Abstract
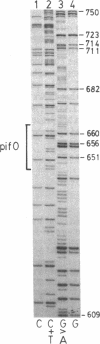
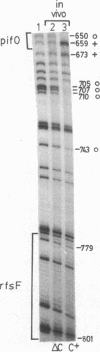
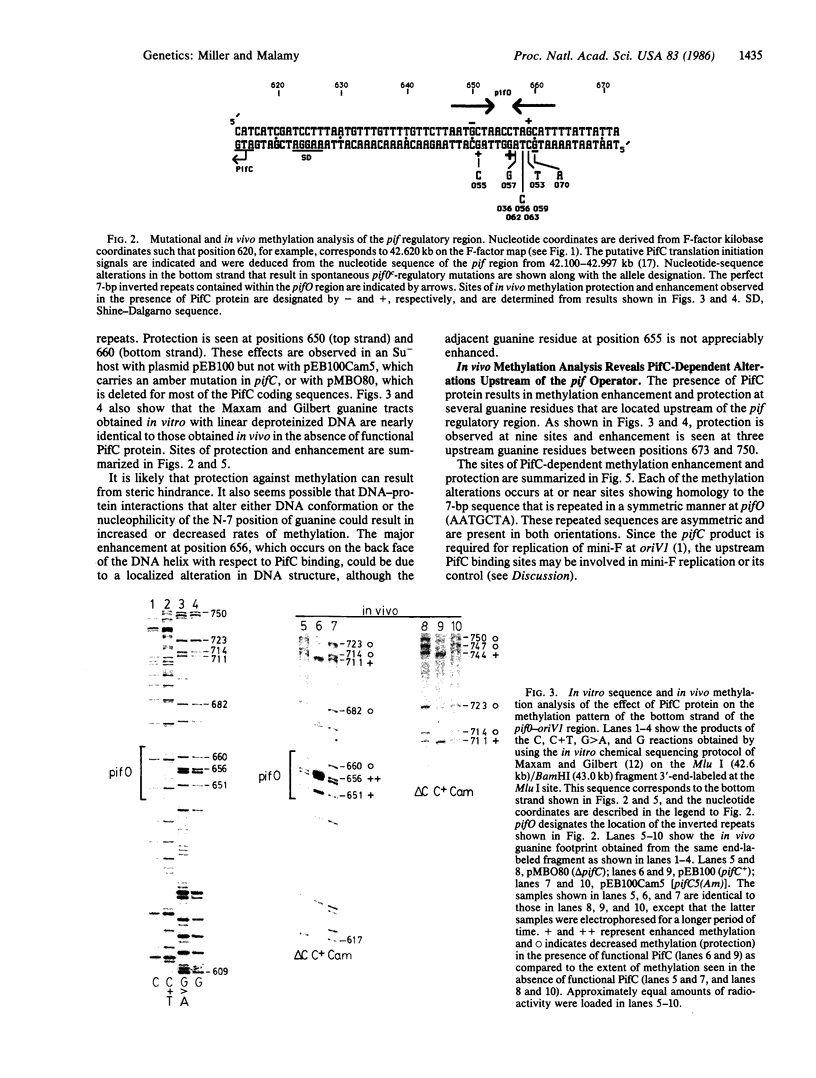
We have used in vivo methods to identify multiple DNA-binding sites for the negatively autoregulated mini-F replication factor PifC. Sequence analysis of pif operator constitutive mutants, isolated as insensitive to repression by PifC, establishes the structure of pifO. This site contains a 17-base-pair (bp) region of dyad symmetry with 7-bp perfect inverted repeats separated by 3 bp. In vivo DNA methylation studies with dimethyl sulfate show that the reactivity of five of six guanine residues in the pifO region is altered in the presence of PifC protein. In addition, there are several sites of PifC-dependent methylation enhancement and protection upstream of pifO within repeated sequences bearing homology to pifO. The significance of the repeated PifC binding sequences and their relationship to the primary origin of mini-F replication (oriV1) are discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blumberg D. D., Mabie C. T., Malamy M. H. T7 protein synthesis in F-factor-containing cells: evidence for an episomally induced impairment of translation and relation to an alteration in membrane permeability. J Virol. 1975 Jan;17(1):94–105. doi: 10.1128/jvi.17.1.94-105.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattoraj D. K., Snyder K. M., Abeles A. L. P1 plasmid replication: multiple functions of RepA protein at the origin. Proc Natl Acad Sci U S A. 1985 May;82(9):2588–2592. doi: 10.1073/pnas.82.9.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattoraj D., Cordes K., Abeles A. Plasmid P1 replication: negative control by repeated DNA sequences. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6456–6460. doi: 10.1073/pnas.81.20.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenlaub R., Figurski D., Helinski D. R. Bidirection replication from a unique origin in a mini-F plasmid. Proc Natl Acad Sci U S A. 1977 Mar;74(3):1138–1141. doi: 10.1073/pnas.74.3.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Lane D., Hill D., Caughey P., Gunn P. The mini-F primary origin. Sequence analysis and multiple activities. J Mol Biol. 1984 Dec 5;180(2):267–282. doi: 10.1016/s0022-2836(84)80004-2. [DOI] [PubMed] [Google Scholar]
- Lee S. B., Bailey J. E. A mathematical model for lambda dv plasmid replication: analysis of wild-type plasmid. Plasmid. 1984 Mar;11(2):151–165. doi: 10.1016/0147-619x(84)90020-9. [DOI] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J Bacteriol. 1973 Jun;114(3):1143–1150. doi: 10.1128/jb.114.3.1143-1150.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- May M. S., Hattaman S. Deoxyribonucleic acid-cytosine methylation by host- and plasmid-controlled enzymes. J Bacteriol. 1975 Apr;122(1):129–138. doi: 10.1128/jb.122.1.129-138.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Chang Z. T., Horiuchi T. Control of cell division by sex factor F in Escherichia coli. II. Identification of genes for inhibitor protein and trigger protein on the 42.84-43.6 F segment. J Mol Biol. 1984 Apr 25;174(4):627–646. doi: 10.1016/0022-2836(84)90087-1. [DOI] [PubMed] [Google Scholar]
- Miki T., Yoshioka K., Horiuchi T. Control of cell division by sex factor F in Escherichia coli. I. The 42.84-43.6 F segment couples cell division of the host bacteria with replication of plasmid DNA. J Mol Biol. 1984 Apr 25;174(4):605–625. doi: 10.1016/0022-2836(84)90086-x. [DOI] [PubMed] [Google Scholar]
- Miller J. F., Lanka E., Malamy M. H. F factor inhibition of conjugal transfer of broad-host-range plasmid RP4: requirement for the protein product of pif operon regulatory gene pifC. J Bacteriol. 1985 Sep;163(3):1067–1073. doi: 10.1128/jb.163.3.1067-1073.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. F., Malamy M. H. Identification of the pifC gene and its role in negative control of F factor pif gene expression. J Bacteriol. 1983 Oct;156(1):338–347. doi: 10.1128/jb.156.1.338-347.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. F., Malamy M. H. Regulation of the F-factor pif operon: pifO, a site required in cis for autoregulation, titrates the pifC product in trans. J Bacteriol. 1984 Oct;160(1):192–198. doi: 10.1128/jb.160.1.192-198.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison T. G., Malamy M. H. Comparisons of F factors and R factors: existence of independent regulation groups in F factors. J Bacteriol. 1970 Jul;103(1):81–88. doi: 10.1128/jb.103.1.81-88.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murotsu T., Tsutsui H., Matsubara K. Identification of the minimal essential region for the replication origin of miniF plasmid. Mol Gen Genet. 1984;196(2):373–378. doi: 10.1007/BF00328075. [DOI] [PubMed] [Google Scholar]
- Nick H., Gilbert W. Detection in vivo of protein-DNA interactions within the lac operon of Escherichia coli. 1985 Feb 28-Mar 6Nature. 313(6005):795–798. doi: 10.1038/313795a0. [DOI] [PubMed] [Google Scholar]
- O'Connor M. B., Malamy M. H. Site-specific recombination in the oriV1 region of the F factor. Cold Spring Harb Symp Quant Biol. 1984;49:421–434. doi: 10.1101/sqb.1984.049.01.048. [DOI] [PubMed] [Google Scholar]
- Ogura T., Hiraga S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4784–4788. doi: 10.1073/pnas.80.15.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Rotman G. S., Cooney R., Malamy M. H. Cloning of the pif region of the F sex factor and identification of a pif protein product. J Bacteriol. 1983 Jul;155(1):254–264. doi: 10.1128/jb.155.1.254-264.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. R. Regulation of plasmid replication. Microbiol Rev. 1984 Mar;48(1):1–23. doi: 10.1016/b978-0-12-048850-6.50006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B. The chemical effects of nucleic acid alkylation and their relation to mutagenesis and carcinogenesis. Prog Nucleic Acid Res Mol Biol. 1975;15(0):219–284. [PubMed] [Google Scholar]
- Sommer S., Bailone A., Devoret R. SOS induction by thermosensitive replication mutants of miniF plasmid. Mol Gen Genet. 1985;198(3):456–464. doi: 10.1007/BF00332939. [DOI] [PubMed] [Google Scholar]
- Søgaard-Andersen L., Rokeach L. A., Molin S. Regulated expression of a gene important for replication of plasmid F in E. coli. EMBO J. 1984 Feb;3(2):257–262. doi: 10.1002/j.1460-2075.1984.tb01794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto K., Iino T. An essential gene for replication of the mini-F plasmid from origin I. Mol Gen Genet. 1984;196(1):59–63. doi: 10.1007/BF00334092. [DOI] [PubMed] [Google Scholar]
- Tolun A., Helinski D. R. Direct repeats of the F plasmid incC region express F incompatibility. Cell. 1981 Jun;24(3):687–694. doi: 10.1016/0092-8674(81)90095-7. [DOI] [PubMed] [Google Scholar]
- Trawick J. D., Kline B. C. A two-stage molecular model for control of mini-F replication. Plasmid. 1985 Jan;13(1):59–69. doi: 10.1016/0147-619x(85)90056-3. [DOI] [PubMed] [Google Scholar]
- Tsutsui H., Fujiyama A., Murotsu T., Matsubara K. Role of nine repeating sequences of the mini-F genome for expression of F-specific incompatibility phenotype and copy number control. J Bacteriol. 1983 Jul;155(1):337–344. doi: 10.1128/jb.155.1.337-344.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]