Abstract
A 42 kb region on human chromosome 9p21 encodes for three distinct tumor suppressors, p16INK4A, p14ARF and p15INK4B, and is altered in an estimated 30–40% of human tumors. The expression of the INK4A-ARF-INK4B gene cluster is silenced by polycomb during normal cell growth and is activated by oncogenic insults and during aging. How the polycomb is recruited to repress this gene cluster is unclear. Here, we show that expression of oncogenic Ras, which stimulates the expression of p15INK4B and p16INK4A, but not p14ARF, inhibits the expression of ANRIL (antisense non-coding RNA in the INK4 locus), a 3.8 kb-long non-coding RNA expressed in the opposite direction from INK4A-ARF-INK4B. We show that the p15INK4B locus is bound by SUZ12, a component of polycomb repression complex 2 (PRC2), and is H3K27-trimethylated. Notably, depletion of ANRIL disrupts the SUZ12 binding to the p15INK4B locus, increases the expression of p15INK4B, but not p16INK4A or p14ARF, and inhibits cellular proliferation. Finally, RNA immunoprecipitation demonstrates that ANRIL binds to SUZ12 in vivo. Collectively, these results suggest a model in which ANRIL binds to and recruits PRC2 to repress the expression of p15INK4B locus.
Keywords: long non-coding RNA, ANRIL, polycomb, p15INK4B, p16INK4A, ARF
Introduction
The INK4A-ARF-INK4B gene cluster occupies a 42 kb stretch on the human chromosome 9p21 and is homozygously deleted or expression silenced in a wide range of human cancers with an estimated frequency of ~40% (Sherr, 1998; Kim and Sharpless, 2006), representing one of the most frequently altered genes in human cancer. Genetic analyses in mice with mutations targeting each gene individually support the tumor suppression function for all three genes (Serrano et al., 1996; Kamijo et al., 1997; Krimpenfort et al., 2001, 2007; Sharpless et al., 2001). Of these three tumor suppressors encoded by this gene cluster, both p15INK4B and p16INK4A function as the inhibitors of CDK4 and CDK6, and thereby retain the growth suppressive activity of RB family proteins, whereas ARF, expressed from a distinct promoter and an alternative reading frame of INK4A, binds to and inhibits the activity of oncoprotein MDM2 and thus stabilizes and activates p53. Clustering three distinct growth and tumor suppressors in such a small region may aid the coordinated integration of different growth insults into two major tumor suppression pathways in mammalian cells, one mediated by RB and the other by p53 (Gil and Peters, 2006).
All the three genes are expressed at a barely detectable low level in young and normal cells, and are accumulated during cell aging or in cells insulted by hyperproliferative oncogenic stimuli (Zindy et al., 1997; Krishnamurthy et al., 2004; Gil and Peters, 2006), suggesting a common cellular mechanism and potentially coordinated regulation of these three genes—to maintain aged cells in senescence and to arrest oncogenic-insulted cells from proliferation.
The repression of this gene cluster involves polycomb proteins and histone H3 lysine27 (H3K27) trimethylation (Jacobs et al., 1999; Itahana et al., 2003; Bracken et al., 2007; Kotake et al., 2007; Kia et al., 2008; Agger et al., 2009). The polycomb proteins form multimeric protein complexes, PRC (polycomb repression complex)-1 and -2 that are involved in the heritable stable repression of genes through histone modification. Previous genetic and biochemical studies suggest a hierarchical recruitment model where PRC2-mediated H3K27 methylation is required for recruitment of PRC1, which causes H2A-K119 ubiquitination on the Hox gene cluster both in fly and human cells (Wang et al., 2004; Cao et al., 2005). A critical issue for better understanding the regulation of the INK4A-ARF-INK4B gene cluster is how PRC1 and PRC2 are recruited to this region.
Several long non-coding RNAs (lncRNAs) have recently been reported to have a direct role in recruiting PRC2 complexes to specific loci and repress gene expression. These include RepA, a 1.6-kb lncRNA involved in X-chromosome inactivation (Zhao et al., 2008), Kcnq1ot1, a 91.5 kb lncRNA required for 10 paternally imprinted genes on 11p15 (Pandey et al., 2008; Terranova et al., 2008; Zhao et al., 2008), and HOTAIR, a 2.2-kb ncRNA involved in the repression of the HOX loci and promoting cancer metastasis (Rinn et al., 2007; Gupta et al., 2010). These findings led us to explore the possibility that ANRIL (antisense non-coding RNA in the INK4 locus), a 3834 bp transcript whose transcription is initiated from the INK4A-ARF-INK4B gene cluster (Figure 1a), may be involved in the repression of INK4A, ARF and/or INK4B. ANRIL contains 19 exons, a polyadenylation site in the last exon and spans over 126 kb of genomic sequence that is deleted in the melanoma–neural system tumor syndrome family analyzed by previous study (Pasmant et al., 2007). Exon 1 of ANRIL locates between the promoter of ARF and INK4B, and is transcribed in the direction opposite from that of p15INK4B. Deletion of a 70-kb sequence in mouse chromosome 4 synteny to a 58-kb non-coding region in human chromosome 9p21 that includes seven exons of ANRIL resulted in a significantly increased expression of both p16INK4A and p15INK4B in several organs and tissues, but had no effect on other neighboring genes (Visel et al., 2010), providing a genetic evidence for the negative regulation of p16INK4A and p15INK4B by the ncRNA sequences expressed in this region. In this report, we show that p15INK4B locus is repressed by PRC2 proteins and that ANRIL is required for the recruitment of PRC2 to and repression of the p15INK4B locus.
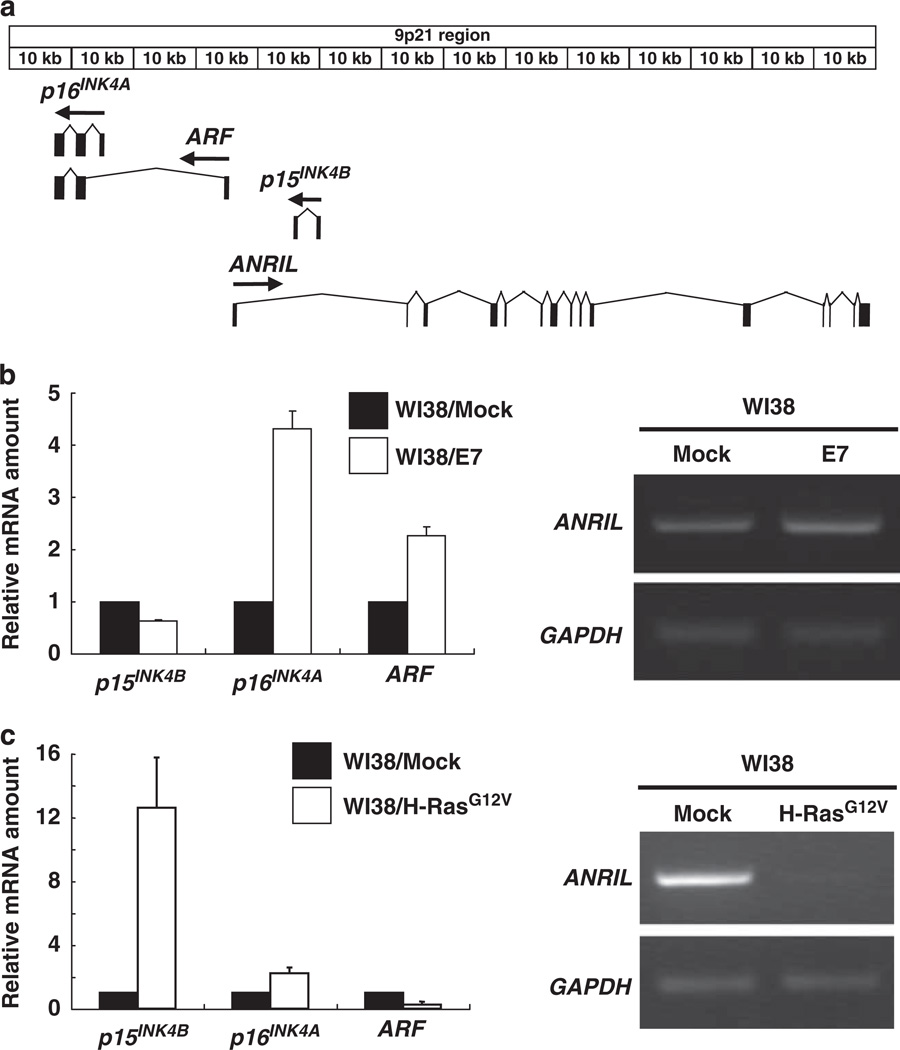
Figure 1. Oncogenic Ras inhibits the expression of ANRIL and stimulates p15INK4B expression.
(a) Schematic representation of INK4 locus. The arrows indicate the direction of transcription of each gene. The black boxes indicate the exons of INK4 and ANRIL genes, respectively. (b) WI38 cells were infected with empty (Mock) or E7-expressing retroviruses and selected by 400 µg/ml G418 treatment as described in Kotake et al. (2007). (Left panel) The levels of p15INK4B, p16INK4A and p14ARF mRNA were determined by quantitative reverse transcriptase PCR, and results are expressed relative to the corresponding values for WI38/Mock cells. The mean values and s.d.s were calculated from triplicates of a representative experiment. The specific PCR pairs for p15INK4B, p16INK4A, ARF and GAPDH are described in Kotake et al. (2007). (Right panel) The expression of ANRIL was determined by reverse transcriptase PCR using primers designed on exon1 (forward primer) and exon2 (reverse primer) as described in the Supplementary Information. (c) WI38 cells were infected with empty (Mock) or oncogenic H-RasG12V-expressing retroviruses, selected by 2 µg/ml puromycin treatment for 3 days and harvested 8 days after initial infection. (Left panel) The levels of p15INK4B, p16INK4A and p14ARF mRNA were determined by quantitative reverse transcriptase PCR, and results are expressed relative to the corresponding values for WI38/Mock cells. The mean values and s.d.s were calculated from triplicates of a representative experiment. (Right panel) The expression of ANRIL was detected by reverse transcriptase PCR using the same primers with (b).
Results and discussion
We first determined the expression of ANRIL in response to two well-characterized oncoproteins, E7 and Ras, which are known to affect the expression of p16INK4A, ARF and/or p15INK4B. Although E7 inactivates RB family proteins and potently activates the expression of p16INK4A and to a lesser extent p14ARF (Kotake et al., 2007, 2009), oncogenic Ras induces the expression of p15INK4B (Malumbres et al., 2000) (Figures 1b and c). We established stable cell lines expressing either E7 or oncogenic RasG12V and determined the expression of these tumor suppressor genes as well as ANRIL. As expected, both p16INK4A and p14ARF mRNAs, but not p15INK4B, were substantially increased in WI38/E7 cells, whereas in WI38/RasG12V cells, p15INK4B and p16INK4A, but not p14ARF, was stimulated by more than 12-fold (p15INK4B) and 2-fold (p16INK4A) (Figures 1b and c). The induction of p16INK4A by RasG12V in WI38 cells is somewhat lower than that seen in some other normal fibroblast such as MEFs or IMR-90. The reason for this is not clear, but may relate to the difference between species or cell lines. The expression of ANRIL was readily detectable in WI38 cells, was not significantly affected by the expression of E7 and was notably reduced by the expression of H-RasG12V (Figure 1c). These data indicate that stimulation of p15INK4B by oncogenic Ras in cultured normal human cells is associated with a decrease of ANRIL expression, suggesting a potential negative regulation of p15INK4B by ANRIL.
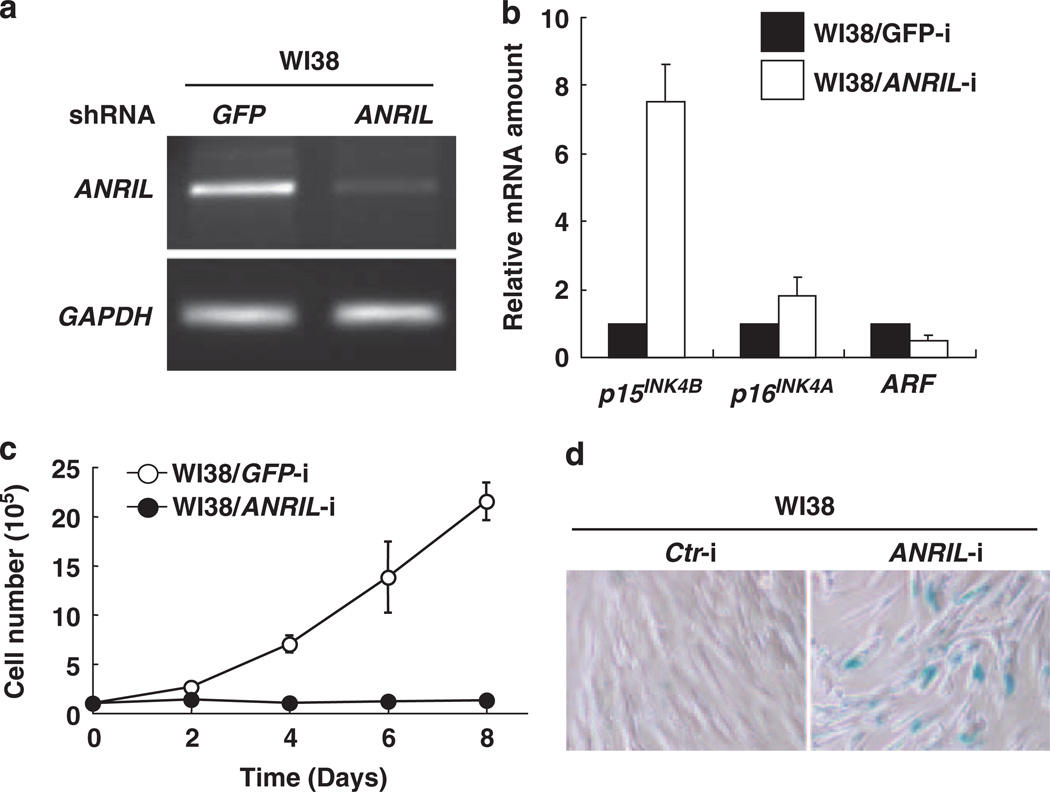
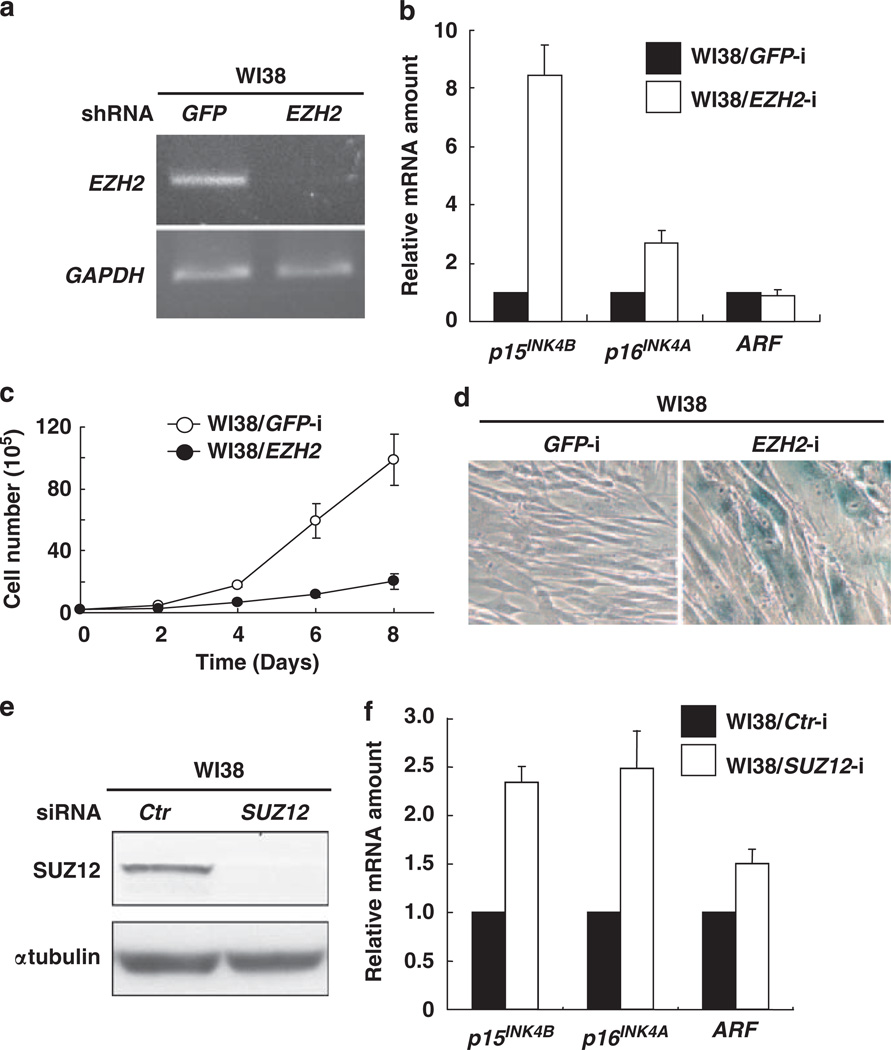
We then designed a retroviral vector encoding a short hairpin RNA that specifically targets the exon1 region of ANRIL. Infection of WI38 cells with ANRIL short hairpin RNA retroviruses efficiently reduced ANRIL level (Figure 2a). Quantitative reverse transcriptase PCR analysis showed that silencing ANRIL resulted in an increase of p15INK4B expression by nearly 8-fold, but had minimal effect on the expression of p16INK4A (increased by 1.8-fold) or p14ARF (reduced by half, Figure 2b). Similar result was also reported very recently by Yap et al. that silencing ANRIL by a different method using antisense DNA increases the p16INK4A expression four-fold in another normal human diploid fibroblasts line, IMR-90 cells (Yap et al., 2010). Associated with p15INK4B increase, ANRIL silencing also resulted in decreased cell growth (Figure 2c) and an increase of cells stained positively for senescence-associated β-galactosidase activity, an indicator of cell senescence (Figure 2d). These results indicate that ANRIL is involved in the selective repression of p15INK4B transcription and prevention of cellular senescence. To search for the mechanism underlying the repression of p15INK4B by ANRIL, we examined the role of PRC2 in p15INK4B regulation. Transduction of WI38 cells with a retroviral vector encoding an short hairpin RNA targeting the catalytic subunit of PRC2 histone methyltransferase, EZH2, efficiently reduced EZH2 level (Figure 3a). Associated with EZH2 decrease is a substantial increase of p15INK4B mRNA by eightfold and p16INK4A mRNA by threfold, but no detectable effect on p14ARF mRNA level (Figure 3b). Associated with increase of p15INK4B and p16INK4A, EZH2 knockdown decreased cell growth (Figure 3c) and induced cell senescence (Figure 3d). To further confirm this, we transfected WI38 cells with small interfering RNA silencing SUZ12, another component of PRC2, and found that silencing SUZ12 (Figure 3e) also increased the expression of both p15INK4B as well as p16INK4A expression (Figure 3f). These results indicate that PRC2 is involved in the p15INK4B repression and cellular senescence.
Figure 2. ANRIL negatively regulates p15INK4B gene expression.
(a) WI38 cells were infected with a retrovirus vector encoding short hairpin RNA (shRNA) against either GFP or ANRIL, selected by 2 µg/ml puromycin treatment for 3 days and harvested 8 days after initial infection. The efficiency of ANRIL silencing was determined by reverse transcriptase PCR. (b) The effects of ANRIL silencing on p15INK4B, p16INK4A and p14ARF were determined by quantitative reverse transcriptase PCR. The results are expressed relative to the corresponding values for WI38/GFP-i cells. The mean values and s.d.s were calculated from triplicates of a representative experiment. (c) The growth curves of WI38 cells infected with a retrovirus expressing shRNA against either GFP or ANRIL. Viable cells were counted by trypan blue staining at indicated days after initial seeding of 1 × 105 cells. (d) WI38 cells with small interfering RNA targeting either control or ANRIL were stained for senescence-associated β-galactosidase by using Senescence Detection Kit (BioVision, Mountain View, CA, USA).
Figure 3. PRC2 binds to and negatively regulates p15INK4B gene expression.
(a) WI38 cells were infected with retrovirus expressing short hairpin RNA (shRNA) against either GFP or EZH2, selected by 2 µg/ml puromycin treatment and harvested 8 days after initial infection. The efficiency of EZH2 silencing was determined by reverse transcriptase PCR. The specific PCR pair for EZH2 is described in the Supplementary Information. (b) The effects of EZH2 silencing on p15INK4B, p16INK4A and p14ARF were determined by quantitative reverse transcriptase PCR, and results are expressed relative to the corresponding values for WI38/GFP-i cells. The mean values and s.d.s were calculated from triplicates of a representative experiment. (c) The growth curves of WI38 cells infected with a retrovirus expressing shRNA against either GFP or EZH2. Viable cells were counted by trypan blue staining at indicated days after initial seeding of 2 × 105 cells. (d) WI38 cells infected with a retrovirus expressing shRNA against either GFP or EZH2 were stained for senescence-associated β-galactosidase. (e) The efficiency of SUZ12 silencing by small interfering RNA (siRNA) was determined by immunoblotting. Antibodies to SUZ12 (Millipore, Billerica, MA, USA) and αtubulin (Sigma, St Louis, MO, USA) were purchased commercially. (f) The effects of SUZ12 silencing on p15INK4B, p16INK4A and p14ARF were determined by quantitative reverse transcriptase PCR.
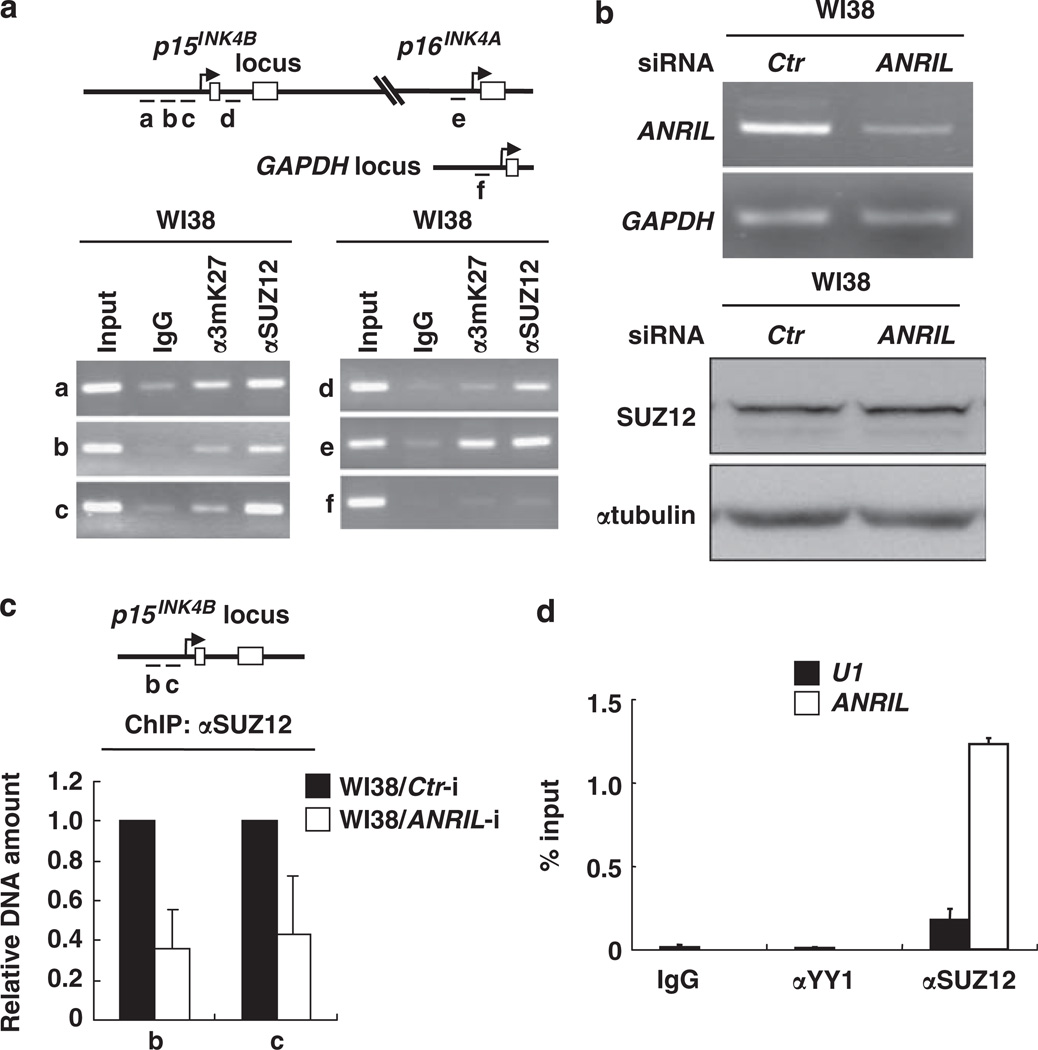
That both ANRIL and PRC2 repress p15INK4B expression led us to determine whether ANRIL is required for PRC2 recruitment to p15INK4B locus. We first carried out a chromatin immunoprecipitation assay to test whether PRC2 directly binds to the p15INK4B locus. This assay demonstrated that the p15INK4B, as well as p16INK4A, locus is bound by SUZ12 and is trimethylated at lys27 of histone H3 (Figure 4a). Chromatin immunoprecipitation–quantitative PCR assay showed that silencing ANRIL causes substantial loss of SUZ12 occupancy on the p15INK4B locus (Figure 4c), although the steady state level of the SUZ12 protein is unchanged (Figure 4b). Finally, we examined whether ANRIL binds directly to SUZ12 by RNA immunoprecipitation assay and found that the SUZ12 immunocomplex, but not the immunocomplexes of immunoglobulin G or another negative control YY1, substantially enriched ANRIL RNA compared with U1 RNA, a non-specific RNA that is expressed in the cells at high levels (Figure 4d). These results demonstrate that ANRIL can bind directly and specifically to SUZ12.
Figure 4. ANRIL binds to and is required for the recruitment of SUZ12 to p15INK4B locus.
(a) A schematic representation of the p15INK4B, p16INK4A and GAPDH gene loci and amplicons (a–f) used for chromatin immunoprecipitation assay. Antibodies against SUZ12, trimethyl-H3K27 and immunoglobulin G (IgG) control were used in the chromatin immunoprecipitation assay. PCR was carried out using primers for each amplicon. The sequence of primer pairs is described in the Supplementary Information. (b) The efficiency of ANRIL silencing by small interfering RNA (siRNA) was determined by reverse transcriptase PCR. The SUZ12 protein level was determined by immunoblotting. (c) A schematic representation of the p15INK4B locus and amplicons (b and c). Abundance of SUZ12 binding on the p15INK4B locus was determined by quantitative PCR following chromatin immunoprecipitation assay with SUZ12 antibody in WI38 cells with siRNA targeting either control or ANRIL. PCR was carried out using primers for each amplicon. (d) Nuclear extracts of WI38 cells were immunoprecipitated by IgG, anti-SUZ12 or anti-YY1. Coprecipitated RNAs were detected by quantitative reverse transcriptase PCR using primers for ANRIL (derived from exons 1 and 2) or U1 small nuclear RNA as described in the Supplementary Information. The results are expressed relative to the corresponding values for input RNAs. The mean values and s.d.s were calculated from triplicates of a representative experiment.
In this report, we showed that mRNA-like ncRNA, ANRIL, and the PRC2 histone methyltransferase complex are involved in the repression of p15INK4B transcription. We further demonstrated that ANRIL binds to SUZ12, a PRC2 component, and is required for the SUZ12 occupancy on the p15INK4B locus. Loss of ANRIL or PRC2 showed the same effects on cellular proliferation and both cause premature senescence, supporting a mechanistic link between PRC2 and ANRIL. The results presented herein provide new insight into the epigenetic control of p15INK4B locus by PRC2 and lncRNA. The detailed biochemical mechanisms underlying the function of ANRIL in the PRC2 recruitment to the p15INK4B locus remain to be determined including, in particular, the interaction between ANRIL and PRC2. The other ncRNA, Xist, directly associates with EZH2 through RepA, a motif comprising 7.5 tandem repeats of a 28-nucleotide sequence that folds into two conserved stem-loop structures (Wutz et al., 2002; Zhao et al., 2008). Although we could not find such a distinctive motif on ANRIL, we showed the direct binding between ANRIL and PRC2. A recent study utilized high-magnification RNA–DNA FISH assay to demonstrate that large ncRNA, Air, interacts with the chromatin of cis-linked target gene promoter and recruits the H3K9 histone methyltransferase to repress the gene expression (Nagano et al., 2008). Based on that, ANRIL may directly interact with the p15INK4B promoter chromatin to recruit PRC2. Very recently, Yap et al. reported that CBX7, an H3k27me3-recognizing component of PRC1, can bind directly to both ANRIL and H3K27me3 via its chromodomain, and both interactions are required for CBX7 to repress the INK4A and INK4B loci (Yap et al., 2010). In the present study, we demonstrated that ANRIL is also involved in the recruitment of another polycomb complex, PRC2, to another gene, p15INK4B, in the INK4A-ARF-INK4B region. Our study thus broadens the scope of ANRIL function. Given that ANRIL encodes an almost 4 kb ncRNA that binds at least two different polycomb proteins, it will be important to determine the responsible regions in ANRIL for the association with each polycomb protein.
Recent genome-wide association studies have linked common single-nucleotide polymorphisms in 9p21 region that is independently associated with both type II diabetes and coronary heart disease (Helgadottir et al., 2007; McPherson et al., 2007; Saxena et al., 2007; Scott et al., 2007). As the ANRIL genomic region spans the risk-associated single-nucleotide polymorphisms (Broadbent et al., 2008), expression of ANRIL might be affected by the unknown causative genetic variant. Therefore, it is possible that the deregulation of ANRIL by such risk-associated single-nucleotide polymorphisms changes the expression level of p15INK4B and/or other target genes, leading to the development of type II diabetes or coronary heart disease in humans.
Supplementary Material
Acknowledgements
We thank Howard Chang for the insightful discussion during this study, Yaxue Zeng and Matt Smith for helpful discussion and reading the manuscript, Mika Matsumoto, Michiyo Hakamata and Harumi Shiratori for technical assistance. This study is supported by grants from the Ministry of Education, Science, Sports, Culture, and Technology of Japan (YK, MK, SS and KK) and NIH grant CA68377 (YX).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, et al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, et al. The polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes & Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbent HM, Peden JF, Lorkowski S, Goel A, Ongen H, Green F, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17:806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- Itahana K, Zou Y, Itahana Y, Martinez JL, Beausejour C, Jacobs JJ, et al. Control of the replicative life span of human fibroblasts by p16 and the polycomb protein Bmi-1. Mol Cell Biol. 2003;23:389–401. doi: 10.1128/MCB.23.1.389-401.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol. 2008;28:3457–3464. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Sharpless NE. The Regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Kotake Y, Cao R, Viatour P, Sage J, Zhang Y, Xiong Y. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4a tumor suppressor gene. Genes Dev. 2007;21:49–54. doi: 10.1101/gad.1499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Zeng YX, Xiong Y. DDB1-CUL4 and MLL1 mediate oncogene-induced p16(INK4a) activation. Cancer Res. 2009;69:1809–1814. doi: 10.1158/0008-5472.CAN-08-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimpenfort P, Ijpenberg A, Song JY, van der Valk M, Nawijn M, Zevenhoven J, et al. p15Ink4b is a critical tumour suppressor in the absence of p16Ink4a. Nature. 2007;448:943–946. doi: 10.1038/nature06084. [DOI] [PubMed] [Google Scholar]
- Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature. 2001;413:83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, et al. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, Perez De Castro I, Hernandez MI, Jimenez M, Corral T, Pellicer A. Cellular response to oncogenic ras involves induction of the Cdk4 and Cdk6 inhibitor p15(INK4b) Mol Cell Biol. 2000;20:2915–2925. doi: 10.1128/mcb.20.8.2915-2925.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Lee H-W, Chin L, Cordon-Cardos C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, Bardeesy N, Lee KH, Carrasco D, Castrillon DH, Aguirre AJ, et al. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- Terranova R, Yokobayashi S, Stadler MB, Otte AP, van Lohuizen M, Orkin SH, et al. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev Cell. 2008;15:668–679. doi: 10.1016/j.devcel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, et al. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464:409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F, Quelle DE, Roussel MF, Sherr CJ. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.