SUMMARY
CUL4B, encoding a scaffold protein for the assembly of Cullin4B-Ring ubiquitin ligase (CRL4B) complexes, is frequently mutated in X-linked mental retardation (XLMR) patients. Here, we show that CUL4B, but not its paralogue CUL4A, targets WDR5, a core subunit of histone H3 lysine 4 (H3K4) methyltransferase complexes, for ubiquitination and degradation in the nucleus. Knocking down CUL4B increases WDR5 and trimethylated H3K4 (H3K4me3) on the neuronal gene promoters and induces their expression. Furthermore, CUL4B depletion suppresses neurite outgrowth of PC12 neuroendocrine cells which can be rescued by co-depletion of WDR5. XLMR-linked mutations destabilize CUL4B and impair its ability to support neurite outgrowth of PC12 cells. Our results identify WDR5 as a critical substrate of CUL4B in regulating neuronal gene expression and suggest epigenetic change as a common pathogenic mechanism for CUL4B-associated XLMR.
INTRODUCTION
The ubiquitin system covalently attaches ubiquitin to specific substrate proteins, resulting in either an alteration of substrate function or, more commonly, degradation by the 26S proteasome (Pickart, 2001). Disruption of this system has been linked to the development of various types of human diseases, including autoimmunity (Bhoj and Chen, 2009), neurological diseases (Tai and Schuman, 2008; Yi and Ehlers, 2007) and cancer (Frescas and Pagano, 2008; Nakayama and Nakayama, 2006). Ubiquitylation begins with the ATP dependent activation of ubiquitin by the E1 enzyme, is followed by the transfer of activated ubiquitin to an E2 ubiquitin conjugating enzyme, and finally an E3 ubiquitin ligase is responsible for recruiting a specific substrate and promoting ubiquitin ligation. Over 1,000 distinct E3 ligases, belonging to two major families, the HECT family and the RING family, are present in higher eukaryotes, implicating a very broad function of the ubiquitin system in cell regulation. Although not containing a RING domain intrinsically themselves, members of the evolutionarily conserved cullin family can bind with a small RING protein, either ROC1 or ROC2 (for RING of Cullins; also known as Rbx and Hrt1), in trans to constitute by far the largest family of E3 ligases, the cullin-RING ligases (CRLs). Unlike other RING E3 ligases, cullins do not bind substrates directly, but rather rely on substrate recruiting receptors that are typically joined to the cullin complex by a linker protein. Remarkably, each cullin can associate with a different family of substrate receptors, potentially constituting as many as 300 – 500 distinct CRLs in vivo (Zimmerman et al., 2010).
CUL4 is one of three founding cullins evolutionarily conserved from yeast to humans. Genetic analyses in various organisms have revealed a wide range of cellular and organismal functions of CUL4, many of which are associated with chromatin such as chromosome condensation, heterochromatin formation, DNA replication and repair (O'Connell and Harper, 2007, Jackson, 2009 #2638). CUL4 presents as a single gene in yeast, plant and invertebrate, but vertebrates express two closely related paralogues, CUL4A and CUL4B. Deletion of Cul4a in mice increased levels of several CRL4 substrates, but did not cause major developmental defects or obvious pathological conditions, (Liu et al., 2009). Cul4b deficiency, on the other hand, resulted in embryonic lethality around E9.5 and defects in nervous system and heart development (Cox et al.), indicating a unique function of Cul4b that cannot be compensated by Cul4a. In human, CUL4A gene amplification is noted in human breast (Chen et al., 1998), liver cancers (Yasui et al., 2002) and pleural mesothelioma (Hung et al., 2009), whereas loss-of-function mutations in CUL4B have been identified in patients with X-linked mental retardation (XLMR) (Badura-Stronka et al., 2010; Isidor et al., 2010; Tarpey et al., 2007; Zou et al., 2007).
Mental retardation (MR) affects approximately 1–3% of the population in the United States (Leonard and Wen, 2002). Genetic factors contribute to a high percentage of severe MR cases, and nearly 300 MR genes have been identified. Among them, about 16% of genes are X-linked genes and affect primarily males (Inlow and Restifo, 2004), leading to XLMR (Chiurazzi et al., 2008; Ropers, 2006), including CUL4B which was identified by a recent chromosome-wide whole exon sequencing study to be one of the most frequently mutated genes in XLMR patients. The molecular mechanisms by which CUL4 regulates chromatin and CUL4B mutations contribute to XLMR are unclear. This study is directed toward these issues.
RESULTS
WDR5 is degraded by DDB1-CUL4B E3 ubiquitin ligase
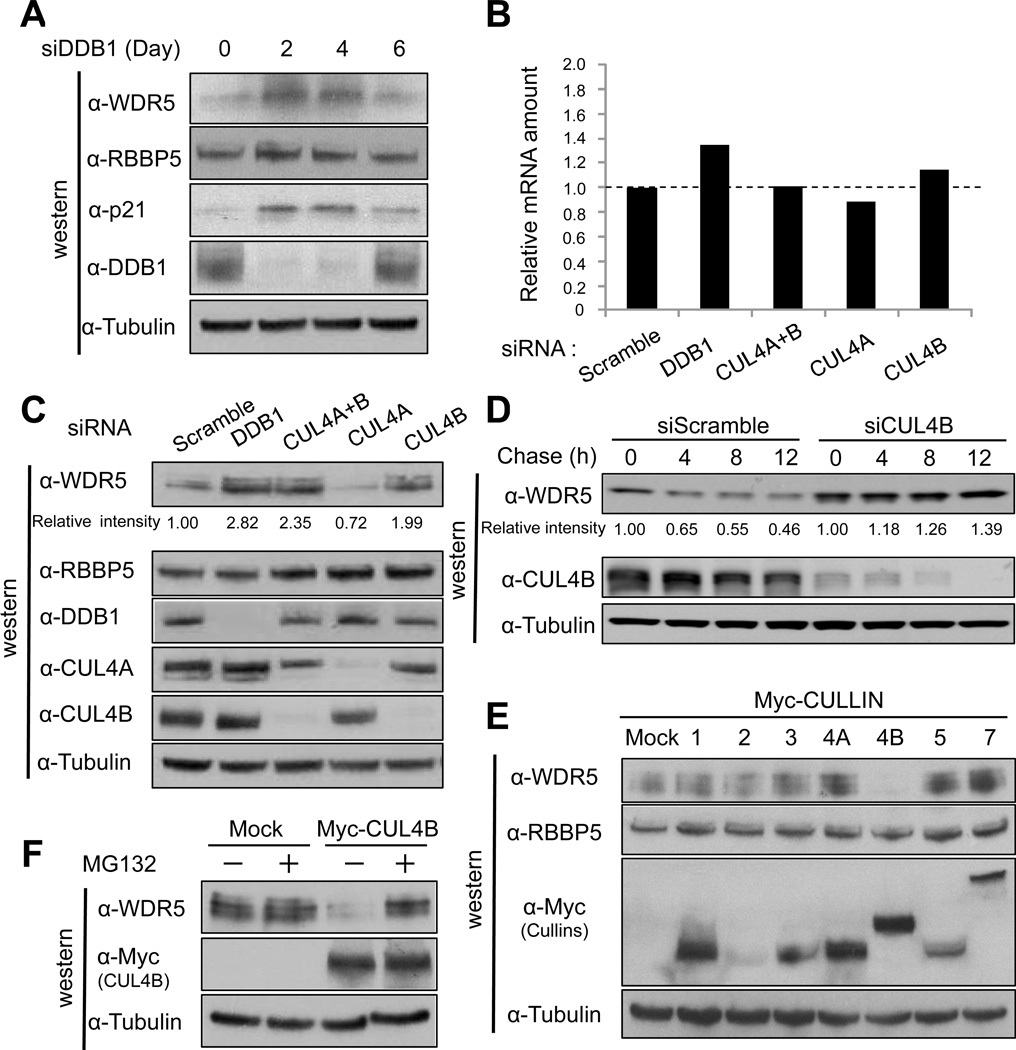
Previously, we and other groups reported that human cells express as many as ninety DDB1-binding WD40 proteins (DWD, also known as DCAF for DDB1- and CUL4-associated factors and CDW for CUL4 and DDB1-associated WD40 repeats) (Angers et al., 2006; He et al., 2006; Higa et al., 2006; Jin et al., 2006). Among these DWD proteins is WDR5, a core subunit of histone H3 lysine 4 (H3K4) methyltransferase complexes (Dou et al., 2006; Trievel and Shilatifard, 2009). Although DWD/DCAF/CDW proteins are believed to function as substrate recognition subunits in CRL4s, we found surprisingly that knocking down DDB1 increased the steady state level of WDR5 protein in either HCT116 (Figure 1A) or HeLa cells (Figure S1A), but had little effect on the level of WDR5 mRNA (Figure 1B). These results suggest that the increase in WDR5 level is achieved via a posttranscriptional mechanism and that WDR5 may be itself a substrate of DDB1-based E3 ligase.
Figure 1. CRL4B degrades WDR5 protein in a proteasome dependent manner.
(A) HCT116 cells were transiently transfected with siRNA for DDB1. Cells were harvested every other day (up to 6 days), and cell lysates were subjected to immunoblot analysis (IB) with indicated antibodies.
(B, C) HCT116 cells were transiently transfected with indicated siRNA. Cells were harvested, and cell lysates were subjected to immunoblot analysis (IB) with indicated antibodies (C) or qRT-PCR analysis to determine the WDR5 mRNA levels in cells 4 days post-transfection (B). Band intensity was measured by NIH Image J (version 1.44k).
(D) HCT116 cells were transiently transfected with siRNA targeting CUL4B. 3 days later, cells were treated with cycloheximide for indicated time and harvested. Cell lysates were subjected to immunoblot analysis (IB) with indicated antibodies.
(E) HCT116 cells were transiently transfected with an expression vector for myc-tagged cullin family members and harvested 2 days later. Cell lysates were subjected to immunoblot analysis (IB) with indicated antibodies.
(F) 293T cells were transiently transfected with an expression vector for myc-tagged CUL4B and treated with MG132 two days later for 6 hours before harvested. Cell lysates were subjected to immunoblot analysis (IB) with indicated antibodies.
See also Figure S1.
DDB1 functions as a linker for both CUL4A and CUL4B ubiquitin ligase complexes. To determine which CUL4 was responsible for the destabilization of WDR5, we treated cells with siRNA targeting CUL4A, CUL4B or combination of both CUL4A and CUL4B, and determined the level of WDR5. We found that knocking down CUL4B, but not CUL4A, increased WDR5 as much as knocking down DDB1 in both HCT116 (Figure 1C) and HeLa cells (Figure S1B), but had little effect on WDR5 mRNA level (Figure 1B). In contrast, RBBP5, another DWD protein and also a core subunit of H3K4 methyltransferase complexes, was not stabilized in DDB1 or CUL4B knockdown cells. Knocking down DDB1 had little effect on the steady state levels of three other DWD proteins (EED, BRWD3 and WDR61) and three XLMR proteins (FMR1, ATRX, BRWD3, Figure S1D), indicating a specificity of DDB1-CUL4B-targeted WDR5 degradation. Cycloheximide chase experiment demonstrates that WDR5 is a unstable protein, with an half-life of about 8 hours, but was stabilized by the CUL4B depletion (Figure 1D). To further confirm the specificity of CUL4B-DDB1 mediated WDR5 degradation, we transiently overexpressed myc-tagged cullin family proteins (CUL1, 2, 3, 4A, 4B, 5, and 7) in HCT116 or 293T cells. The ectopic expression of CUL4B, but not the other 6 cullins, reduced WDR5 in HCT116 cells to a barely-detectable low level (Figure 1E), which was inhibited by proteasome inhibitor treatment (Figure 1F), indicating a CUL4B- and DDB1-promoted, proteosome-dependent degradation of WDR5. In 293T cells, however, not only CUL4B but also CUL1 overexpression decreased WDR5 (Figure S1C), suggesting the possibility that in different cell types, WDR5 may be regulated by multiple E3 ligases. We did not investigate further CUL1-mediated WDR5 degradation and instead focused this study on elucidating the mechanism and significance in XLMR development of CUL4B-mediated WDR5 degradation.
WDR5 is ubiquitylated by CRL4B E3 ubiquitin ligase in vivo and in vitro
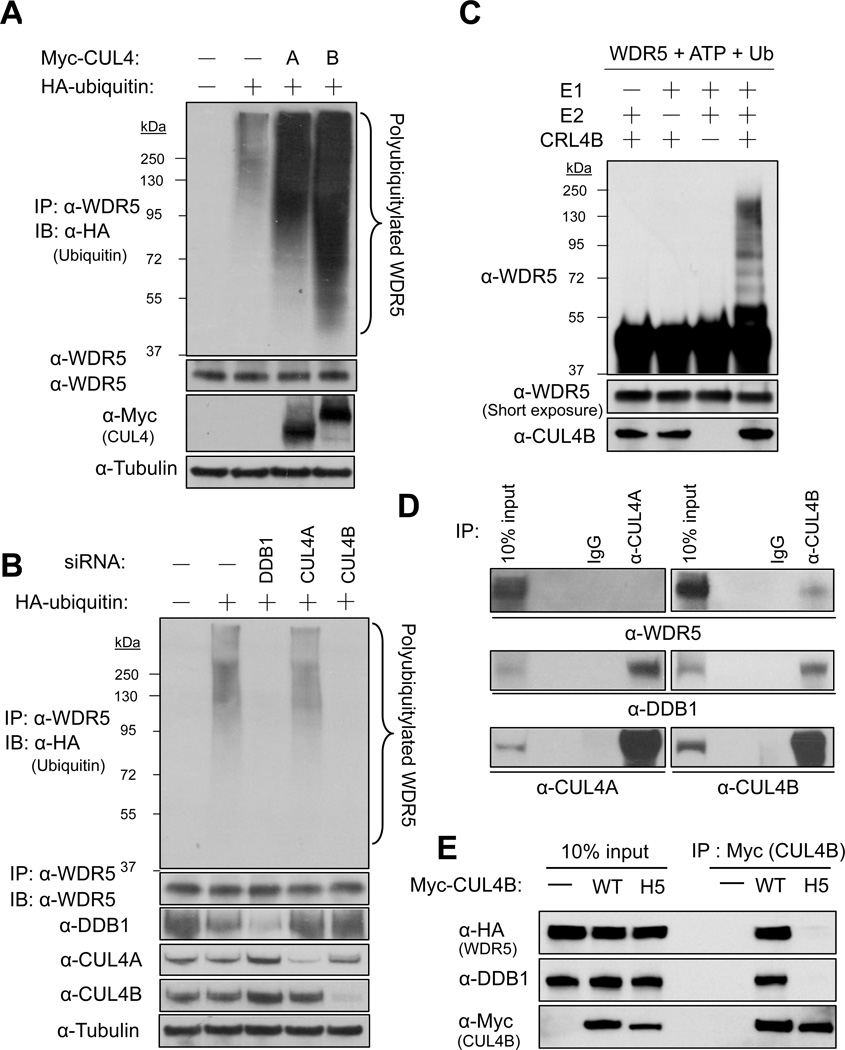
To further determine the hypothesis that WDR5 is a substrate of CRL4B, we performed both in vivo and in vitro ubiquitination assays. HCT116 cells were transiently transfected with either myc-tagged CUL4A or CUL4B alone with hemagglutinin (HA)-tagged ubiquitin and WDR5 was immunoprecipitated under a denaturing condition, followed by direct immunoblotting analyses. This experiment revealed that endogenous WDR5 is ubiquitinated in vivo, which is enhanced by either CUL4A or CUL4B overexpression with the latter being more potent (Figure 2A). Conversely, to demonstrate WDR5 ubiquitylation by CUL4B-DDB1 in a more physiological settings, we transfected HCT116 cells with siRNA targeting DDB1, CUL4A or CUL4B, followed by transient transfection of HA-ubiquitin and in vivo ubiquitylation assay. Although CUL4A knockdown did not affect the ubiquitylation of endogenous WDR5, DDB1 and CUL4B knockdown, both efficiently inhibited the ubiquitylation of endogenous WDR5 (Figure 2B). To directly test whether WDR5 is a substrate of CRL4B, we set up the in vitro ubiquitination assay and found that WDR5 is ubiquitinated by CRL4B in a E1-, E2- and E3(CRL4B)-dependent manner (Figure 2C). Next, we determined the binding of endogenous WDR5 with CUL4 proteins and detected a clear association of WDR5 with CUL4B, but not CUL4A in HCT116 cells after treatment with a proteasome inhibitor (Figure 2D). To determine whether the CUL4B-WDR5 association is bridged by DDB1, we constructed a CUL4B mutant which harbors four amino acids substitution in the H5 helix region (293WQNH296 to 293AAAA296, Figure S5A), that are invariably conserved in orthologues, but are different in paralogues and required for interact with linker proteins (Zheng et al., 2002). Like the mutation in the H5 helix of CUL4A which disrupted its binding with DDB1 (Hu et al., 2004), the mutation in the H5 helix of CUL4B also completely disrupted its binding to not only DDB1 but also WDR5 (Figure 2E). Taken together, these results demonstrate that DDB1 recruits WDR5 to the CUL4B-ROC1 E3 ligase for ubiquitylation.
Figure 2. WDR5 is ubiquitinated by CRL4B in vivo and in vitro.
(A) HCT116 cells transfected with indicated plasmids were lysed and subjected to immunoprecipitation (IP) with antibody to WDR5 under denaturing condition. The resulting precipitates were subjected to western analysis (IB) with antibodies to HA or to WDR5. A portion of the input lysates was also subjected directly to immunoblot analysis with antibodies to myc or tubulin.
(B) HCT116 cells transfected with plasmids for HA-ubiquitin along with siRNA targeting DDB1, CUL4A or CUL4B were lysed and subjected to immunoprecipitation (IP) and immunoblot analysis (IB) as in (A).
(C) Recombinant WDR5 was incubated with different combinations of E1, E2 and CRL4B (E3) along with ubiquitin and ATP. Reactants were subjected to immunoblot analysis (IB) with indicated antibodies.
(D) Lysates of HEK293T cells treated with MG132 for 6 hours were subjected to immunoprecipitation with indicated antibodies and the resulting precipitates, as well as a portion (10% of the input for immunoprecipitation) of the cell lysates, were subjected to immunoblot analysis (IB) with indicated antibodies.
(E) HEK293T cells were transiently transfected with indicated vectors, treated with MG132 for 6 hours and harvested. Protein bindings were determined by IP-western analysis using indicated antibodies.
Nuclear localization is critical for CRL4B to degrade WDR5
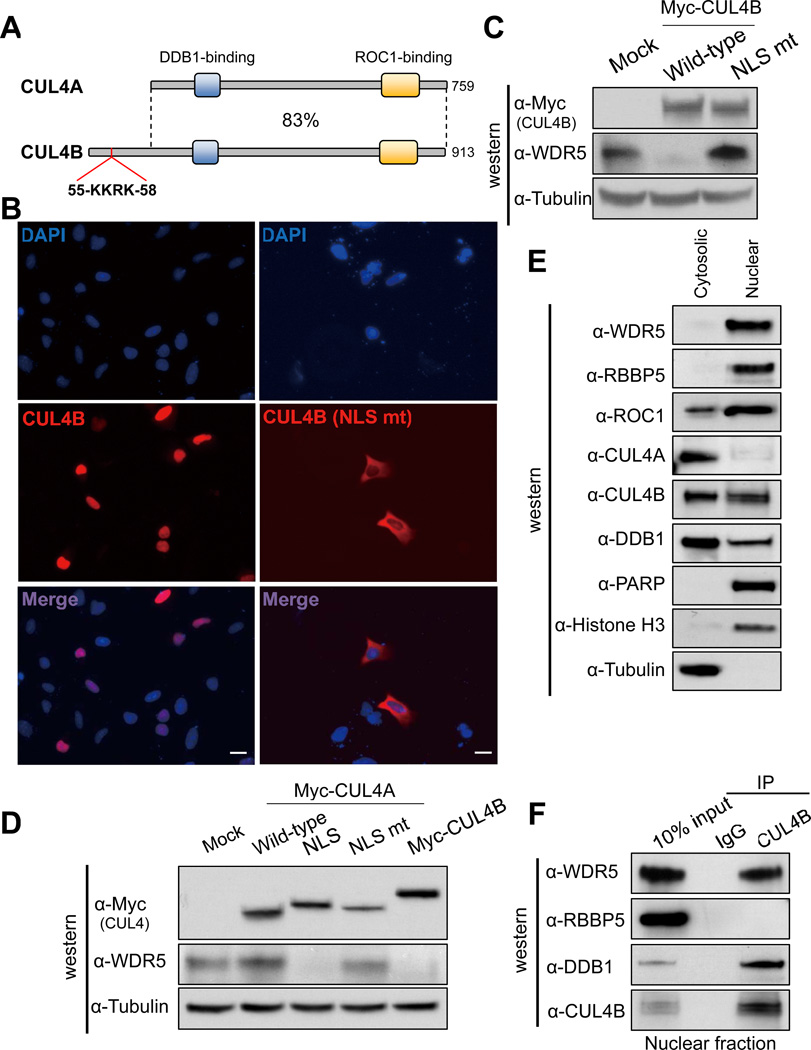
Human CUL4A and CUL4B share 80% amino acid identity throughout the entire sequences and are often functionally redundant. It is therefore intriguing to see that WDR5 is only ubiquitylated by CUL4B, but not CUL4A. To determine the structural basis of this difference, we noted that CUL4B contains a unique N-terminal sequence that contains a putative nuclear localization signal (NLS, 55KKRK58, Figure S5A), and is not present in CUL4A (Figure 3A), which is also reported by another group recently (Zou et al., 2009). Given that WDR5 is also localized in the nucleus [(Wysocka et al., 2005), and our confirmatory result in Figure 3E], we tested the possibility that CUL4B specifically targets WDR5 degradation in the nucleus. We constructed NLS mutant of CUL4B (K56A/R57A) and found that while wild type CUL4B is localized predominantly in the nucleus, the CUL4BK56A/R57A is localized mainly in the cytoplasm (Figure 3B). Importantly, overexpression of NLS mutant CUL4BK56A/R57A failed to degrade WDR5 and instead increased WDR5 level (Figure 3C), indicative of possible dominant negative effect over the endogenous wild type CUL4B. Conversely, to test whether the lack of NLS in CUL4A is responsible for its inability to degrade WDR5, we transplanted N-terminal 60 amino acids from CUL4B containing the NLS to the N-terminus of CUL4A. We found that fusion with this 60-mer sequence (Myc-CUL4A-NLS), but not the same 60-mer containing K56A/R57A substitution in the NLS (NLS mut), conferred CUL4A the ability to promote WDR5 degradation (Figure 3D). Finally, we separated PC12 lysate into nuclear and cytosolic fractions, and found that while CUL4B is localized in both the nucleus and the cytoplasm, CUL4A is localized predominantly in the cytoplasm (Figure 3E). A stronger CUL4B-WDR5 association was seen in the nuclear fraction (Figure 3F). In contrast, RBBP5, although also enriched in the nucleus, did not bind to CUL4B. Together, these results demonstrate that CUL4B, but not CUL4A, is localized in the nucleus, binds to and targets nuclear localized WDR5 for degradation.
Figure 3. Nuclear localization of CUL4B is critical for degrading WDR5.
(A) Schematic illustration comparing human CUL4A and CUL4B proteins. DDB1 and ROC1 binding regions conserved in both proteins and an NLS (55KKRK58) unique to CUL4B are indicated.
(B) U2OS cells were transiently transfected with an expression vector for myc-tagged wild type CUL4B or NLS mutant CUL4B, and the cells were fixed and immunostained with anti-myc antibody. Nucleus was visualized by DAPI. Scale bar, 10 µm.
(C) HEK293T cells transiently transfected with plasmid expressing myc-tagged wild type CUL4B or NLS mutant CUL4B were lysed and subjected to immunoblot analysis (IB).with indicated antibodies.
(D) HEK293T cells were transiently transfected with plasmid expressing indicated proteins. The steady state levels of individual ectopically expressed protein and endogenous WDR5 were determined by immunoblot analysis (IB).
(E) Cytosolic and nuclear fractions of PC12 cells were subjected to immunoblot analysis (IB) with indicated antibodies.
(F) Nuclear extract of PC12 cells were subjected to IP-western analysis for the association of CUL4B with WDR5.
See also Figure S2.
CUL4B regulates neuronal gene expression through H3K4 modification in a WDR5 dependent manner
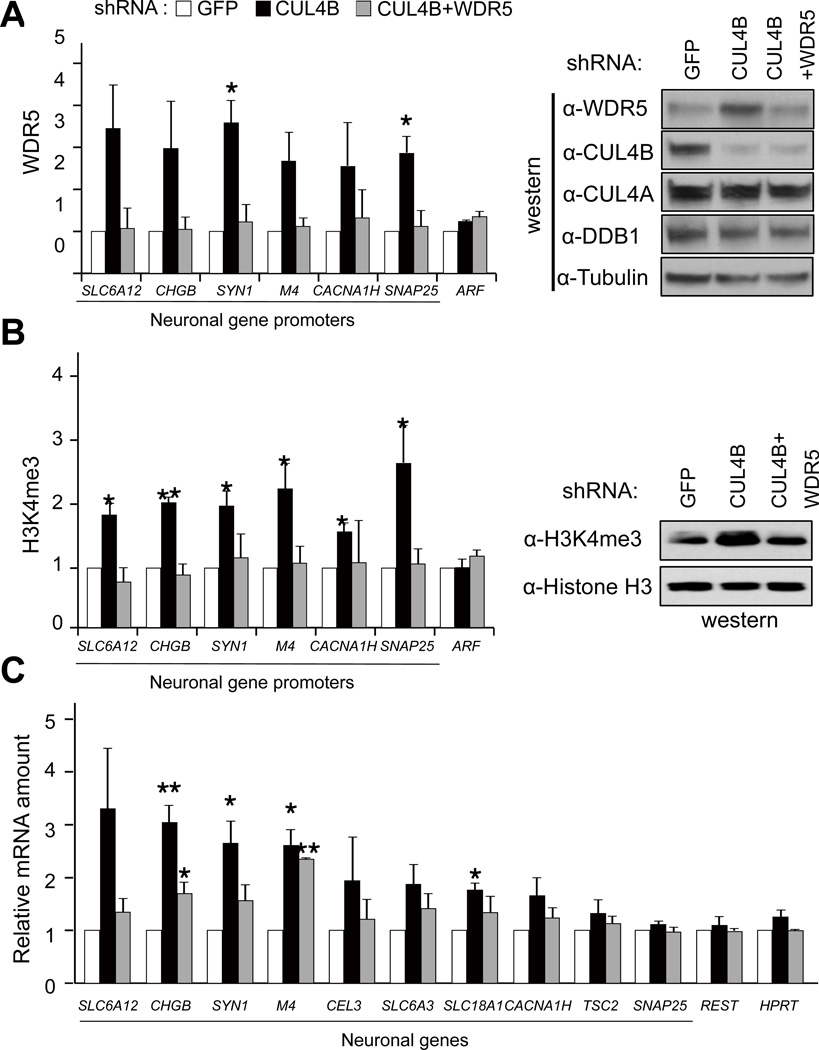
In searching for the mechanism underlying the contribution of CUL4B mutations to the development of XLMR, we noted that another XLMR gene product, KDM5C/SMCX/JARID1C, is a H3K4 demethylase and its demethylase activity is abolished or reduced by XLMR mutations, leading to the abnormal activation of a group of neuronal genes (Iwase et al., 2007; Tahiliani et al., 2007). Given the critical function of WDR5 for the H3K4 methylation by MLL1 and other related methyltransferases, we hypothesized that CUL4B mutation may contribute to XLMR development similarly because of abnormal accumulation of WDR5 and activation of neuronal genes. To test this hypothesis, we transduced HeLa cells with retrovirus expressing encoding control (GFP) or CUL4B shRNA, and performed chromatin immunoprecipitation (ChIP) analyses with WDR5, RBBP5 or H3K4me3 antibodies. Stable knockdown of CUL4B increased the steady state level of WDR5 protein, the binding of both WDR5 and RBBP5 and H3K4me3 on several neuronal gene promoters (Figures 4A, 4B and S4A), but not the binding to a control gene, ARF tumor suppressor which is known to be activated by H3K4 methylation in a WDR5-dependent manner following stimulation by oncogene E7 (Kotake et al., 2009). Next, we examined whether expression of these neuronal genes are induced by CUL4B knockdown. To this end, we isolated RNA from control (GFP) or CUL4B knockdown cells and performed quantitative reverse-transcribed PCR (qRT-PCR) analyses. Consistent with the ChIP assay, mRNA levels of these neuronal genes were increased in CUL4B knockdown cells (Figure 4C). Importantly, these phenotypes are suppressed by simultaneous knocking-down of WDR5 (Figures 4A, 4B and 4C). Furthermore, global level of H3K4me3 was also increased after CUL4B knocking down in WDR5 dependent manner (Figure 4B). Taken together, these results suggest that CUL4B negatively regulates H3K4me3 level by degrading WDR5 and controls the expression of neuronal genes.
Figure 4. Knocking-down CUL4B abnormally activate neuronal gene expression in a WDR5 dependent manner.
(A) HeLa cells were transduced with retrovirus encoding shRNA targeting GFP (control), CUL4B or WDR5, and selected by puromycin treatment. DNA/protein complexes were crosslinked, and subjected to ChIP analysis with a WDR5 antibody. Immunoprecipitates were analyzed by quantitative PCR (qPCR) using primers for indicated neuronal gene promoters (left panel). Global level of WDR5 and several other proteins were determined by immunoblotting (IB) using indicated antibodies (right panel).
(B) ChIP and immunoblotting (IB) was performed with anti-H3K4me3 antibody as in (A). Chromatin fractions were used for immunoblotting to determine nucleosomal H3K4me3 level.
(C) RNA from the same cells as in (A) and (B) was isolated, reverse-transcribed and quantitative PCR (RT-qPCR) were performed using primers for indicated neuronal genes.
Error bars represent S.E. from three independent experiments. * indicates p<0.05 and ** indicate p<0.01 by t test.
See also Figure S3.
Cul4b regulates NGF-induced neurite extension of PC12 cells in a Wdr5 dependent manner
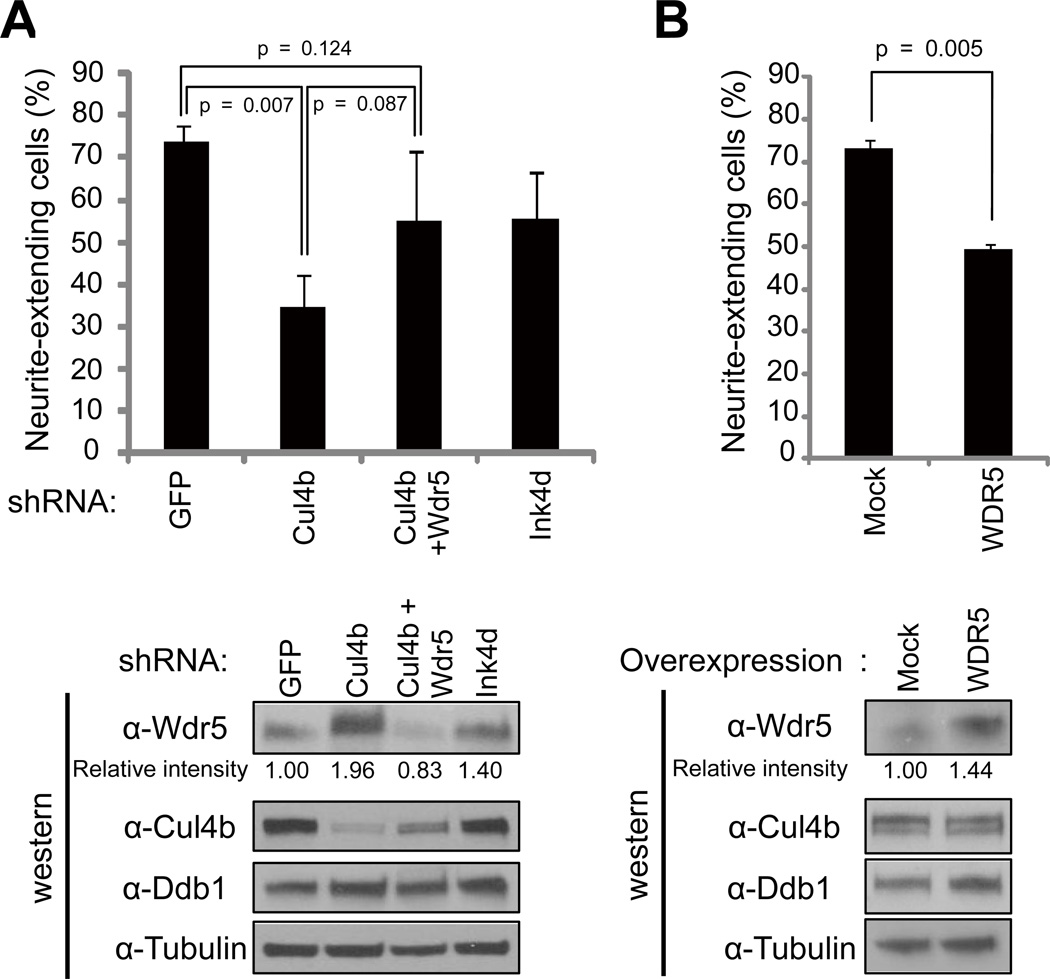
Exact cellular phenotypes caused by XLMR mutations are yet to be defined and are likely multifaceted. However, a previous study showed that reduced expression of XLMR gene JARID1C in rat primary cortical neurons suppresses dendritic growth (Iwase et al., 2007). Furthermore, both IL1RAPL1 and ATP6AP2, two XLMR genes, are reported to regulate NGF-induced neurite extension in rat neuroendocine cell line PC12 cells (Contrepas et al., 2009; Gambino et al., 2007). To investigate whether CUL4B has a function in neuronal cell differentiation and if this function is dependent on WDR5, we made Cul4b stably knocked down PC12 cells by transduction of retrovirus encoding rat Cul4b shRNA. Knockdown Cul4b increased both Wdr5 mRNA (Figure S4C) and protein (Figure 5A, lower panel), confirming the down-regulation of Wdr5 by Cul4b in rat neuroendocrine cells and also suggesting the possibility that Wdr5 activates its own transcription. We treated PC12 cells with nerve growth factor (NGF) and observed time-dependent neurite extension. We found that NGF-induced neurite extension was suppressed by Cul4b knockdown (Figure 5A, Figures S4A and S4B). To determine whether Wdr5 accumulation is responsible for this effect, we made Cul4b and Wdr5 double knockdown cells and found that suppression of neurite extension could be partially rescued by co-knockdown of Wdr5, restoring the neurite extension to a level that is not significantly different from GFP control population (p=0.124) (Figure 5A, Figure S4B). The reason that co-depletion of both Cul4b and Wdr5 resulted in only partial instead of complete rescue is likely caused by too much decrease of WDR5. Simultaneous knocking down both Cul4b and WDR5, instead of restoring WDR5 protein to its normal level, decreased it to a level lower (by 17%) than that normally expressed in cells (lower panel, Figure 5A), suggesting that WDR5 protein must be tightly regulated and neither too high nor too low level of WDR5 is appropriate for neural differentiation. In addition, overexpression of human WDR5, which is identical to rat Wdr5, suppressed NGF-induced neurite extension (Figure 5B). These results demonstrate that Cul4b promotes neurite extension by down-regulating Wdr5 protein.
Figure 5. Knockdown of Cul4b suppresses NGF-induced neurite elongation of PC12 cells in a Wdr5 dependent manner.
(A) PC12 cells were infected with retrovirus encoding shRNA for GFP, Cul4b, Wdr5 or p19Ink4d and infected cells were selected by puromycin treatment. Neurite-extending cells were counted 4 days after NGF treatment (upper panel) or cells were lysed and subjected to immunoblot analysis (IB) with indicated antibodies (lower panel).
(B) PC12 cells were infected with retrovirus encoding Flag-WDR5-HA, and infected cells were selected by puromycin treatment. Neurite extension and immunoblot analysis were performed as in (A).
Error bars represent S.E. from three independent experiments. Band intensity was measured by NIH Image J (version 1.44k). p values were measured by t test.
See also Figure S4.
XLMR-derived mutations impair the ability of CUL4B to degrade WDR5 and support neurite extension of PC12 cells
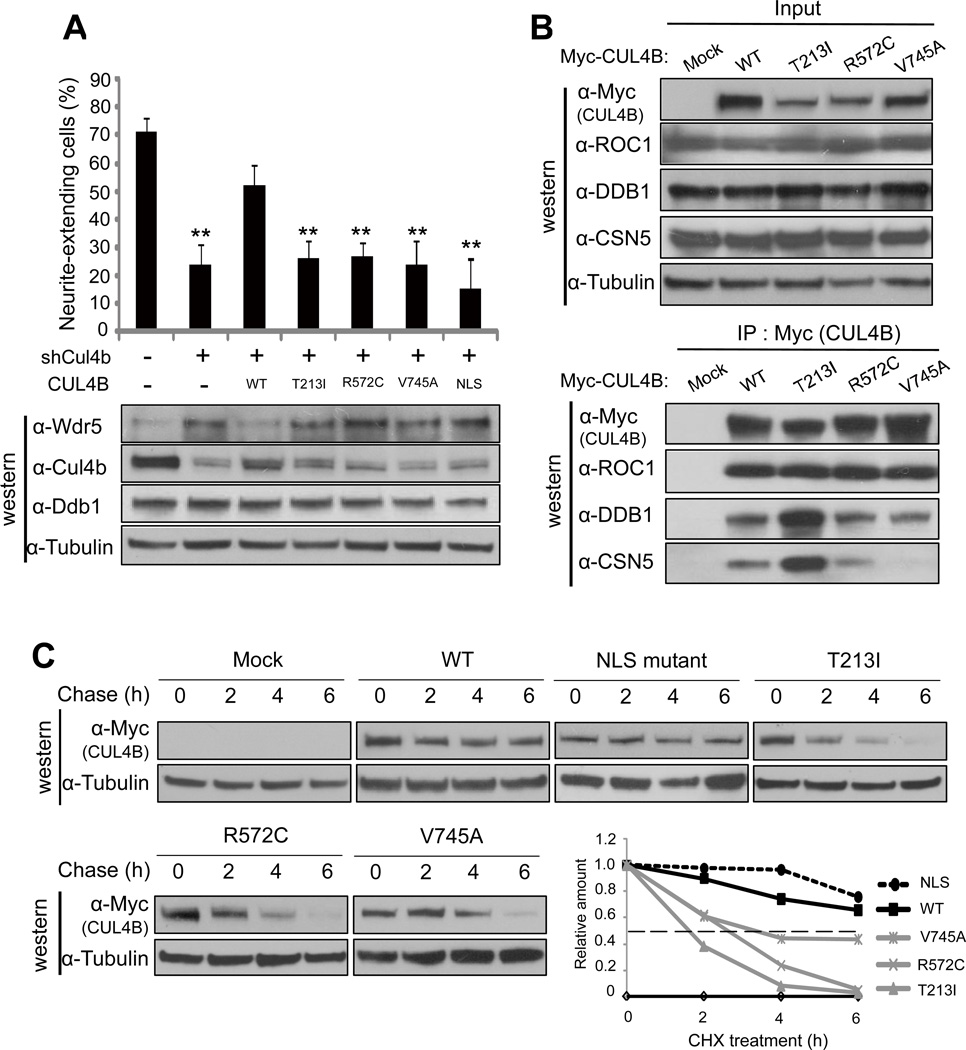
Nine different mutations in CUL4B were identified from ten XLMR families and six caused truncation of CUL4B, indicating a loss-of-function alteration (Badura-Stronka et al., 2010; Tarpey et al., 2007; Zou et al., 2007). The three remaining are missense mutations, T213A, R572C and V745A, whose effect on CUL4B are not clear. To determine this, we created these three XLMR-derived CUL4B point mutants and first examined their function in supporting NGF-induced neurite outgrowth of PC12 cells. Rat Cul4b protein is 96% identical to human CUL4B, including the NLS and three missense XLMR mutations (Figure S5A). We transduced PC12 cells with retrovurus expressing shRNA targeting rat Cul4b alone or with retrovirus expressing wild type, XLMR-derived, or NLS mutant human CUL4B that are resistant to the Cul4b shRNA. Treatment of these cells with NGF demonstrate that neurite extension was suppressed by Cul4b knockdown and this suppression was partially rescued by the ectopic expression of wild type CUL4B, but not three XLMR-derived CUL4B mutants or the NLS mutant (Figure 6A, Figures S6B, S6C and S6D). Consistent with this functional assay, immunoblot analyses revealed that Cul4b knockdown increased Wdr5 protein and this increase is almost completely blocked by the ectopic expression of wild type human CUL4B, but not three XLMR-derived CUL4B mutants nor the NLS mutant. These results demonstrate that XLMR-derived mutations significantly impaired the ability of CUL4B in promoting WDR5 degradation and in supporting neurite extension of PC12 cells. Furthermore, these results also demonstrate that nuclear localization of CUL4B is critical for the function of CUL4B in supporting PC12 cell differentiation.
Figure 6. XLMR patients-derived CUL4B mutants cannot rescue Cul4b knockdown phenotype.
(A) PC12 cells were transduced with retrovirus encoding wild type or mutants CUL4B along with retrovirus encoding shRNA targeting Cul4b, and transduced cells were selected by puromycin treatment. Neurite-extending cells were counted 4 days after NGF treatment (upper panel) or protein expression was determined by immunoblot analysis (lower panel). Error bars represent S.E. from three independent experiments. ** indicate p<0.01 by t test.
(B) HEK293T cells were transiently transfected with an expression vector for myc-tagged wild type or mutant CUL4B. The expression of ROC1, DDB1 and CSN5 (upper panel) and their binding with wild type or mutant CUL4B (lower panel) were determined by direct immunoblotting or IP-western analysis using indicated antibodies.
(C) HEK293T cells were transiently transfected with an expression vector for myc-tagged wild type or mutant CUL4B and treated with cycloheximide for indicated time. Cell lysates were subjected to immunoblot analysis (IB) with indicated antibodies. Band intensity was measured by NIH Image J (version 1.44k).
See also Figure S5.
XLMR-derived mutations disrupt CSN binding and destabilize CUL4B protein
To determine the mechanism by which XLMR-derived mutations impair the function of CUL4B, we examined the binding of CUL4B with three essential subunits of CRL4 E3 complexes, ROC1, DDB1 and CSN5/JAB1. No significant effect was seen on the DDB1 or ROC1 binding by any of three missense XLMR mutations, but both R572C and V745A mutations clearly reduced the binding of CUL4B with CSN5, the catalytic subunit of COP9/signalosome (CSN) complex that plays an essential function in the activation of CRL E3 complexes by promoting the cleavage of covalent linkage of NEDD8 from cullins (Wei et al., 2008). R572 resides within the central region of CUL4B whose function, as is the case for other cullins, has not been clearly defined and our finding thus suggests that this region is involved in the binding with CSN complex. The V745 locates within the ROC1-binding site. Interestingly, while V745A mutation had little effect on ROC1 binding, it severely impaired the CSN5 binding. The exact mechanism underlying the disruption of CSN5-CUL4B by the V745A mutation has not been determined, but deletion of a 6-residue sequence in CUL1 that includes the equivalent Val residue has previously been noted to disrupt Nedd8 modification and CUL1 function (Furukawa et al., 2000).
No obvious effect on DDB1, ROC1 or CSN5 binding was detected by T213I mutation. Unlike R572 and V745, both of which are conserved in other cullins, T213 is only conserved in between two CUL4, and no known function or subunit binding is known to locate in this region. In searching for the mechanism underlying the impairment of CUL4B function by T213I mutation, we noted that in repeated ectopic expression experiments, CUL4BT213I mutant was consistently expressed at a lower level than the wild type CUL4B. We therefore determined the mRNA level and protein stability of CUL4BT213I mutant. qRT-PCR analyses demonstrated that the mRNA of CUL4BT213I and the other two XLMR mutants were expressed at similar levels as both the wild type and NLS mutant of CUL4B (Figure S5D), excluding the possibility of reduced mRNA stability by the XLMR mutations. We then determined the half-life of different CUL4B mutants and found that the turnover of three XLMR mutants of CUL4B was much faster than either wild type or NLS mutant CUL4B (Figure 6B). The half-life of CUL4B was reduced from more than 6 h of the experimental duration for wild type protein to 3.7 h by V745A mutation, 2.7 h by R572C mutation and 1.8 h by T213I mutation. Treatment of cells with proteasome inhibitor resulted in the accumulation of three XLMR mutants to a similar level as the wild type CUL4B (Figure S5E), providing additional support for the destabilizing effects conferred by T213I and the other two XLMR mutations. Together, these findings provide molecular explanations—reduced CSN binding and activation and protein stability—for impaired function of CUL4B.
DISCUSSION
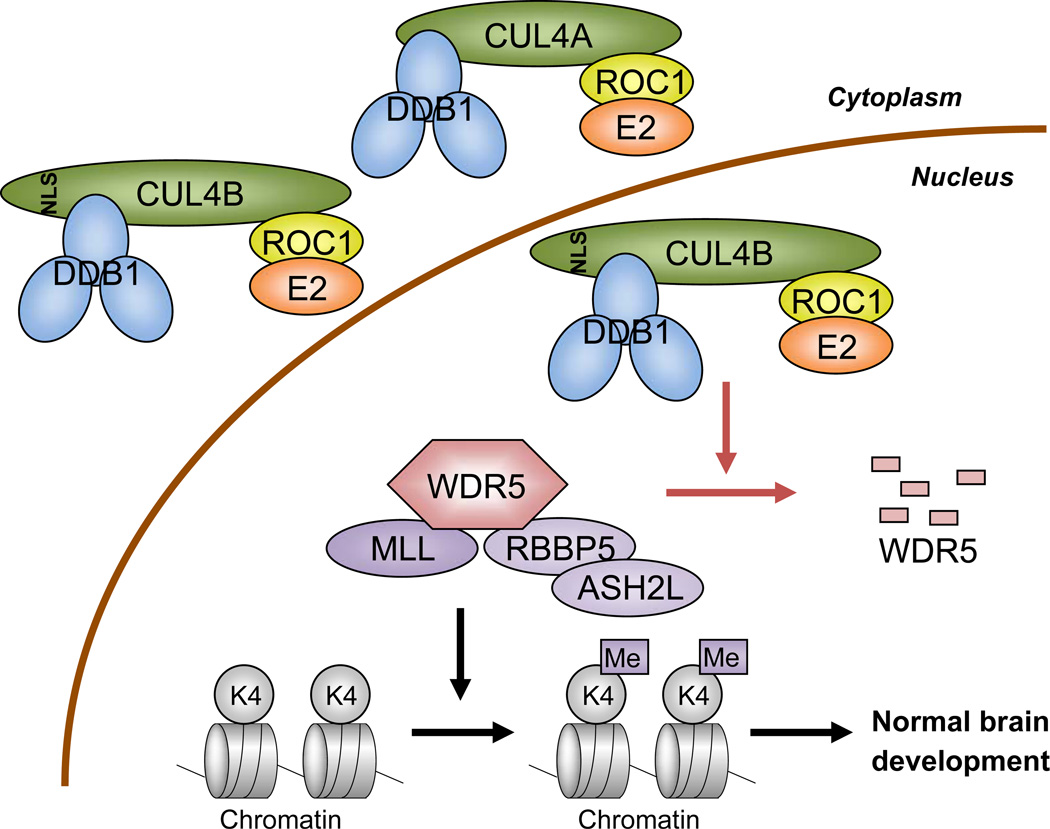
CUL4 is one of three founding cullins conserved from yeast to humans. Genetic analyses in various organisms have previously revealed a wide range of cellular and organismal function of CUL4, with many of them associated with chromatin such as chromosomal condensation, heterochromatin formation, DNA replication and repair. At least seven CRL4 substrates have thus far been identified in mammalian cells that are linked to DNA metabolism, including DNA repair proteins DDB2 and XPC, histones H2A, H3 and H4, DNA replication licensing factor CDT1 [reviewed in (Hannah and Zhou, 2009; Jackson and Xiong, 2009)] and very recently histone H4 monomethylase PR-Set7 (Abbas et al., 2010; Oda et al., 2010). Our finding that WDR5 is a substrate of CRL4B E3 ligase and that CRL4B-mediated WDR5 degradation plays a critical function in the regulation of neural gene expression (summarized in Figure 7) provides the first example where a CRL4 E3 ligase controls cell differentiation through targeting the ubiquitylation of a histone modifying enzyme to regulate gene expression, providing new insight into the function of CUL4 in chromatin regulation. In addition to serving as an essential factor for H3K4 methyltransferases of MLL1 family, WDR5 is present in many other chromatin modifying or transcriptional complexes, including NIF-1 histone methyltransferase complex (Garapaty et al., 2009), ATAC histone acetyltransferase complex (Wang et al., 2008), RING1b histone ubiquitin ligase complex (Sanchez et al., 2007), CHD8 chromatin remodeling complex (Dou et al., 2005; Thompson et al., 2008), H1.2 transcriptional repressive complex (Kim et al., 2008), and clock proteins PERIOD (PER)-mediated transcriptional repression (Brown et al., 2005). As a result of such a broad function of WDR5, CRL4B may play even more extensive roles in chromatin control and gene regulation. In fact, we observed that the levels of acetyl-H4 (H4Ac) on those neuronal genes regulated by WDR5 was increased after knocking down CUL4B and restored after co-depletion of both CUL4B and WDR5 (Figure S3B).
Figure 7. Schematic representation of our finding.
CRL4B E3 ligase targets WDR5 ubiquitylation in the nucleus and regulates neuronal gene expression. See Discussion for details.
The physiological condition(s) signaling WDR5 ubiquitylation by DDDB1-CUl4B-ROC1 remains to be elucidated. In NGF-treated PC12 cells, no significant change of the steady state level of CRL4B components (Cul4b, Ddb1 and Roc1) nor WDR5 was observed, indicating a constitutive, rather than an NGF-regulated degradation in this setting (Figure S4D). We speculate that DDB1-CUL4B may function to regulate the homeostasis of WDR5 complexes. By maintaining a dynamic regulation of WDR5, cells can ensure continued responsiveness of different WDR5 complexes to different growth conditions and signaling pathways.
At the molecular level, CRL4B-meidated WDR5 ubiquitylation has two unique features—ubiquitylating a DWD protein and functioning in a specific subcellular compartment. DDB1-binding WD40 (DWD, or DCAF or CDW) proteins are believed to recognize and recruit other substrates to the CUL4-ROC1 catalytic core for ubiquitylation. Our finding indicates that DWD proteins themselves can be a direct substrate of CRL4 E3 ligase. Similar observations have previously been made for F-box protein Cdc4 and BTB protein RhoBTB2 which are ubiquitylated and degraded by their associated CRL1 and CRL3 E3 ligases, respectively (Wilkins et al., 2004; Zhou and Howley, 1998). How a CRL E3 ligase distinguishes a protein being as a substrate recognition subunit or a substrate is an interesting and important issue that is yet to be determined.
CUL4B-targeted WDR5 ubiquitylation occurs specifically in the nucleus. There have been relatively few examples of localized ubiquitylation reported thus far. Two notable examples are p53 and β-catenin, transcription factors that can be exported out of the nucleus for degradation in the cytoplasm. This nuclear export and cytoplasmic ubiquitylation ensures that the function of the transcription factor is geographically separated from its degradation to avoid promiscuous degradation and to allow rapid activation of the factor in response to signaling events by simply blocking export and ubiquitylation (Xiong, 2010). Our finding provides another example of targeted ubiquitylation in a specific subcellular compartment, but the significance of having the function and degradation of WDR5 in the same compartment is unclear at present.
Mutations of as many as 90 genes have been linked to XLMR, a pathologically heterogeneous disease that has thus far not been causally linked to one single clear or major molecular or cellular defect. The finding reported here provides a mechanism—CRL4B-mediated ubiquitylation and degradation of WDR5—for the cause of CUL4B mutations to the development of XLMR. Supporting its function in normal brain development and suppression of XLMR, Cul4b is highly expressed in the cerebrum and hippocampus of mouse brain, both affected in mental retardation patients (Lein et al., 2007). It should be pointed out that it is not clear whether the development of XLMR in CUL4B mutated cells was mainly by the defects in WDR5 ubiquitylation or also involved additional substrates. It is worthwhile to note that there are two other XLMR-mutated genes, KDM5C/SMCX/JARID1C which encodes a H3K4 demethylase and PHF8 which binds to tri-methylated H3K4 to catalyze the demethylation of H3K9 (Iwase et al., 2007; Kleine-Kohlbrecher et al., 2010; Qi et al., 2010; Qiu et al., 2010; Tahiliani et al., 2007). We demonstrate here that the degradation of a core subunit of H3K4 methyltransferase, WDR5, is critically important for the function of XLMR gene product CUL4B in the regulation of neural gene expression and PC12 cell differentiation, as seen by the suppression of these defects in CUL4B knocking down cells by the co-depletion of WDR5. This finding points to a common molecular defect—abnormal activation of neural gene expression resulting from altered histone modification—underlying these three otherwise biochemically unrelated XLMR genes, adding further to the notion that mutations targeting a major part of XLMR genes deregulate epigentic control. Recent genome-wide association study suggested that WDR5 is also one of the candidate genes affecting general cognitive ability (Davis et al., 2010), suggesting that CRL4B-mediated WDR5 ubiquitylation may be more broadly involved in learning and memory.
EXPERIMENTAL PROCEDURES
Transfection and retroviral infection
Fugene 6 transfection reagent (Roche) or Lipofectamine 2000 reagent (Invitrogen) was used for transient overexpression or transient knocking down by siRNA, respectively. The retroviral production and transduction were performed as previously described (Kotake et al., 2007). A detailed description of the experimental procedures is available in the Supplemental Experimental Procedures. Target sequences of siRNAs are shown in Table S1.
Plasmid Construction
Expression constructs of human cullins and ubiquitin were previously described (Hu et al., 2008; Michel and Xiong, 1998). Expression construct of human WDR5 was kindly provided by Dr. Yali Dou (University of Michigan, Ann Arbor, MI). shRNA construct for human WDR5 was previously described (Kotake et al., 2009). A detailed description of other expression constructs is available in the Supplemental Experimental Procedures. Target sequences of shRNA constructs for human CUL4B, rat Cul4b, rat Wdr5, rat p19Ink4d are described in Table S1.
Nuclear/cytosolic and chromatin fractionation
A detailed description of the experimental procedures is available in the Supplemental Experimental Procedures.
Immunoprecipitation and Immunoblotting
A detailed description of the experimental procedures is available in the Supplemental Experimental Procedures.
Ubiquitination assays
In vivo and in vitro ubiquitination assays were performed as described previously (Liu et al., 2002, Furukawa et al., 2003, Hu et al., 2004, Kaneko-Oshikawa et al., 2005). For in vitro ubiquitination assay, WDR5 was produced by bacteria and CRL4B complex was purified from HEK293T cells. A detailed description of the experimental procedures is available in the Supplemental Experimental Procedures.
Cycloheximide chase experiment
293T cells were transfected with CUL4B constructs, treated with cycloheximide (100 µg /ml), and then subjected to immunoblot analysis. A detailed description of the experimental procedures is available in the Supplemental Experimental Procedures.
Quantitative reverse-transcription PCR (qRT-PCR)
qRT-PCR was performed as previously described (Kotake et al., 2009). PCR primers used in this study are summarized in Table S3.
Chromatin Immunoprecipitation assay
Chromatin Immunoprecipitation assay was performed as previously described (Kotake et al., 2009). Antibodies and PCR primers used in this study are summarized in Table S2 and Table S4, respectively.
PC12 Neurite Extension Assay
PC12 Neurite Extension assay was performed as previously described with some modifications (Bai et al., 2005; Greene, 1978; Saita et al., 2009). A detailed description of the experimental procedures is available in the Supplemental Experimental Procedures.
Immunofluorescence Staining
U2OS cells were fixed and incubated with lab-made anti-myc antibody, followed by incubated with rhodamine-labeled secondary antibody. After washing, 5 µg/ml 4’,6-diamidino-2-phenylindole (DAPI) was treated and cells were examined with fluorescence microscope. A detailed description of the experimental procedures is available in the Supplemental Experimental Procedures.
Highlights.
CRL4B is a nuclear E3 ligase and targets WDR5 for ubiquitylation and degradation
CUL4B controls WDR5 and H3K4me3 on the neuronal gene promoters
CUL4B is required for NGF-induced neurite extension
XLMR mutations destabilize CUL4B and impair its ability in neurite extension
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Steve Crews, Bob Duronio, Joe Pearson, Ben Philpot and members of Xiong laboratory for discussions, technical assistance and encouragement. This study is supported by an NIH grant (GM067113) to Y. X.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell. 2010;40:9–21. doi: 10.1016/j.molcel.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- Badura-Stronka M, Jamsheer A, Materna-Kiryluk A, Sowinska A, Kiryluk K, Budny B, Latos-Bielenska A. A novel nonsense mutation in CUL4B gene in three brothers with X-linked mental retardation syndrome. Clin Genet. 2010;77:141–144. doi: 10.1111/j.1399-0004.2009.01331.x. [DOI] [PubMed] [Google Scholar]
- Bai S, Ghoshal K, Datta J, Majumder S, Yoon SO, Jacob ST. DNA methyltransferase 3b regulates nerve growth factor-induced differentiation of PC12 cells by recruiting histone deacetylase 2. Mol Cell Biol. 2005;25:751–766. doi: 10.1128/MCB.25.2.751-766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458:430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- Brown SA, Ripperger J, Kadener S, Fleury-Olela F, Vilbois F, Rosbash M, Schibler U. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science. 2005;308:693–696. doi: 10.1126/science.1107373. [DOI] [PubMed] [Google Scholar]
- Chen L-C, Manjeshwar S, Lu Y, Moore D, Ljung B-M, Kuo W-L, Dairkee SH, Wernick M, Collins C, Smith HS. The human homologue for the Caenorhabditis elegans cul-4 gene is amplified and overexpressed in primary breast cancers. Cancer Res. 1998;58:3677–3683. [PubMed] [Google Scholar]
- Chiurazzi P, Schwartz CE, Gecz J, Neri G. XLMR genes: update 2007. Eur J Hum Genet. 2008;16:422–434. doi: 10.1038/sj.ejhg.5201994. [DOI] [PubMed] [Google Scholar]
- Contrepas A, Walker J, Koulakoff A, Franek KJ, Qadri F, Giaume C, Corvol P, Schwartz CE, Nguyen G. A role of the (pro)renin receptor in neuronal cell differentiation. Am J Physiol Regul Integr Comp Physiol. 2009;297:R250–R257. doi: 10.1152/ajpregu.90832.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox BJ, Vollmer M, Tamplin O, Lu M, Biechele S, Floss T, Gertsenstein M, van Campenhout C, Kuhn R, Wurst W, et al. Phenotypic annotation of the mouse X chromosome. Genome Res. 2010;20:1154–1184. doi: 10.1101/gr.105106.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis OS, Butcher LM, Docherty SJ, Meaburn EL, Curtis CJ, Simpson MA, Schalkwyk LC, Plomin R. A three-stage genome-wide association study of general cognitive ability: hunting the small effects. Behav Genet. 2010;40:759–767. doi: 10.1007/s10519-010-9350-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa M, Yanping Z, McCarville J, Ohta T, Xiong Y. The C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol. Cell. Biol. 2000;20:8185–8197. doi: 10.1128/mcb.20.21.8185-8197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambino F, Pavlowsky A, Begle A, Dupont JL, Bahi N, Courjaret R, Gardette R, Hadjkacem H, Skala H, Poulain B, et al. IL1-receptor accessory protein-like 1 (IL1RAPL1), a protein involved in cognitive functions, regulates N-type Ca2+-channel and neurite elongation. Proc Natl Acad Sci U S A. 2007;104:9063–9068. doi: 10.1073/pnas.0701133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garapaty S, Xu CF, Trojer P, Mahajan MA, Neubert TA, Samuels HH. Identification and characterization of a novel nuclear protein complex involved in nuclear hormone receptor-mediated gene regulation. J Biol Chem. 2009;284:7542–7552. doi: 10.1074/jbc.M805872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene LA. Nerve growth factor prevents the death and stimulates the neuronal differentiation of clonal PC12 pheochromocytoma cells in serum-free medium. J Cell Biol. 1978;78:747–755. doi: 10.1083/jcb.78.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah J, Zhou P. Regulation of DNA damage response pathways by the cullin-RING ubiquitin ligases. DNA Repair (Amst) 2009;8:536–543. doi: 10.1016/j.dnarep.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YJ, McCall CM, Hu J, Zeng Y, Xiong Y. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes & Dev. 2006;20:2949–2954. doi: 10.1101/gad.1483206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat. Cell Biol. 2006;8:1277–1283. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol. 2004;6:1003–1009. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- Hu J, Zacharek S, He YJ, Lee H, Shumway S, Duronio RJ, Xiong Y. WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. Genes & Dev. 2008;22:866–871. doi: 10.1101/gad.1624008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung MS, Mao JH, Xu Z, Yang CT, Yu JS, Harvard C, Lin YC, Bravo DT, Jablons DM, You L. Cul4A is an oncogene in malignant pleural mesothelioma. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2009.00971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inlow JK, Restifo LL. Molecular and comparative genetics of mental retardation. Genetics. 2004;166:835–881. doi: 10.1534/genetics.166.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isidor B, Pichon O, Baron S, David A, Le Caignec C. Deletion of the CUL4B gene in a boy with mental retardation, minor facial anomalies, short stature, hypogonadism, and ataxia. Am J Med Genet A. 2010;152A:175–180. doi: 10.1002/ajmg.a.33152. [DOI] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci. 2009;34:562–570. doi: 10.1016/j.tibs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Kaneko-Oshikawa C, Nakagawa T, Yamada M, Yoshikawa H, Matsumoto M, Yada M, Hatakeyama S, Nakayama K, Nakayama KI. Mammalian E4 is required for cardiac development and maintenance of the nervous system. Mol. Cell. Biol. 2005;25:10953–10964. doi: 10.1128/MCB.25.24.10953-10964.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Choi J, Heo K, Kim H, Levens D, Kohno K, Johnson EM, Brock HW, An W. Isolation and characterization of a novel H1.2 complex that acts as a repressor of p53-mediated transcription. J Biol Chem. 2008;283:9113–9126. doi: 10.1074/jbc.M708205200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Kohlbrecher D, Christensen J, Vandamme J, Abarrategui I, Bak M, Tommerup N, Shi X, Gozani O, Rappsilber J, Salcini AE, Helin K. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol Cell. 2010;38:165–178. doi: 10.1016/j.molcel.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Cao R, Viatour P, Sage J, Zhang Y, Xiong Y. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4a tumor suppressor gene. Genes & Dev. 2007;21:49–54. doi: 10.1101/gad.1499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Zeng Y, Xiong Y. DDB1-CUL4 and MLL1 mediate oncogene-induced p16INK4a activation. Cancer Res. 2009;69:1809–1814. doi: 10.1158/0008-5472.CAN-08-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Leonard H, Wen X. The epidemiology of mental retardation: challenges and opportunities in the new millennium. Ment Retard Dev Disabil Res Rev. 2002;8:117–134. doi: 10.1002/mrdd.10031. [DOI] [PubMed] [Google Scholar]
- Liu L, Lee S, Zhang J, Peters SB, Hannah J, Zhang Y, Yin Y, Koff A, Ma L, Zhou P. CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol. Cell. 2009;34:451–460. doi: 10.1016/j.molcel.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel JJ, Xiong Y. Human CUL-1, but not other cullin family members, selectively interacts with SKP1 to form a complex with SKP2 and cyclin A. Cell Growth Differ. 1998;9:435–449. [PubMed] [Google Scholar]
- Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- O'Connell BC, Harper JW. Ubiquitin proteasome system (UPS): what can chromatin do for you? Curr. Opin. Cell Biol. 2007;19:206–214. doi: 10.1016/j.ceb.2007.02.014. [DOI] [PubMed] [Google Scholar]
- Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell. 2010;40:364–376. doi: 10.1016/j.molcel.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms Underlying Ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, et al. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010;466:503–507. doi: 10.1038/nature09261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Shi G, Jia Y, Li J, Wu M, Dong S, Wong J. The X-linked mental retardation gene PHF8 is a histone demethylase involved in neuronal differentiation. Cell Res. 2010;20:908–918. doi: 10.1038/cr.2010.81. [DOI] [PubMed] [Google Scholar]
- Ropers HH. X-linked mental retardation: many genes for a complex disorder. Curr Opin Genet Dev. 2006;16:260–269. doi: 10.1016/j.gde.2006.04.017. [DOI] [PubMed] [Google Scholar]
- Saita S, Shirane M, Natume T, Iemura S, Nakayama KI. Promotion of neurite extension by protrudin requires its interaction with vesicle-associated membrane protein-associated protein. J Biol Chem. 2009;284:13766–13777. doi: 10.1074/jbc.M807938200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C, Sanchez I, Demmers JA, Rodriguez P, Strouboulis J, Vidal M. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, Shi Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- Tai HC, Schuman EM. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat Rev Neurosci. 2008;9:826–838. doi: 10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]
- Tarpey PS, Raymond FL, O'Meara S, Edkins S, Teague J, Butler A, Dicks E, Stevens C, Tofts C, Avis T, et al. Mutations in CUL4B, which encodes a ubiquitin E3 ligase subunit, cause an X-linked mental retardation syndrome associated with aggressive outbursts, seizures, relative macrocephaly, central obesity, hypogonadism, pes cavus, and tremor. Am. J. Hum. Genet. 2007;80:345–352. doi: 10.1086/511134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BA, Tremblay V, Lin G, Bochar DA. CHD8 is an ATP-dependent chromatin remodeling factor that regulates beta-catenin target genes. Mol Cell Biol. 2008;28:3894–3904. doi: 10.1128/MCB.00322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trievel RC, Shilatifard A. WDR5, a complexed protein. Nat Struct Mol Biol. 2009;16:678–680. doi: 10.1038/nsmb0709-678. [DOI] [PubMed] [Google Scholar]
- Wang YL, Faiola F, Xu M, Pan S, Martinez E. Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J Biol Chem. 2008;283:33808–33815. doi: 10.1074/jbc.M806936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Serino G, Deng XW. The COP9 signalosome: more than a protease. Trends Biochem Sci. 2008;33:592–600. doi: 10.1016/j.tibs.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Wilkins A, Ping Q, Carpenter CL. RhoBTB2 is a substrate of the mammalian Cul3 ubiquitin ligase complex. Genes Dev. 2004;18:856–861. doi: 10.1101/gad.1177904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Xiong Y. Targeting p21 degradation locally. Dev Cell. 2010;19:641–643. doi: 10.1016/j.devcel.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui K, Arii S, Zhao C, Imoto I, Ueda M, Nagai H, Emi M, Inazawa J. TFDP1, CUL4A, and CDC16 identified as targets for amplification at 13q34 in hepatocellular carcinomas. Hepatology. 2002;35:1476–1484. doi: 10.1053/jhep.2002.33683. [DOI] [PubMed] [Google Scholar]
- Yi JJ, Ehlers MD. Emerging roles for ubiquitin and protein degradation in neuronal function. Pharmacol Rev. 2007;59:14–39. doi: 10.1124/pr.59.1.4. [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- Zhou P, Howley PM. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol. Cell. 1998;2:571–580. doi: 10.1016/s1097-2765(00)80156-2. [DOI] [PubMed] [Google Scholar]
- Zimmerman ES, Schulman BA, Zheng N. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr Opin Struct Biol. 2010;20:714–721. doi: 10.1016/j.sbi.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Liu Q, Chen B, Zhang X, Guo C, Zhou H, Li J, Gao G, Guo Y, Yan C, et al. Mutation in CUL4B, which encodes a member of cullin-RING ubiquitin ligase complex, causes X-linked mental retardation. Am. J. Hum. Genet. 2007;80:561–566. doi: 10.1086/512489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Mi J, Cui J, Lu D, Zhang X, Guo C, Gao G, Liu Q, Chen B, Shao C, Gong Y. Characterization of nuclear localization signal in the N terminus of CUL4B and its essential role in cyclin E degradation and cell cycle progression. J Biol Chem. 2009;284:33320–33332. doi: 10.1074/jbc.M109.050427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.