Abstract
We have found that IS10 transposase is synthesized in tiny amounts, about 0.15 polypeptide chain per cell per generation on average, as judged from the beta-galactosidase activity of a single chromosomal copy of a suitable transposase-lacZ gene fusion. Enzymatic activity from the fusion gene is a factor of 10 lower in a permeabilized whole cell assay than in cell extracts. Probably, most cells contain fewer than four polypeptide chains, and these chains can assemble into active tetramers only after cell disruption. This interpretation permits formulation of two equations relating enzyme activities to transcription and translation rates, solution of which reveals that the fusion gene is expressed at the average rate of only 0.25 transcript per cell per generation, with an average of only 0.58 translation product per transcript. This methodology is generally applicable to analysis of any gene from which fewer than four polypeptide chains are synthesized per cell per generation.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Casadaban M. J. Regulation of the regulatory gene for the arabinose pathway, araC. J Mol Biol. 1976 Jul 5;104(3):557–566. doi: 10.1016/0022-2836(76)90120-0. [DOI] [PubMed] [Google Scholar]
- Contaxis C. C., McAfee D., Goodrich R., Reithel F. J. Studies on protein multimers. V. Further characterization of beta-galactosidase self-association behavior with observations on apparent specific volume. Int J Pept Protein Res. 1973;5(4):207–213. [PubMed] [Google Scholar]
- Contaxis C. C., Reithel F. J. Studies on protein multimers. The association-dissociation behaviour of -galactosidase in glycerol. Biochem J. 1971 Sep;124(3):623–632. doi: 10.1042/bj1240623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debarbouille M., Schwartz M. The use of gene fusions to study the expression of malT the positive regulator gene of the maltose regulon. J Mol Biol. 1979 Aug 15;132(3):521–534. doi: 10.1016/0022-2836(79)90273-0. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Chandler M. Structure and stability of Tn9-mediated cointegrates. Evidence for two pathways of transposition. J Mol Biol. 1982 Jan 15;154(2):245–272. doi: 10.1016/0022-2836(82)90063-8. [DOI] [PubMed] [Google Scholar]
- Gilbert W., Müller-Hill B. Isolation of the lac repressor. Proc Natl Acad Sci U S A. 1966 Dec;56(6):1891–1898. doi: 10.1073/pnas.56.6.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halling S. M., Simons R. W., Way J. C., Walsh R. B., Kleckner N. DNA sequence organization of IS10-right of Tn10 and comparison with IS10-left. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2608–2612. doi: 10.1073/pnas.79.8.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R. L., Yanofsky C. Trp aporepressor production is controlled by autogenous regulation and inefficient translation. Proc Natl Acad Sci U S A. 1982 May;79(10):3120–3124. doi: 10.1073/pnas.79.10.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennell D., Riezman H. Transcription and translation initiation frequencies of the Escherichia coli lac operon. J Mol Biol. 1977 Jul;114(1):1–21. doi: 10.1016/0022-2836(77)90279-0. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Maloney P. C., Rotman B. Distribution of suboptimally induces -D-galactosidase in Escherichia coli. The enzyme content of individual cells. J Mol Biol. 1973 Jan;73(1):77–91. doi: 10.1016/0022-2836(73)90160-5. [DOI] [PubMed] [Google Scholar]
- Maurer R., Meyer B., Ptashne M. Gene regulation at the right operator (OR) bacteriophage lambda. I. OR3 and autogenous negative control by repressor. J Mol Biol. 1980 May 15;139(2):147–161. doi: 10.1016/0022-2836(80)90302-2. [DOI] [PubMed] [Google Scholar]
- Mitra S., Pal B. C., Foote R. S. O6-methylguanine-DNA methyltransferase in wild-type and ada mutants of Escherichia coli. J Bacteriol. 1982 Oct;152(1):534–537. doi: 10.1128/jb.152.1.534-537.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisato D., Way J. C., Kim H. J., Kleckner N. Tn10 transposase acts preferentially on nearby transposon ends in vivo. Cell. 1983 Mar;32(3):799–807. doi: 10.1016/0092-8674(83)90066-1. [DOI] [PubMed] [Google Scholar]
- Oliver D. B., Beckwith J. E. coli mutant pleiotropically defective in the export of secreted proteins. Cell. 1981 Sep;25(3):765–772. doi: 10.1016/0092-8674(81)90184-7. [DOI] [PubMed] [Google Scholar]
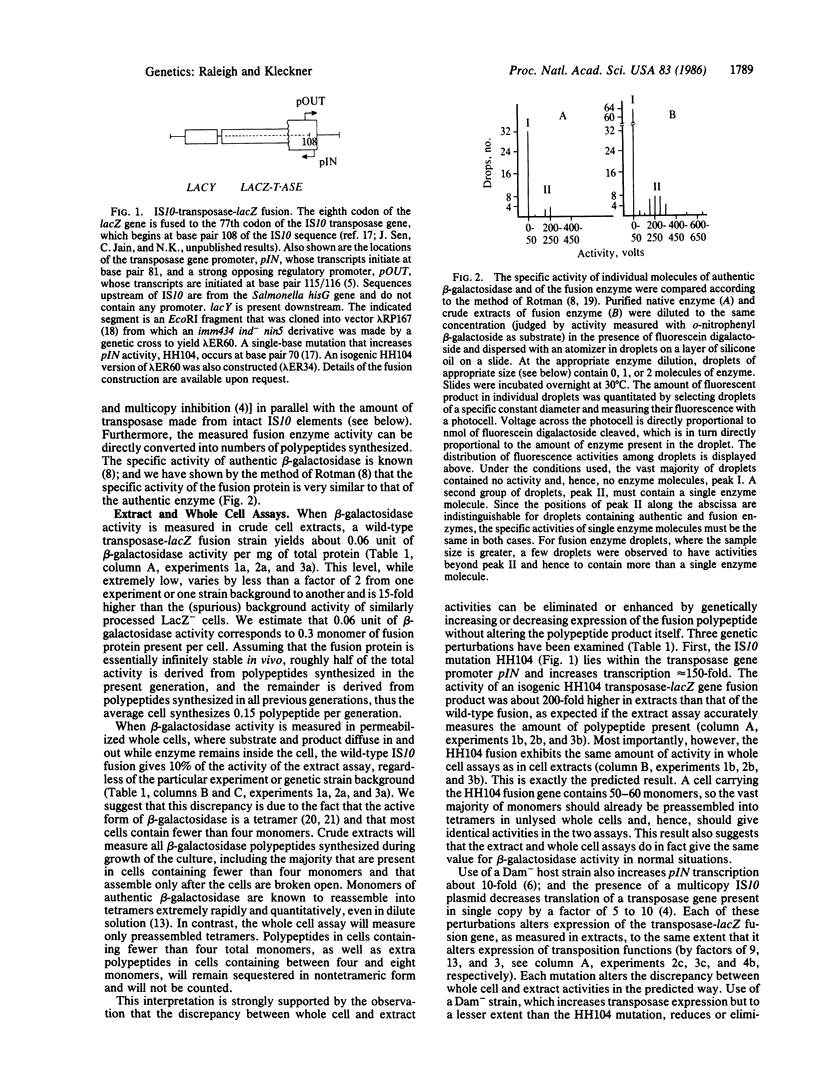
- ROTMAN B. Measurement of activity of single molecules of beta-D-galactosidase. Proc Natl Acad Sci U S A. 1961 Dec 15;47:1981–1991. doi: 10.1073/pnas.47.12.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt L., Kaiser A. D. Control of lambda repressor synthesis. Proc Natl Acad Sci U S A. 1971 Sep;68(9):2185–2189. doi: 10.1073/pnas.68.9.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigney D. R. Note on the kinetics and stochastics of induced protein synthesis as influenced by various models for messenger RNA degradation. J Theor Biol. 1979 Jul 21;79(2):247–257. doi: 10.1016/0022-5193(79)90250-9. [DOI] [PubMed] [Google Scholar]
- Rigney D. R. Stochastic model of constitutive protein levels in growing and dividing bacterial cells. J Theor Biol. 1979 Feb 21;76(4):453–480. doi: 10.1016/0022-5193(79)90013-4. [DOI] [PubMed] [Google Scholar]
- Roberts D., Hoopes B. C., McClure W. R., Kleckner N. IS10 transposition is regulated by DNA adenine methylation. Cell. 1985 Nov;43(1):117–130. doi: 10.1016/0092-8674(85)90017-0. [DOI] [PubMed] [Google Scholar]
- Rowen L., Kornberg A. Primase, the dnaG protein of Escherichia coli. An enzyme which starts DNA chains. J Biol Chem. 1978 Feb 10;253(3):758–764. [PubMed] [Google Scholar]
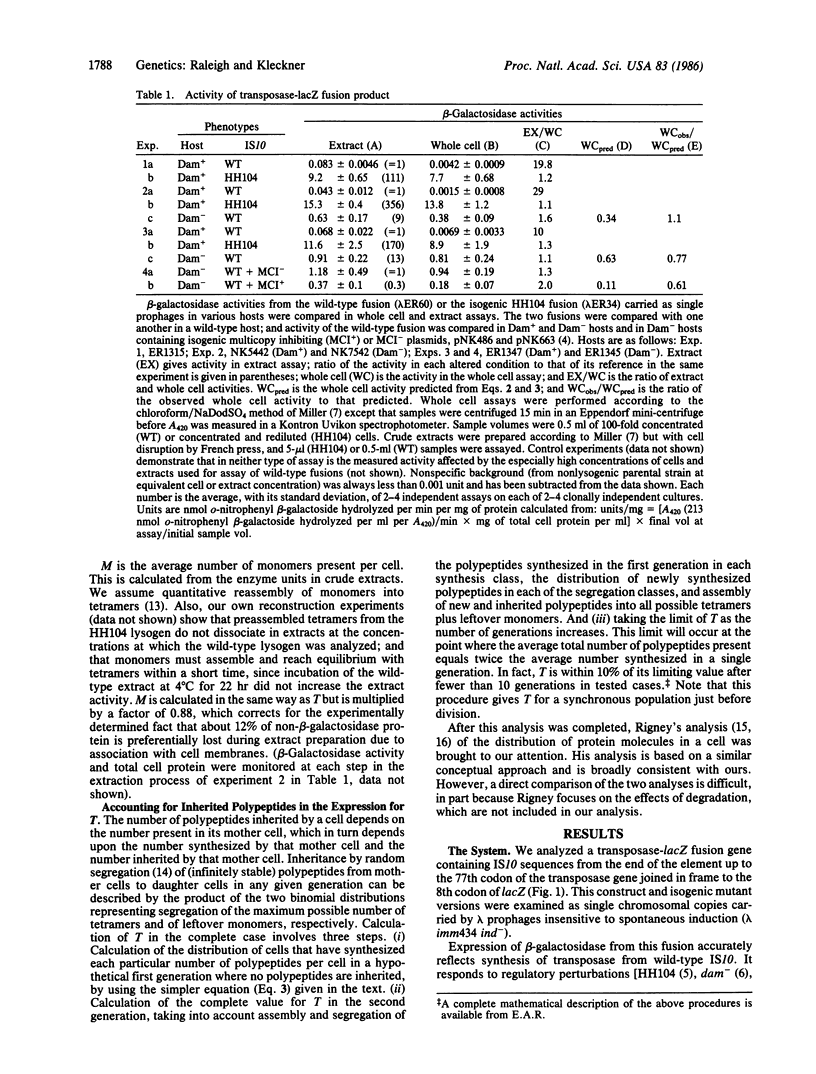
- Simons R. W., Hoopes B. C., McClure W. R., Kleckner N. Three promoters near the termini of IS10: pIN, pOUT, and pIII. Cell. 1983 Sep;34(2):673–682. doi: 10.1016/0092-8674(83)90400-2. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Kleckner N. Translational control of IS10 transposition. Cell. 1983 Sep;34(2):683–691. doi: 10.1016/0092-8674(83)90401-4. [DOI] [PubMed] [Google Scholar]
- Stamatoyannopoulos G., Kurnit D. M., Papayannopoulou T. Stochastic expression of fetal hemoglobin in adult erythroid cells. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7005–7009. doi: 10.1073/pnas.78.11.7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Derbyshire K. M., Hughson F. M., Grindley N. D. Replicative and conservative transpositional recombination of insertion sequences. Cold Spring Harb Symp Quant Biol. 1984;49:251–260. doi: 10.1101/sqb.1984.049.01.029. [DOI] [PubMed] [Google Scholar]


