catherine Belliveau and Miguel Lopez Ferber.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References and Notes
- 1.Theilmann D.A., Blissard G.W., Bonning B., Jehle J., O’Reilly D.R., Rohrman G.F., Thiem S., Vlak J.M. Baculoviridae. In: Fauquet C.M., Mayo M.A., Maniloff J., Desselberger U., Ball L.A., editors. Virus Taxonomy, VIIIth Report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press; London, UK: 2005. pp. 177–185. [Google Scholar]
- 2.Lucarotti C.J., Morin B., Graham R.I., Lapointe R. Production, application and field performance of Abietiv™ the balsam fir sawfly nucleopolyhedrovirus. Virologica Sinica. 2007;22:163–172. [Google Scholar]
- 3.Moscardi F. Assessment of the application of baculovirus for control of Lepidoptera. Ann. Rev. Entomol. 1999;44:257–289. doi: 10.1146/annurev.ento.44.1.257. [DOI] [PubMed] [Google Scholar]
- 4.Erlandson M. Insect pest control by viruses. In: Mahy B.W.J., van Regenmortel M.H.V., editors. Encyclopedia of Virology. Elsevier/Academic Press; London, UK: 2008. pp. 125–133. [Google Scholar]
- 5.Aucoin M.G., Mena J.A., Kamen A.A. Bioprocessing of baculovirus vectors: A review. Curr. Gene Ther. 2010;10:3. doi: 10.2174/156652310791321288. [DOI] [PubMed] [Google Scholar]
- 6.Yin H.F., Zhou X., Wu H.F., Li B., Zhang Y.F. Baculovirus vector-mediated transfer of NIS gene into colon tumor cells for radionuclide therapy. World J. Gastroenterol. 2010;16:5367–5374. doi: 10.3748/wjg.v16.i42.5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jehle J.A., Blissard G.W., Bonning B.C., Cory J.S., Herniou E.A., Rohrmann G.F., Theilmann D.A., Thiem S.M., Vlak J.M. On the classification and nomenclature of baculoviruses: A proposal for revision. Arch. Virol. 2006;151:1257–1266. doi: 10.1007/s00705-006-0763-6. [DOI] [PubMed] [Google Scholar]
- 8.Blissard G.W., Wenz J.R. Baculovirus gp64 envelope glycoprotein is sufficient to mediate pH-dependent membrane fusion. J. Virol. 1992;66:6829–6835. doi: 10.1128/jvi.66.11.6829-6835.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bulach D.M., Kumar C.A., Zaia A., Liang B., Tribe D.E. Group II nucleopolyhedrovirus subgroups revealed by phylogenetic analysis of polyhedrin and DNA polymerase gene sequences. J. Invertebr. Pathol. 1999;73:59–73. doi: 10.1006/jipa.1998.4797. [DOI] [PubMed] [Google Scholar]
- 10.Jiang Y., Deng F., Rayner S., Wang H., HU Z. Evidence of major role of GP64 in group I alphabaculovirus evolution. Virus Resear. 2009;142:85–91. doi: 10.1016/j.virusres.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Maruniak A., Maruniak J.E., Zanotto P.M.A., Doumbouya A.E., Liu J-C., Merritt T.M., Lanoie J.S. Sequence analysis of the genome of the Neodiprion sertifer nucleopolyhedrovirus. J. Virol. 2004;78:7036–7051. doi: 10.1128/JVI.78.13.7036-7051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lauzon H.A.M., Lucarotti C.J., Krell P.J., Feng Q., Retnakaran A., Arif B.M. Sequence and Organization of the Neodiprion lecontei nucleopolyhedrovirus genome. J. Virol. 2004;78:7023–7035. doi: 10.1128/JVI.78.13.7023-7035.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Frankenhuyzen K., Ebling P., Thurston G., Lucarotti C., Royama T., Goscott R., Georgeson E., Silver J. Incidence and impact of Entomophaga aulicae (Zygomycetes: Entomophthorales) and a nucleopolyhedrovirus in an outbreak of the whitemarked tussock moth (Lepidoptera: Lymantriidae) Can. Entomol. 2002;134:825–845. [Google Scholar]
- 14.Cunningham J.C., Kaupp W.J. Insect viruses. In: Amstrong J.A., Ives W.G.H., editors. Forest Pest Insects in Canada, Natural Resources Canada. Natural Resources Canada; Ottawa, Ontario, Canada: 1995. pp. 327–340. [Google Scholar]
- 15.Hayakawa T., Ko R., Okano K., Seong S., Goto C., Maeda S. Sequence analysis of the Xiestia c-nigrum granulovirus genome. Virology. 1999;262:277–297. doi: 10.1006/viro.1999.9894. [DOI] [PubMed] [Google Scholar]
- 16.Miele S.A.B., Garavaglia M.J., Belaich M.N., Ghiringhelli P.D. Baculovirus: Molecular insights on their diversity and conservation. Int. J. Evol. Biol. 2011;10 doi: 10.4061/2011/379424. 4061/2011/379424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ayres M.D., Howard S.W., Kuzio J., Ferber-Lopez M., Possee R.D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 18.Herniou E.A., Oleszewski J.A., Cory J.S., O’Reilly D.R. The genome sequence and evolution of baculoviruses. Annu. Rev. Entomol. 2003;48:211–234. doi: 10.1146/annurev.ento.48.091801.112756. [DOI] [PubMed] [Google Scholar]
- 19.Herniou E.A., Luque T., Chen X., Vlak J.M., Winstanley D., Cory J., O’Reilly D.R. Use of whole genome sequence data to infer baculovirus phylogeny. J. Virol. 2001;75:8117–8126. doi: 10.1128/JVI.75.17.8117-8126.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Z.H., Arif B.M., Jin F., Martens W.M., Chen X.W., Sun J.S., Zuidema D., Goldbach R.W., Vlak J.M. Distinct gene arrangement in the Bazura suppressaria single-nucleocapsid nucleopolyhedrovirus genome. J. Gen. Virol. 1998;79:2841–2851. doi: 10.1099/0022-1317-79-11-2841. [DOI] [PubMed] [Google Scholar]
- 21.Zhu S.Y., Yi J.P., Shen W.D., Wang L.Q., He H.G., Wang Y., Li B., Wang W.B. Genomic sequence, organization and characteristic of a new nucleopolyhedrovirus isolated from Clanis bilineata larva. BMC Genomics. 2009;10:91. doi: 10.1186/1471-2164-10-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pang Y., YU J., Wang L., Hu X., Bao W., Li G., Chen C., Han H., Hu S., Yan H. Sequence analysis of the Spodoptera litura multicapsid nucleopolyhedrovirus genome. Virology. 2001;287:391–404. doi: 10.1006/viro.2001.1056. [DOI] [PubMed] [Google Scholar]
- 23.Leisy D.J., Rohrmann G.F. Characterization of the replication of plasmids containing hr sequences in baculovirus-infected Spodoptera frugiperda cells. Virology. 1993;196:722–730. doi: 10.1006/viro.1993.1529. [DOI] [PubMed] [Google Scholar]
- 24.Kool M., Ahrens CH., Vlak J.M., Rohrmann G.F. Replication of baculovirus DNA. J. Gen. Virol. 1995:76, 2103–2118. doi: 10.1099/0022-1317-76-9-2103. [DOI] [PubMed] [Google Scholar]
- 25.Theilmann D.A., Stewart S. Tandemly repeated sequence at the 3′ end of the baculovirus Orygia pseudotsugata multicapsid nuclear polyhedrosis virus is an enhancer element. Virology. 1992;187:97–106. doi: 10.1016/0042-6822(92)90298-4. [DOI] [PubMed] [Google Scholar]
- 26.Landais I., Vincet R., Bouton M., Devauchelle G., Duonor-Cerutti M., Ogliastro M. Functional analysis of evolutionary conserved clusterinng of bZIP binding sites in the baculovirus homologous regions (hrs) suggests a cooperativity between host and viral transcriptional factors. Virology. 2006;334:421–431. doi: 10.1016/j.virol.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 27.Rohrmann G.F. Baculovirus Molecular Biology. 2nd ed. National Centre for Biotechnology Information (NCBI); Bethesda, MD, USA: 2011. Early events in infection: Virus transcription. [Google Scholar]
- 28.Ma X.C., Shang J.Y., Yang Z.N., Bao Y.Y., Xiao Q., Zhang C.X. Genome sequence and organization of a nucleopolyhedrovirus that infects the tea looper caterpiller Ectropis obliqua. Virology. 2007;360:235–246. doi: 10.1016/j.virol.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 29.Ahrens C.H., Russell R.L.Q., Funk C.J., Evans J.T., Harwood S.H., Rohrmann G.F. The sequence of the Orygia pseudotsuguta multilcapsid nuclear polyhedrosis virus genome. Virology. 1997;229:381–399. doi: 10.1006/viro.1997.8448. [DOI] [PubMed] [Google Scholar]
- 30.de Jong J.G., Lauzon H.A., Dominy C., Poloumienko A., Carstens E.B., Arif B.M., Krell P.J. Analysis of the Choristoneura fumiferana nucleopolyhedrovirus genome. J. Gen. Virol. 2005;86:929–943. doi: 10.1099/vir.0.80490-0. [DOI] [PubMed] [Google Scholar]
- 31.Hayakawa T., Rohrmann G.F., Hashimoto Y. Patterns of genome organization and content in lepidopteran baculoviruses. Virology. 2000;278:1–12. doi: 10.1006/viro.2000.0668. [DOI] [PubMed] [Google Scholar]
- 32.Lee H., Krell P.J. Reiterated DNA fragments in defective genomes of Autographa californica nuclear polyhedrosis virus are competent for AcMNPV-dependent DNA replication. Virology. 1994;202:418–429. doi: 10.1006/viro.1994.1358. [DOI] [PubMed] [Google Scholar]
- 33.Habib S., Hasnain S.E. Differential activity of two non-hr origins during replication of the baculovirus Autographa californica nuclear polyhedrosis virus genome. J. Virol. 2000;74:5182–5189. doi: 10.1128/jvi.74.11.5182-5189.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearson M., Bjornson R., Pearson G., Rohrman G. The Autographa californica baculovirus genome: evidence for multiple replication origins. Science. 1992;257:1382–1384. doi: 10.1126/science.1529337. [DOI] [PubMed] [Google Scholar]
- 35.Kool M., Goldbach R.W., Vlak J.M. A putative non-hr origin of DNA replication in the HindIII-K fragment of the Autographa californica multiple nucleocapsid nuclear polyhedrosis virus. J. Gen. Virol. 1994;75:3345–3352. doi: 10.1099/0022-1317-75-12-3345. [DOI] [PubMed] [Google Scholar]
- 36.Heldens J.G., Broer R., Zuidema D., Goldbach R.W., Vlak J.M. Identification and functional analysis of a non-hr origin of DNA replication in the genome of Spodoptera exigua multicapsid nucleopolyhedrovirus. J. Gen. Virol. 1997;78:1497–1506. doi: 10.1099/0022-1317-78-6-1497. [DOI] [PubMed] [Google Scholar]
- 37.Huang J., Levin D.B. Identification and functional analysis of a putative non-hr origin of DNA replication from the Spodoptera littorris type B multinucleocapsid nucleopolyhedrovirus. J. Gen. Virol. 1999;80:2263–2274. doi: 10.1099/0022-1317-80-8-2263. [DOI] [PubMed] [Google Scholar]
- 38.Carstens E.B., Wu Y. No single homologous repeat region is essential for DNA replication of the baculovirus Autographa californica multiple nucleopolyhedrovirus. J. Gen Virol. 2007;88:114–112. doi: 10.1099/vir.0.82384-0. [DOI] [PubMed] [Google Scholar]
- 39.Bideshi D.K., Renault S., Stasiak K., Federici B.A., Bigot Y. Phylogenetic analysis and possible functions of bro-like genes, a multigene family widespread among double-stranded DNA viruses of invertebrates and bacteria. J. Gen Virol. 2003;84:2531–2544. doi: 10.1099/vir.0.19256-0. [DOI] [PubMed] [Google Scholar]
- 40.Li Z., Pan L., Yu H., Li L., Gong Y., Yang K., Pang Y. Characterization of Spodoptera litura multicapsid nucleopolyhedrovirus 38.7 K protein, which contain a conserved BRO domain. Virus Res. 2006;115:185–191. doi: 10.1016/j.virusres.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 41.Zemskov E., Kang W., Maeda S. Evidence for nucleic acid binding ability and nucleosome association of Bombyx mori nucleopolyhedrovirus BRO proteins. J. Virol. 2000;74:6784–6789. doi: 10.1128/jvi.74.15.6784-6789.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang W., Kurihara M., Matsumoto S. The BRO proteins of Bombyx mori nucleopolyhedrovirus are nucleocytoplasmic shuttling proteins that utilize the CRM1-mediated nuclear export pathway. Virology. 2006;350:184–191. doi: 10.1016/j.virol.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 43.Kuzio J., Pearson M.N., Harwood S.H., Funk C.J., Evans J.T., Slavicek J.M., Rohrmann G.F. Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar. Virology. 1999;253:17–34. doi: 10.1006/viro.1998.9469. [DOI] [PubMed] [Google Scholar]
- 44.Braunagel S.C., Williamson S.T., Saksena S., Zhong Z., Russell W.K., Russell D.H., Summers M.D. Trafficking of ODV-E66 is mediated via a sorting motif and other viral proteins: Facilitated trafficking to the inner nuclear membrane. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8372–8377. doi: 10.1073/pnas.0402727101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ijkel W.F.J., van Strien E.A., Heldens J.G.M., Broer R., Zuidema D., Golbach R.W., Vlak J.M. Sequence and organization of the Spodoptera exigua multinucleocapsid nucleopolyhedrovirus genome. J. Gen. Virol. 1999;80:3289–3304. doi: 10.1099/0022-1317-80-12-3289. [DOI] [PubMed] [Google Scholar]
- 46.Li L., Donly C., Li Q., Willis L.G., Keddie B.A., Erlandson M.A., Theilmann D.A. Identification and genomic analysis of a second species of nucleopolyhedrovirus isolated from Mamestra configurata. Virology. 2002;297:226–244. doi: 10.1006/viro.2002.1411. [DOI] [PubMed] [Google Scholar]
- 47.Li Q., Donly C., Li L., Willis L.G., Theilmann D.A., Erlandson M. Sequence and organization of the Mamestra configurata nucleopolyhedro-virus genome. Virology. 2002;294:106–121. doi: 10.1006/viro.2001.1313. [DOI] [PubMed] [Google Scholar]
- 48.Nai Y.S., Wu C.Y., Wang T.C., Chen Y.R., Lau W.H, Lo C.F. Genomic sequencing and analysis of Lymantria xylina multiple nucleopolyhedrovirus. BMC Genomics. 2010;11:116. doi: 10.1186/1471-2164-11-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mikhailov V.S., Vanarsdall A.L., Rohrmann G.F. Isolation and characterization of the DNA-binding protein (DBP) of the Autographa californica multiple nucleopolyhedrovirus. Virology. 2008;370:415–429. doi: 10.1016/j.virol.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vanarsdall A.L., Mikhailov V.S., Rohrmann G.F. Characterization of a baculovirus lacking the DBP (DNA-binding protein) gene. Virology. 2007;364:475–485. doi: 10.1016/j.virol.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller L.K., Kaiser W.J., Seshagiri S. Baculovirus regulation of apoptosis. Semin. Virol. 1998;8:445–452. [Google Scholar]
- 52.Ikeda M., Yanagimoto K., Kobayashi M. Identification and functional analysis of Hyphantria cunea nucleopolyhedrovirus iap genes. Virology. 2004;321:359–371. doi: 10.1016/j.virol.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 53.Yan F., Deng X., Yan J., Wang J., Yao L., Lu S., Qi Y., Xu H. Functional analysis of the inhibitor of apoptosis genes in Antheraea pernyi nucleopolyhedrovirus. J. Microbiol. 2010;48:199–205. doi: 10.1007/s12275-010-9108-y. [DOI] [PubMed] [Google Scholar]
- 54.Kim Y.S., Xiao H.Z., Du E.Q., Cai G.S., Lu S.Y., Qi Y.P. Identification and functional analysis of LsMNPV Anti-apoptosis genes. J. Biochem. Mol. Biol. 2007;40:571–576. doi: 10.5483/bmbrep.2007.40.4.571. [DOI] [PubMed] [Google Scholar]
- 55.Crook N.E., Clem R.J., Miller L.K. An apoptosis inhibiting baculovirus gene with a zinc finger-like motif. J. Virol. 1993;67:2168–2174. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Afonso C.L., Tulman E.R., Lu Z., Balinsky C.A., Moser B.A., Becnel J.J., Rock D.L., Kutish G.F. Genome sequence of a baculovirus pathogenic for Culex nigripalpus. J. Virol. 2001;75:11157–11165. doi: 10.1128/JVI.75.22.11157-11165.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Oers M.M., Abma-Henkens M.H.C., Herniou E.A., de Groot J.C.W., Peters S., Vlak J.M. Genome sequence of Chrysodeixis chalcites nucleopolyhedrovirus, a baculovirus with two DNA photolyase genes. J. Gen. Virol. 2005;86:2069–2080. doi: 10.1099/vir.0.80964-0. [DOI] [PubMed] [Google Scholar]
- 58.Herniou E.A., Olszewski J.A., O’Reilly D.R., Cory J.S. Ancient coevolution of baculoviruses and their insects hosts. J. Virol. 2004;78:3244–3252. doi: 10.1128/JVI.78.7.3244-3251.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H., Bonning B.C. Methods in Molecular Biology. Baculovirus and Insect Cell Expression Protocols. 2nd ed. Vol. 338. Humana Press Inc; Totowa, NJ, USA: 2007. Evaluation of the insecticidal efficacy of wild-type and recombinant baculoviruses. [DOI] [PubMed] [Google Scholar]
- 60.O’Reilly D.R., Miller L.K. In: Baculovirus Expression Vectors: A Laboratory Manual. XII. Freeman W.H., editor. Oxford University Press; New York, NY, USA: 1992. [Google Scholar]
- 61.Escasa S.R., Lauzon H.A.M., Mathur A.C., Krell P.J., Arif B.M. Sequence analysis of the Choristoneura occidentalis granulovirus genome. J. Gen. Virol. 2006;87:1917–1933. doi: 10.1099/vir.0.81792-0. [DOI] [PubMed] [Google Scholar]
- 62.Rice P., Longden I., Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 63.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 64.Crook G.E., Hon G., Chandonja J.M., Brenner S.E. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jensen S.T., Lui J.S. BioOptimizer: A Bayesian scoring function approach to motif discovery. Bioinformatics. 2004;20:1557–1564. doi: 10.1093/bioinformatics/bth127. [DOI] [PubMed] [Google Scholar]
- 66.Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kurtz S., Choudhuri J.V., Ohlebusch E., Schleiermacher C., Stoye J., Giegerich R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 69.Nie Z.M., Zhang Z.F., Wang D., He P.A., Jiang C.Y., Song L., Chen F., Xu J., Yang L., Yu L., et al. Complete sequence and organization of Antheraea pernyi nucleopolyhedrovirus, a dr-rich baculovirus. BMC Genomics. 2007;8:248. doi: 10.1186/1471-2164-8-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fan Q., Li S., Wang L., Zhang B., Ye B., Zhao Z., Cui L. The genome sequence of the multinucleocapsid nucleopolyhedrovirus of the Chinese oak silkworm Antheraea pernyi. Virology. 2007;366:304–315. doi: 10.1016/j.virol.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 71.de Castro Oliveira J.V., Wolff J.L.C., Garcia-Maruniak A., Ribeiro B.M., de Castro M.E.B., de Souza M.L., Moscardi F., Maruniak J.E., de Andrea Zanotto P.M. Genome of the most widely used viral biopesticide: Anticarsia gemmatalis multiple nucleopolyhedrovirus. J. Gen. Virol. 2006;87:3233–3250. doi: 10.1099/vir.0.82161-0. [DOI] [PubMed] [Google Scholar]
- 72.Gomi S., Majima K., Maeda S. Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J. Gen. Virol. 1999;80:1323–1337. doi: 10.1099/0022-1317-80-5-1323. [DOI] [PubMed] [Google Scholar]
- 73.Xu Y.P., Ye Z.P., Niu C.Y., Bao Y.Y., Wang W.B., Shen W.D., Zhang C.X. Comparative analysis of the genomes of Bombyx mandarina and Bombyx mori nucleopolyhedroviruses. J. Microbiol. 2010;48:102–110. doi: 10.1007/s12275-009-0197-4. [DOI] [PubMed] [Google Scholar]
- 74.Lauzon H.A.M., Jamieson P.B., Krell P.J., Arif B.M. Gene organization and sequencing of Choristoneura fumiferana defective nucleopolyhedrovirus genomes. J. Gen. Virol. 2005;86:945–961. doi: 10.1099/vir.0.80489-0. [DOI] [PubMed] [Google Scholar]
- 75.Hyink O., Dellow R.A., Olsen M.J., Caradoc-Davies K.M.B., Drake K., Herniou E.A., Cory J.S., O’Reilly D.R., Ward V.K. Whole genome analysis of the Epiphyas postvittana nucleopolyhedrovirus. J. Gen. Virol. 2002;83:957–971. doi: 10.1099/0022-1317-83-4-957. [DOI] [PubMed] [Google Scholar]
- 76.Ikeda M., Shikata M., Shirata N., Chaeychomsri S., Kobayashi M. Gene organization and complete sequence of the Hyphantria cunea nucleopolyhedrovirus genome. J. Gen Virol. 2006;87:2549–2562. doi: 10.1099/vir.0.81930-0. [DOI] [PubMed] [Google Scholar]
- 77.Chen Y.R., Wu C.Y., Lee S.T., Wu Y.J., Lo C.F., Tsai M.F., Wang C.H. Genomic and host range studies of Maruca vitrata nucleopolyhedrovirus. J. Gen. Virol. 2008;89:2315–2330. doi: 10.1099/vir.0.2008/001412-0. [DOI] [PubMed] [Google Scholar]
- 78.Harrison R.L, Lynn D.E. Genomic sequence analysis of a nucleopolyhedrovirus isolated from the diamondback moth Plutella xylostella. Virus Genes. 2007;35:857–873. doi: 10.1007/s11262-007-0136-6. [DOI] [PubMed] [Google Scholar]
- 79.Harrison R.L., Bonning B.C. Comparative analysis of the genomes of Rachiplusia ou and Autographa californica multiple nucleopolyhedroviruses. J. Gen. Virol. 2003;84:1827–1842. doi: 10.1099/vir.0.19146-0. [DOI] [PubMed] [Google Scholar]
- 80.Nakai M., Goto C., Kang W., Shikata M., Luque T., Kunimi Y. Genome sequence and organization of a nucleopolyhedrovirus isolated from the smaller tea totrix Adoxophyes honmai. Virology. 2003;316:171–183. doi: 10.1016/j.virol.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 81.Hilton S., Winstanley D. Genomic sequence and biological characterization of a nucleopolyhedrovirus isolated from the summer fruit tortrix Adoxopheys orana. J. Gen. Virol. 2008;89:2898–2908. doi: 10.1099/vir.0.2008/002881-0. [DOI] [PubMed] [Google Scholar]
- 82.Harrison R.L. Genomic sequence analysis of the Illinois strain of the Agrotis ipsilon multiple nucelopolyhedrovirus. Virus Genes. 2009;38:155–170. doi: 10.1007/s11262-008-0297-y. [DOI] [PubMed] [Google Scholar]
- 83.Jakubowska A.K., Peters S.A., Ziemnicka J., Vlak J.M., van Oers M.M. Genome sequence of an enhancin gene-rich nucleopolyhedrovirus (NPV) from Agrotis segetum: Collinearity with Spodoptera exigua multiple NPV. J. Gen. Virol. 2006;87:537–551. doi: 10.1099/vir.0.81461-0. [DOI] [PubMed] [Google Scholar]
- 84.Tang X.D., Xiao Q., Ma X.C., Zhu Z.R., Zhang C.X. Morphology and genome of Euproctis pseudoconspersa nucleopolyhedrovirus. Virus Genes. 2009;38:495–506. doi: 10.1007/s11262-009-0355-0. [DOI] [PubMed] [Google Scholar]
- 85.Chen X., Ijkel W.F.J., Tarachini R., Wang H., Peters S., Zuidema D., Lankhorst R.K., Vlak J.M., Hu Z. The sequence of the Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus genome. J. Gen. Virol. 2001;82:241–257. doi: 10.1099/0022-1317-82-1-241. [DOI] [PubMed] [Google Scholar]
- 86.Zhang C.X., Ma X.C., Guo Z.J. Comparison of the complete genome sequence between C1 and G4 isolates of the Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus. Virology. 2005;333:190–199. doi: 10.1016/j.virol.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 87.Ogembo J.G., Caoili B.L., Shikata M., Chaeychomsri S., Kobayashi M., Ikeda M. Comparative genomic sequence analysis of novel Helicoverpa armigera nucleopolyhedrovris (NPV) isolated from Kenya and three other previously sequenced Helicoverpa spp. NPVs. Virus Genes. 2009;39:261–272. doi: 10.1007/s11262-009-0389-3. [DOI] [PubMed] [Google Scholar]
- 88.Chen X., Zhang W.-J., Wong J., Chun G., Lu A., McCutchen B.F., Presnail J.K., Herrmann R., Dolan M., Tingey S., et al. Comparative analysis of the complete genome sequences of Helicoverpa zea and Helicoverpa armigera single-nucleocapsid nucleopolyhedroviruses. J. Gen. Virol. 2002;83:673–684. doi: 10.1099/0022-1317-83-3-673. [DOI] [PubMed] [Google Scholar]
- 89.Xiao H., Qi Y. Genome sequence of Leucania seperata nucleopolyhedrovirus. Virus Genes. 2007;35:845–856. doi: 10.1007/s11262-007-0106-z. [DOI] [PubMed] [Google Scholar]
- 90.Li Q., Li L., Willis G., Erlandson M., Theilmann D.A., Donly C. Complete comparative genomic analysis of two field isolates of Mamestra configurata nucleopolyhedrovirus-A. J. Gen. Virol. 2005;86:91–105. doi: 10.1099/vir.0.80488-0. [DOI] [PubMed] [Google Scholar]
- 91.Harisson R.L., Puttler B., Popham H.J.R. Genomic sequence analysis of a fast-killing isolate of Spodoptera frugiperda multiple nucleopolyhedrovirus. J. Gen. Virol. 2008;89:775–790. doi: 10.1099/vir.0.83566-0. [DOI] [PubMed] [Google Scholar]
- 92.Wolff J.L.C., Valicente F.H., Martins R., Oliveira J.V., Zanotto P.M. Analysis of the genome of Spodoptera frugiperda nucleopolyhedrovirus (SfMNPV-19) and of the high genomic heterogeneity in group II nucleopolyhedroviruses. J. Gen. Virol. 2009;89:1202–1211. doi: 10.1099/vir.0.83581-0. [DOI] [PubMed] [Google Scholar]
- 93.Willis L.G., Siepp R., Stewart T.M., Erlandson M.A., Theilmann D.A. Sequence analysis of the complete genome of Trichoplusia ni single nucleopolyhedrovirus and identification of of a baculoviral photolyase gene. Virology. 2005;338:209–226. doi: 10.1016/j.virol.2005.04.041. [DOI] [PubMed] [Google Scholar]
- 94.Wormleaton S., Kuzio J., Winstanley D. The complete sequence of the Adoxophyes orana granulovirus genome. Virology. 2003;331:350–365. doi: 10.1016/s0042-6822(03)00149-1. [DOI] [PubMed] [Google Scholar]
- 95.Laing Z., Zhang X., Yin X. Genomic sequencing and analysis of Clostera anachoreta granuloviruses. Arch Virol. 2011;156:1185–1198. doi: 10.1007/s00705-011-0977-0. [DOI] [PubMed] [Google Scholar]
- 96.Lange M., Jehle J.A. The genome of the Cryptophlebia leucotreta granulovirus. Virology. 2003;317:220–236. doi: 10.1016/s0042-6822(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 97.Luque T., Finch R., Crook N., O’Reilly D.R., Winstanley D. The complete sequence of the Cydia pomonella granulovirus genome. J. Gen. Virol. 2001;82:2531–2547. doi: 10.1099/0022-1317-82-10-2531. [DOI] [PubMed] [Google Scholar]
- 98.Harrison R.L., Popham H.J.R. Genomic sequence analysis of a granulovirus isolated from the Old World bollworm Helicoverpa armigera. Virus Genes. 2000;21:147–155. doi: 10.1007/s11262-008-0218-0. [DOI] [PubMed] [Google Scholar]
- 99.Wang X.F., Zhang B.Q., Xu H.J., Cui Y.J., Xu Y.P., Zhang M.J., Han Y.S., Lee Y.S., Bao Y.Y., Zhang C.X. ODV-associated proteins of the Pieris rapae granulovirus. J. Proteome Res. 2011;10:2817–2827. doi: 10.1021/pr2000804. [DOI] [PubMed] [Google Scholar]
- 100.Hashimoto Y., Hayakawa T., Ueno Y., Fujita T., Sano Y., Matsumoto T. Sequence analysis of the Plutella xylostella granulovirus genome. Virology. 2000;275:358–372. doi: 10.1006/viro.2000.0530. [DOI] [PubMed] [Google Scholar]
- 101.Wang Y., Choi J.Y., Roh J.Y., Woo S.D., Jin B.R., Je Y.H. Molecular and phylogenetic characterization of Spodoptera litura granulovirus. J. Microbiol. 2008;46:704–708. doi: 10.1007/s12275-008-0133-z. [DOI] [PubMed] [Google Scholar]
- 102.Duffy S.P., Young A.M., Morin B., Lucarotti C.J., Koop B.F., Levin D.B. Sequence analysis and organization of the Neodiprion abietis nucleopolyhedrovirus genome. J. Virol. 2006;80:6952–6963. doi: 10.1128/JVI.00187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]