Abstract
Estrogen receptor (ER) biology reflects the actions of estrogens through the two receptors, ERα and ERβ, although little is known regarding the preference for formation of ER homo- vs. heterodimers, and how this is affected by the level of ligand occupancy and preferential ligand affinity for one of the ER subtypes. In this report, we use a split optical reporter-protein complementation system to demonstrate the physical interaction between ERα and ERβ in response to different ER ligands in cells and, for the first time, by in vivo imaging in living animals. The genetically encoded reporter vectors constructed with the ligand-binding domains of ERα and ERβ, fused to split firefly or Renilla luciferase (Fluc or hRluc) fragments, were used for this study. This molecular proteomic technique was used to detect ERα/ERα or ERβ/ERβ homodimerization, or ERα/ERβ heterodimerization induced by ER subtype-selective and nonselective ligands, and selective ER modulators (SERM), as well as in dimers in which one mutant monomer was unable to bind estradiol. The SERM-bound ERα and ERβ form the strongest dimers, and subtype-preferential homodimerization was seen with ERα-selective ligands (methyl piperidino pyrazole/propyl pyrazole triol) and the ERβ-selective ligands (diarylpropionitrile/tetrahydrochrysene/genistein). We also demonstrated that a single ligand-bound monomer can form homo- or heterodimers with an apo-monomer. Xenografts of human embryonic kidney 293T cells imaged in living mice by bioluminescence showed real-time ligand induction of ERα/ERβ heterodimerization and reversal of dimerization upon ligand withdrawal. The results from this study demonstrate the value of the split luciferase-based complementation system for studying ER-subtype interactions in cells and for evaluating them in living animals by noninvasive imaging. They also probe what combinations of ERα and ERβ dimers might be the mediators of the effects of different types of ER ligands given at different doses.
The estrogen receptor (ER), a major therapeutic target in ER-positive breast and endometrial cancers, functions as a ligand-regulated transcription factor. The estrogen response in target tissues is further modulated by posttranslational modifications and interactions that ER has with other nuclear proteins. Initially, only one type of ER (ERα) was thought to be involved in estrogen hormone signaling. The discovery of a second ER subtype, ERβ, including its various isoforms (β1, β2, β4, and β5), has added another degree of complexity to the manner in which the estrogen response might be regulated (1, 2). Indeed, ER biology now appears much more multifaceted than has hitherto been thought (3–6).
ERα and ERβ share 55% sequence identity in their ligand-binding domains (LBD) and approximately 97% sequence identity in their DNA-binding domains (7). Reflecting the high degree of similarity in their DNA-binding domains, both receptors might be expected to interact with the same estrogen response elements, either as ERα/ERα or ERβ/ERβ homodimers, or as ERα/ERβ heterodimers, although this issue needs further investigation (8–11). Both ERs bind with high affinity to the endogenous ligand estradiol (E2) and to other steroidal estrogens, but they vary in their ability to bind to other natural and synthetic nonsteroidal ligands; they also vary significantly in the types of responses elicited upon ligand binding (7, 12–14). In addition, the tissue-specific expression and coexpression of the ER subtypes suggest that ER homo- and heterodimers might mediate distinct hormone responses (4, 7). Moreover, the discovery of ERβ variants with different structural and functional characteristics, and varying tissue distribution further highlights the potential complexity of the interactions between the ER subtypes and the mechanisms by which estrogens mediate their responses.
A number of recent studies have focused on understanding the distinct biology of ERα and ERβ (4, 5, 12), but there are still many aspects regarding the interaction between these two subtypes and the respective biological roles of homo- vs. heterodimers that need exploration. For example, is there a preference for formation of homo- or heterodimers that is regulated by ER ligands of different structure and different ER subtype binding selectivity? Do dimers form when only one monomer is ligand occupied as might be the case at low hormone levels or in heterodimers with ER subtype-selective ligands? Is there ever a dominance of homo vs. heterodimerization? Does one ER subtype dominate over the other?
To unravel some of the biological consequences of the individual and combined interactions of ERα and ERβ, we have used in this study new versions of our split reporter-based luciferase complementation system (15, 16) to explore by bioluminescence the patterns of ligand-induced dimerization of ERα and ERβ in cells and also in xenografts expressing the complementation system in living mice. Using these complementation systems, we were able to evaluate the role of ligands of varying biocharacter (agonists and partial agonist/antagonists, such as selective ER modulators or SERM), finding that SERM-bound receptors formed the most stable homo- and heterodimers. Additionally, we investigated the effects of ER subtype-selective ligands in promoting ERα and ERβ homo- vs. heterodimerization and noted a preference for homodimerization. Our results also demonstrate that a single ligand-bound monomer can form a stable homo- or heterodimer. Using noninvasive optical bioluminescence imaging, we can monitor ligand-regulated induction of ERα/ERβ heterodimerization in living animals. In addition to highlighting the value of our luciferase complementation systems for studying ERα/ERβ dimerization, our study has identified novel aspects of ligand-mediated ER-subtype interactions that could underline the diversity of estrogen-mediated regulation of different cellular functions as mediated by ligands of different biocharacter, subtype selectivity, and dose.
Results
The LBD of ERα and ERβ are sufficient to effect ligand-induced dimerization, as studied by a split-luciferase complementation system
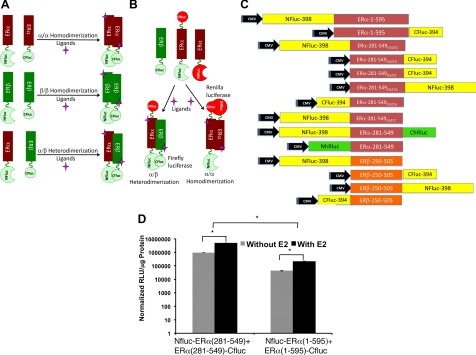
To evaluate ligand-induced homo- and heterodimerizations of ERα and ERβ (Fig. 1, A and B), we constructed eukaryotic plasmid vectors expressing fusion proteins of full-length ERα or ERβ, or their LBD, linked at either their N- or C-termini to the N- and C-terminal fragments of split-firefly luciferase in various arrangements (Fig. 1C). Pairs of these fusion proteins, each bearing complementing fragments of the split-firefly luciferase, were transiently cotransfected in human embryonic kidney (HEK)293T cells and assayed for complemented firefly luciferase activity 24 h after exposure to 1 μm of the ligand E2. The results showed significant levels (P < 0.05) of homodimerizations (ERα/ERα and ERβ/ERβ), as measured by the complemented luciferase signals, from both full-length and the LBD of the ER. However, the absolute signal levels were 20 ± 3-fold higher in cells engineered to express split-luciferase fragments linked to the LBD of the ER (Fig. 1D, note logarithmic scale) rather than to the full-length ER. This is most likely because the smaller and more structured LBD position the N- and C-terminal ends of luciferase fragments in a manner that facilitates more efficient complementation. These findings indicate that the LBD of both receptors are sufficient to mediate ligand-induced dimerizations, which is consistent with the large dimer interface seen in x-ray structures of the ER LBD. Although portions of the ER that are outside of the LBD have some modulatory effects on estrogen action, interactions in the ER LBD are the dominant determinants of ligand binding affinity and structural discrimination, subtype selectivity, and dimerization. Therefore, because of the larger bioluminescence signal, which we were able to obtain with the ER LBD constructs, we used these in our subsequent experiments.
Fig. 1.
A and B, Scheme of independent and simultaneous split firefly and Renilla luciferase reporter protein complementation systems designed for studying ER-ligand induced homo- and heterodimerizations (ERα and ERβ) in cells, and noninvasively imaging in living animals by optical bioluminescence imaging. In this system, the LBD of ER (ERα and ERβ) are linked to the N- and C-terminal portions of luciferases, which then complement when the cells in which they are coexpressed are treated with ligands having different specificity and biocharacters for these receptors. The orientation (NH2 to COOH) of fusion proteins in different constructs were represented by labeling the different ends of the ER-LBD. C, Schematic diagram of different eukaryotic expression vectors constructed for the study in pcDNA (3.1) vector backbone. The schemes show the order of fragments of reporters (firefly luciferase: Fluc and Renilla luciferase: hRluc: N- and C- indicates the NH2 and COOH-terminal fragments of reporter proteins) and the ER (α and β) with respective sizes cloned to express different fusion proteins. D, The sensitivity of ER-dimerization system evaluated by vectors constructed to express fusion proteins of split-firefly luciferase fragments with either full-length or the LBD of ERα. HEK293T cells cotransfected with the vectors assayed for the complemented luciferase activity after exposure to ligand E2 (y-axis in log scale; *, P < 0.01). CMV, Cytomegalovirus.
Fusion proteins of ER-LBD monomers with split-luciferase fragments can be used to measure ligand-induced ER dimerization and also to distinguish the effects of ligands of different biocharacter on ER homo- or heterodimer stability
In our previous work (17), we designed single receptor monomers to which we attached both the N- and C-terminal portions of the split-luciferase enzymes. In those experiments, the split-luciferase complementation signal output was a measure of regional conformational changes near the C-terminus of the LBD that acts as a molecular switch in response to the binding of ligands of differing biocharacter. In this prior intramonomer split-luciferase complementation configuration, there might also have been a small contribution from complementation occurring between monomers within a dimer, as well as from changes in global receptor structural stability, especially when the split proteins were placed near the far C-terminus (F-domain) of the receptor (17).
By contrast, to study receptor dimerization in this report, we designed each monomer to contain only a single portion of the split-luciferase protein but to be used as pairs, each member of which would contribute either the N- or the C-terminal fragment of the split luciferase. The split-luciferase complementation signal in these experiments has three potential sources: 1) Dimerization leading to complementation; 2) global receptor structural stability allowing for more efficient complementation; and 3) regional molecular switch conformational changes near the C-terminal end of the LBD.
To find the optimum pairs, we transiently cotransfected HEK293T cells with all of the possible combinations of either N- or C-terminal split-luciferase ERα and ERβ monomers and assayed them for complementation signal after exposure to ligands E2, genistein (Gen), 4-hydroxytamoxifen (OHT) and raloxifene (Ral), as well as dimethylsulfoxide as solvent control. Ligands were used at 1 μm to ensure that both ER would be fully saturated, despite differences in ligand binding affinities. Significant levels of complemented luciferase signals through heterodimerization (P < 0.01 for all ligands in comparison with solvent control) were seen with all combinations. We selected for further study the combination coexpressing NFluc-ERβ-LBD and ERα-LBD-CFluc; although all eight combinations gave robust signals with some of the ligands, the selected pair gave both high induction signal level through heterodimerization with various ligands, while at the same time maintaining a trend of signal patterns that reflected the varying biocharacter of different ligands (E2: agonist; Gen: ERβ-specific partial agonist/antagonist; OHT and Ral: SERM) (Supplemental Fig. 1 published on The Endocrine Society's Journals Online web site at http://mend.endojournals.org).
The chosen combination of vectors was used for making stable cell lines and in all other cell culture studies, and for subsequent imaging in animals. Of note, the lack of change in the signal trend across ligands of various biocharacter with heterodimer split-luciferase complementation using these monomer constructs is a good outcome, because it indicates that the regional molecular switch conformational changes (mechanism c) plays a minimal role during the production of this cross-dimer complementation signal. Thus, the major contributors to the complementation bioluminescence signal in the current experiments appear to be receptor dimerization, with perhaps some contribution from global receptor structural stability.
We also used our previously reported ERα single-mutation receptor G521T. This mutant has essentially no affinity for E2, but shows minimal loss of affinity for OHT and diethylstilbestrol (DES), and has increased affinity for the pyrazole-based ligands propyl pyrazole triol (PPT) and methyl piperidino pyrazole (MPP) (18). It proved useful in our studies on half-occupied ER dimer stability.
Western blot analysis of HEK293T cells engineered to express ERα/ERβ heterodimerization system identifies no substantial change in the expression levels of different monomers before and after exposure to different ER ligands
HEK293T cells engineered to stably express fusion proteins NFluc-ERβ-LBD and ERα-LBD-CFluc were tested for the expression level of fusion monomers by Western blot analysis, after exposure to different ER ligands [E2, OHT, Ral, MPP, PPT, and diarylpropionitrile (DPN)] by using anti-Fluc antibody. We found no significant change in the expressed levels of monomers in response to the treatment of different ER ligands (Supplemental Fig. 2).
SERM-bound receptors form the strongest homo- and heterodimers
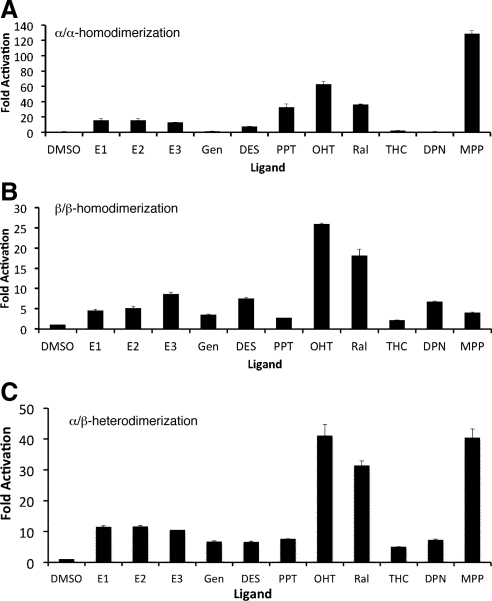
The SERM-bound ERα and ERβ receptors lead to the highest split-luciferase complementation signal within homo- and heterodimers (Fig. 2, A–C). Within the ERα homodimers, we note the highest dimer stability signal from OHT, Ral, MPP-bound receptors (Fig. 2A), and within the ERβ homodimers, the highest dimer stability signal came from OHT and Ral-bound receptors (Fig. 2B). Finally, within the ERα/ERβ heterodimers, we note the highest dimer stability signal from OHT, Ral, and MPP (Fig. 2C). Our previous work using small molecule fluorophore-labeled receptors also demonstrated similar pronounced SERM-bound ERα homodimer stability (19).
Fig. 2.
ER ligand-induced dimerizations [ERα/ERα-homodimerization (panel A); ERβ/ERβ-homodimerization (panel B); and ERα/ERβ-heterodimerization (panel C) studied in HEK293T cells cotransfected with respective combination of vectors (ERα/ERα: NFluc-ERα-LBD/ERα-LBD-CFluc; ERα/ERβ: NFluc-ERα-LBD/ERβ-LBD-CFluc; ERβ/ERβ: NFluc-ERβ-LBD/ERβ-LBD-CFluc) after exposure to several ligands including ERα-selective (MPP and PPT), ERβ-selective (DPN, THC, and Gen), and ER subtype nonselective ligands [ERα and ERβ; estrone (E1), E2, estriol (E3), OHT, and Ral]. The results showed significant level of luciferase signal induced by all ligands (P < 0.05). The subtype-specific ligands induced higher level of signals with their respective homodimerization along with the heterodimerization. DMSO, Dimethylsulfoxide.
Split luciferase complementation assay reveals ERα and ERβ homodimerization induced by ER subtype-selective ligands
We next studied the ligand-mediated regulation of ER homodimerization (ERα/ERα and ERβ/ERβ) using HEK293T cells stably expressing the appropriate sets of NFluc-ER-LBD and ER-LBD-CFluc subtypes. The complemented luciferase signal was measured after 24-hr exposure to ligands that are either subtype-selective for ERα [propyl pyrazole triol (PPT), 410-fold selective (20)], methyl piperidino pyrazole [MPP, 220-fold selective (21)], or subtype-selective for ERβ [diarylpropionitrile (DPN), 70-fold selective (22)], Gen (∼400-fold selective) (23)], tetrahydrochrysene [THC, 16-fold selective (24)]. We also used a series of subtype-nonselective ligands as controls.
Subtype-nonselective ligands (estrone, E2, estriol, and DES, OHT, and Ral) induced significant (P < 0.02) levels of complemented luciferase signal in both ERα/ERα and ERβ/ERβ homodimers, demonstrating that they could effect homodimerization of both pure subtypes (Fig. 2, A and B). By contrast, subtype-selective ligands were more effective in inducing homodimerization interactions of their cognate ER subtype, to which they bound preferentially. For example, the highly ERα-selective ligands PPT and MPP were more effective in homodimerizing ERα than ERβ, whereas the ERβ-selective ligands, Gen, DPN, and THC, more effectively dimerized ERβ than ERα as homodimers.
A single ligand-occupied monomer can form stable homo- and heterodimers with an apo-monomer
The biology of an ER homo- and heterodimer in which only one monomer component is occupied by a ligand is not well understood, despite the fact that in biological settings with low ligand concentrations, such half-ligand-occupied ER dimers are undoubtedly present and are likely to make important contributions to the observed patterns of ligand-regulated gene expression. To evaluate these biologically significant, yet poorly understood, molecular interactions, we used our split-luciferase complementation assay together with subtype-selective ligands. As shown in Fig. 2C, we found that a receptor monomer bound with a subtype-selective ligand (ERα with PPT or MPP, or ERβ with Gen or DPN) formed stable heterodimers with an apo-ERβ or apo-ERα monomer, respectively.
We also investigated formation of singly liganded ER heterodimers using the ERαG521T mutant that we previously identified as having no affinity for E2 but with retained affinity for all other nonsteroidal ligands, including DES, PPT, and OHT (18). In transfected cells, one or both of the two ERα fusion proteins in the ERα homodimerization system, and the ERα protein in the ERα/ERβ heterodimerization system, were replaced by the fusion protein containing the mutant form of ERα-LBD (ERG521T), and then assayed for complemented luciferase activity after exposure to different ligands, including E2 (Fig. 3, A and B).
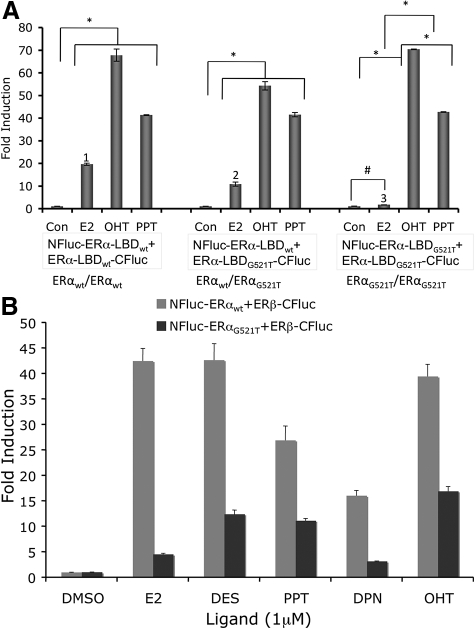
Fig. 3.
A, ERα/ΕRα homodimerization studied in HEK293T cells cotransfected with vectors expressing fusion proteins of split-luciferase fragments with LBD derived from the wild-type ERα (ERαwt), and E2-nonresponsive mutant-ERα (ERαG521T). The cells cotransfected to express both wild-type ER-LBD (NFluc-ERα-LBDwt and ERα-LBDwt-CFluc), wild type with a mutant ER-LBD (NFluc-ERα-LBDwt and ERα-LBDG521T-CFluc), or both mutant ER-LBD (NFluc-ERα-LBDG521T and ERα-LBDG521T-CFluc), were assayed for luciferase activity after exposure to E2, OHT, and PPT, along with solvent dimethylsulfoxide as control. The results showed significant (*, P < 0.01; #, P < 0.05) level of inductions from all ligands in all three combinations, except cells that were cotransfected to express fusion proteins with both mutant ERα-LBD, which showed significantly lower level of complementation for ligand E2 (1 and 2 in comparison to 3; P < 0.01). B, ERα/ERβ-heterodimerization was studied with either wild-type ERα-LBD (NFluc-ERα-LBDwt + ERβ-LBD-CFluc) or mutant ERα-LBD (NFluc-ERα-LBDG521T and ERβ-LBD-CFluc). The cells cotransfected with the mutant ERα-LBD showed considerably lower luciferase complementation signal than those with wild-type ERα-LBD, especially when exposed to ligand E2. Con, Control.
In the ERα/ERα homodimerization system, we observed high or equivalent dimerizations induced by OHT and PPT when none, one, or both of the ERα-LBD fusion protein partners were replaced with mutant-ERα-LBD (ERG521T), consistent with the good affinity that these ligands have for both wild-type and G521T mutant ER. By contrast, with E2, good dimerization was observed in the wild-type (ERαwt/ERαwt) combination, and the wild type with mutant (ERαwt/ERαG521T), but when both ERα partners were mutant (ERαG521T/ERαG521T), no dimerization was evident, because neither of the mutant monomers can bind E2 (Fig. 3A). This suggests that ERα homodimers can form even if only one of the monomer partners is occupied by E2, but, as expected, dimers do not form with two apo-ER monomers.
There is a general loss of signal across all ligand-bound ERβ/ERαG521T heterodimers (Fig. 3B). Because this occurs regardless of whether the monomer components are singly or doubly occupied, it is likely due to a somewhat decreased dimer stability and/or decreased global structural rigidity of the ERαG521T mutant receptor that leads to less efficient complementation. In this regard, it is notable that when ERα/ERβ heterodimers are singly occupied with subtype-selective ligands (Fig. 2C), there is no evident loss of signal intensity compared with the signals from the corresponding doubly occupied homodimers (e.g., PPT and MPP in Fig. 2, panel C vs. panel A, and DPN in Fig. 2, panel C vs. panel B).
Simultaneous monitoring of ERα/ERα homo- and ERα/ERβ heterodimerization studied using a split-Renilla and split-firefly luciferase dual reporter complementation system
The split-protein complementation technologies allow for simultaneous evaluation of multiple and competing protein-protein interactions within a mixed heterogeneous cellular milieu. Potentially, this technology can be used in a multi-modality imaging approach to monitor specific protein interactions via bioluminescence or positron emission tomography signals in living animals (25).
We used a combination of split-Renilla (Rluc) and split-firefly (Fluc) luciferase-based bioluminescence complementation systems as a proof-of-concept experiment to investigate how ligands regulate the competition between ERα/ERα homodimerization vs. ERα/ERβ heterodimerization. In this more involved protocol, HEK293T cells were cotransfected to express three fusion proteins: NFluc-ERα-LBD-ChRluc, ERβ-LBD-CFluc, and NhRluc-ERα-LBD; dimerization of the first and second proteins would enable complementation of firefly luciferase and produce a signal unique for ERα/ERβ heterodimerization, whereas dimerization of the first and third proteins would complement Renilla luciferase and produce a signal unique for ERα/ERα homodimerization.
The results identified significant (P < 0.01) levels of complemented activities from both luciferases after exposure to different ER ligands, including subtype-selective ones (Fig. 4), indicating that both ERα/ERα homodimers and ERα/ERβ heterodimers were forming (Fig. 4A). Nevertheless, despite the quantitative differences in the magnitude of the Fluc and hRluc signals, these can be readily distinguished because they are measured independently after the addition of different bioluminescence substrates. The Western blot analysis for the expressed level of all three-fusion proteins showed no substantial change in response to the treatment of different ligands, further confirming the specificity of our system (Fig. 4B).
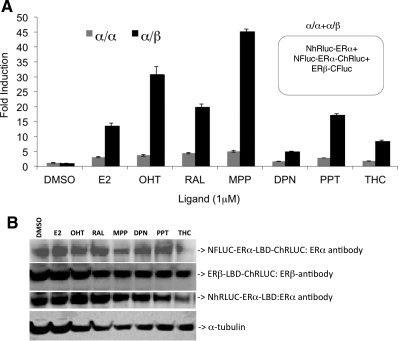
Fig. 4.
Simultaneous monitoring of ligand-induced ERα/ERα homo- and ERα/ERβ heterodimerization by dual reporter complementation system. A, The cells cotransfected to express fusion proteins NFluc-ERα-LBD-ChRluc, ERβ-LBD-Cfluc, and NhRluc-ERα-LBD were assayed for ERα/ERα homodimerization (Renilla luciferase) and ERα/ERβ heterodimerization (firefly luciferase) 24 h after exposure to different ER ligands including subtype-selective ligands. The results showed significant levels of αα-homodimerization (P < 0.05) and αβ-heterodimerization (P < 0.01), as measures of complemented luciferases activities. B, Western blot analysis of cell lysates from panel A assayed for the expressed fusion proteins using ERα- and ERβ-specific antibodies. DMSO, Dimethylsulfoxide.
These are early results from a technically challenging but uniquely informative approach for monitoring two competing and simultaneous protein-protein interactions of the ER. The higher signal seen with the heterodimer complexes compared with homodimers may be related to different levels of activation upon Fluc vs. hRluc complementation or to conformational changes within these fusion constructs, and not necessarily to differences in receptor dimer stability. Nevertheless, all of the changes in bioluminescence signals are statistically significant; they again show a dependence on ligand structure that is similar to that seen in the two-component complementation systems (Fig. 2, A and C). Further investigations with specific controls are needed to more fully characterize this NFluc-ERα-LBD-ChRluc double-split fusion ER construct and its influence upon the native receptor homo- and heterodimerization.
Ligand-induced ERα/ERβ heterodimerization studied in living animals
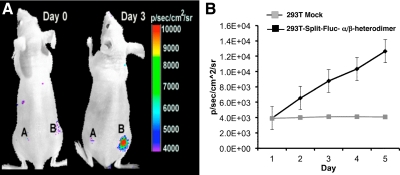
ER ligand-induced heterodimerization (ERα/ERβ) was studied in vivo in living animals by bioluminescence imaging. Parental HEK293T cells and HEK293T cells engineered to stably express fusion proteins of ERα/ERβ heterodimerization system (NFluc-ERβ-LBD and ERα-LBD-CFluc) were used for the study. Mice (n = 5 each for solvent control and the animals receiving ligand MPP), implanted with 5 × 106 parental HEK293T cells as a control (site A) or HEK293T cells stably expressing the heterodimerization system (site B), were imaged after ip injection of substrate D-luciferin (120 mg/kg body weight). Imaging was repeated every day after injection of ligand (MPP, 50 μg) or solvent. Cells at the control site (A) in both sets of mice showed no change in luciferase signal before and after ligand treatment, whereas cells stably expressing fusion proteins for the ERα/ERβ heterodimerization system (site B) showed a progressive increase in the level of luciferase signal that reached 3.5 ± 2-fold above control by day 5 (P < 0.05) (Fig. 5, A and B).
Fig. 5.
In vivo imaging of ligand-induced ERα/β-heterodimerization in living mice. A, Mice implanted with HEK293T cells stably coexpressing NFluc-ERβ-LBD and ERα-LBD-CFluc (site B), and control HEK293T cells (site A) were imaged immediately and after every day injection of 50 μg of ligand MPP. The results showed significant level of complemented luciferase signal from the site implanted with the cells coexpressing fusion proteins, only after the injection of ligand MPP (3.5 ± 2-fold on d 3; P < 0.05). B, The results from a group of animals imaged every day after injecting 50 μg of ligand MPP. Results are the average of five animals, and the error bars are sem of five mice. Although intact mice were used in this experiment, endogenous E2 levels are insufficient to fully occupy ER, and our ERα/ERβ complementation system is more responsive to MPP than E2.
Discussion
The pharmacology of estrogens is complex, with their actions often resulting in both desired and undesired effects, and the tissue-selective nature of this pharmacology has prompted the development of SERMs that are useful in breast cancer treatment and in postmenopausal hormone replacement therapy but are less than ideal in their level of activity and selectivity. More recently, the recognition that estrogens act through two receptors, ERα and ERβ (4, 7), has stimulated the development of ER subtype-selective ligands (5), with the hope that the balance of desired vs. undesired effects of estrogens could be further improved. It has proved challenging to understand, however, what effects of estrogens of diverse structure are mediated by ERα or by ERβ as homodimers, or by ERα/ERβ heterodimers, and how these different ER dimer combinations might respond to ligands of different structures, biocharacter, subtype selectivity, doses, and mixtures. Although ERα and ERβ-knockout mice have proved useful in identifying responses that are mediated by ERα or ERβ alone, the full deletion of one ER subtype abrogates responses that might rely on the combined action of ERα/ERα and ERβ/ERβ homodimers, or of ERα/ERβ heterodimers (26), and it does permit investigation of how these three types of ER dimers form at different ligand doses or in the context of ligands with selectivity for either ERα or ERβ.
Our study has begun to address these as yet unanswered questions by using the split luciferase complementation system we have developed. With this new bioluminescence technology, we have been able to examine in greater detail how formation of the three complements of ER dimers, ERα/ERα and ERβ/ERβ homodimers and ERα/ERβ heterodimers, is regulated as a function of ligand dose, ligand structure, and SERM or subtype selectivity, in both a cellular context and in vivo. We find that both ER homo- and heterodimers can form, sometimes under conditions in which one might not expect them to form.
Split luciferase complementation provides a uniquely powerful approach to study specific ER dimerization events in cells and in vivo
To begin to unravel the role of the ER subtypes and their homo- and heterodimers as mediators of estrogen biology, we adapted a split-luciferase reporter protein complementation system that could be used to monitor various ligand-regulated attributes of the ER (15, 17, 27). Our previous ER constructs were carefully engineered to contain the two complementary portions of the luciferase protein fused to the N- and C-termini of a single receptor monomer (17). That earlier work focused on ligand regulation of ER regional conformations with respect to the well-evaluated molecular switch at the C-terminal portion of the LBD. Here, we have focused on how ligands regulate interactions of ER proteins, namely the formation of ERα and ERβ homo- and heterodimers. This required that the N- and C-terminal complementary fragments of luciferase be attached separately to different ER monomers, either to two ERα monomers to study ERα homodimerization, two ERβ monomers to study ERβ homodimerization, or one ERα monomer and one ERβ monomer to study ERα/ERβ heterodimerization (18). It is notable that the two complementing luciferase fragments provide unique tags of individual ER subtypes (or of an ERα mutant), and these unique tags then enable the formation of specific dimers to be monitored uniquely through complementation, even in the presence of other untagged ER. In principle, such luciferase complementation can be extended in a dual fashion, using three firefly and Renilla luciferase-complementing fragments, and in this regard we provide results from preliminary experiments that establish the feasibility of this experiment.
We found that the LBD of ERα and ERβ were sufficient to maintain the ligand-mediated homo- and heterodimerization properties of these receptors, and the luciferase complementation assays were more sensitive and more convenient when the ER LBD rather than the full-length ER were used, consistent with the more structured nature of the LBD portion of the ER. We also optimized both the positioning and orientation of the N- and C-terminal luciferase fragments on ERα and ERβ. Importantly, the signal output from the split-luciferase complementation appears to be reflective of dimer stability and not the regional conformational changes within the C-terminal portion of the LBD.
Ligands of different pharmacological activity and ER subtype selectivity have different effects on ER dimer strength and ER subtype dimer selectivity
We found that ligand regulation of ER homo- and heterodimer formation, as sensed by luciferase complementation, demonstrates that SERM-bound ER dimers give the highest signal output, suggesting the formation of the strongest dimer complexes. Earlier, in a purely in vitro system, we also found that SERM binding afforded the greatest stability of ERα/ERα homodimers (19). This increased dynamic stability of SERM-bound ER homo- and heterodimers may reflect mechanistic aspects of partial antagonist-bound receptor biology and may be related as well to the well-known slower cellular turnover of SERM-bound ERα (28).
In this regard, it is of note that the THC compound, which has a mixed ERα and ERβ biocharacter (ERα agonist/ERβ antagonist), demonstrates no significant dimer stabilization within either ERα/ERα or ERβ/ERβ homodimers. Again, we reported this unique lack of dimer stabilization with THC within ERα/ERα homodimers in our prior in vitro study (19). This lack of dimer stabilization may reflect the unique biological mechanism and mixed pharmacological profile of THC, as is seen in cell-based assays and in crystallographic studies (24, 29). Similarly, the homodimer stability of ER subtypes reflect the binding preferences of the ER subtype-selective ligands [ERα-selective PPT (20) and MPP (21), and ERβ-selective Gen (23), DPN (22), and THC (24)].
ER dimers only partially occupied by ligands can still dimerize. We studied whether ER dimers form when only one monomer component is occupied with ligand by using subtype-selective ligands or a mutant ERα (G521T) that does not bind E2 but retains good affinity for several other ERα ligands (e.g. DES, OHT, and PPT). Previously, this ERα mutant was used to show that half-occupied ERα/ERα homodimers could also form when one monomer was unoccupied (18). By both approaches we found that ERα and ERβ homodimers and ERα/ERβ heterodimers form between a single ligand-bound monomer and an apo-monomer.
This cross talk across the dimer interface represents the influence of a single ligand on the stability of a dimer and is suggestive of a novel aspect of ER action that is difficult to study by other means. For example, when ligand concentrations are insufficient to saturate both monomers in a dimer, dimers could still form; it is interesting to consider what sort of activity these half-occupied dimers might have. Also, if a cell that contains both ERα and ERβ is treated with a subtype-selective ligand, capable of binding to only one ER subtype, ERα/ERβ heterodimers in which only one monomer is occupied could form. The formation of ligand-occupied ER/apo-ER dimers is reminiscent of the behavior of type II nuclear hormone receptors, such as thyroid hormone receptor, peroxisomal proliferator-activated receptor, retinoic acid receptor, and vitamin D receptor, that form heterodimers with unoccupied retinoid X receptor (30).
Simultaneous monitoring of ERα/ERα homo- and ERα/ERβ heterodimerization by dual-split reporters
We noted that, in principle, it should be possible to simultaneously monitor two complementation events by suitable deployment of the split fragments of Renilla-luciferase and firefly luciferase in a three-component system. As a proof-of-principle, in this study we have evaluated the technical feasibility of using such a dual reporter system. We used ERα and ERβ expressing different combinations of split Renilla and firefly luciferase fragments, and we successfully demonstrated ligand-induced homo- and heterodimerization in response to subtype-selective and nonselective ligands. Simultaneous measurement of two events, in a same cell, is useful in extracting significantly valuable information executed by the interacting partners at micro levels.
Bioluminescence imaging of ER dimer formation in vivo
Optical methods have previously been used to study ER homo- and heterodimerization in cell-free in vitro systems and in intact cells (24, 29); however, none of these approaches could potentially be used to study the same process in living animals (31). In earlier work, we used fluorescence resonance energy transfer and excimer-emission methods to study the effect of ligand binding on the kinetic and thermodynamic stability of ERα homodimers in vitro and the additional stabilizing effect due to coactivator binding to ER-agonist complexes (19, 32, 33). Studies in cells using bioluminescence resonance energy transfer methods have also revealed that ERα plays a dominant role in ERα/ERβ heterodimerization (34–36). Similarly, we have found a similar dominance of ERα in the selection of ERs for binding to chromatin (8).
We used bioluminescence imaging to monitor formation of ERα/ERβ heterodimers in vivo: a strong emission signal from a xenograft growing in an athymic mouse could be followed with time, indicating the progressive formation of this heterodimer. It is of note that the specific complementing luciferase fragments used to tag ERα and ERβ in this experiment enabled formation of ERα/ERβ heterodimers to be monitored specifically. Tagging the ER subtypes in different manners could enable formation of ER homodimers and potentially half-occupied ER dimers to be monitored specifically, as well as the effects of ER ligands of different structure, SERM, and subtype-selective ligands.
Conclusions
Our findings demonstrate that the split-luciferase-based complementation system is useful in studying the interactions between ER subtypes in cells and living animals, and in tracking ER dimer formation using this method we have identified several important features of ER biology: SERM-bound receptors form dimers with high stability; the formation of ER homo- vs. heterodimers is differentially regulated by subtype-selective ligands; and homo- and heterodimers dimers form when only one monomer is ligand occupied, as might be the case for all types of dimers at low hormone levels or in heterodimers with ER subtype-selective ligands. In addition, we have validated a two-channel luciferase complementation experiment by which two distinct dimerization events can be monitored simultaneously. Finally, we have demonstrated ligand-regulated ERα/ERβ heterodimerization in living animals. This complementation system could also be used efficiently for identifying and characterizing new ER subtype-specific ligands by high throughput screening, with subsequent confirmation of their activity in cells and living animals. In closing, we are currently investigating further the use of the two-channel luciferase complementation approach that simultaneously and specifically monitors two distinct dimerization events for identifying in greater detail new ways in which cross talk can occur across the dimer interface within the three complements of ER subtype dimers. These results will be reported in due course.
Materials and Methods
Chemicals, enzymes and reagents
The plasmids pCMV-hRluc, encoding the full-length synthetic Renilla luciferase (hRluc), and the pCMV-Fluc, encoding the full-length firefly luciferase (Fluc), LARII substrate for firefly luciferase assay, and 5× passive lysis buffer, were purchased from Promega Corp. (Madison, WI). Restriction and modification enzymes and T4-DNA ligase were purchased from New England Biolabs (Beverly, MA). Triple-Master mix Taq-DNA polymerase for PCR amplification was purchased from Brinkmann Eppendorf (Hamburg, Germany). Site-directed mutagenesis kit was purchased from Stratagene (La Jolla, CA). Ampicillin for bacterial culture, dimethyl sulfoxide, and different ER ligands used in this study were purchased from Sigma (St. Louis, MO). Bacterial culture media were purchased from BD Diagnostic Systems (Sparks, MD). The plasmid extraction kit and DNA gel extraction kit were purchased from QIAGEN (Valencia, CA). Coelenterazine was purchased from Nanolight Technology (Pinetop, AZ). All of the animal cell culture media, fetal bovine serum, the antibiotics streptomycin and penicillin, and plastic ware for cell cultures and Lipofectamine 2000 transfection reagents and pcDNA3.1(+) eukaryotic expression vector for constructing different plasmid vectors, were purchased from Invitrogen (Carlsbad, CA).
Construction of plasmids
The NH2-terminal portion (NFluc, amino acids 1–398) of the Fluc gene was PCR amplified using the forward primer designed with NheI and the start codon, and the reverse primer designed with BamHI with and without a peptide linker sequence (GGGGSGGGGS) using pCMV-Fluc plasmid (Promega) as a template. Similarly, the COOH-terminal portion (CFluc, amino acids 394–550) of the Fluc gene was amplified using the forward primer designed with BamHI and the reverse primer designed with XhoI and a stop codon. The restriction enzyme-digested fragments were cloned into a corresponding enzyme-digested vector backbone of pcDNA3.1(+) (Invitrogen), modified to express the puromycin-resistant gene as a eukaryotic selection marker. The PCR product of full-length [ERα (1–595); ERβ (1–530)] or the LBD [ERα (281–549); ERβ (250–505)] of human ERα and ERβ both upstream (NheI-BamHI sites) and downstream (BamHI-XhoI) of N- and C-terminal firefly luciferase fragments were cloned into the corresponding restriction-digested vectors. The nucleotide sequence coding for linker peptide (GGGGS)3 was introduced in between the ER and the luciferase fragments in all the vectors. Site-directed mutagenesis was used for constructing different vectors containing the G521T mutant ERα. A similar strategy was used for constructing different vectors containing N- and C-terminal Renilla luciferase (hRluc) fragments (NhRluc: amino acids 1–229 and ChRluc: amino acids 230–310). The sequence-confirmed vectors were used for different experiments. See Fig. 1C for the complete list of plasmid vectors constructed and used in this study.
Cell culture
Human 293T embryonic kidney cancer cells (HEK293T) were grown in MEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin solutions.
Cell transfection, firefly, and Renilla luciferase assays
Transfections were performed in 80% confluent 24 h-old cultures of HEK293T cells. For transfection, 250 ng/well of pcDNA molar equivalent of each split hRluc/Fluc reporter, and full-length hRluc/Fluc were used in 12-well tissue culture plates, and DNA and Lipofectamine-2000 in 1:3 ratio were used for different transfection experiments, following the manufacturer's instructions. A vector (1 ng) expressing full-length Renilla luciferase (phRluc-CMV; Promega) was cotransfected per well to normalize for transfection efficiency. The cells were assayed for luciferase activity after a 24-h incubation, either with or without ligands, by keeping the culture at 37 C in 5% CO2. The luminometer assays for Fluc and hRluc activity were performed as previously described (37, 38). In brief, transfected cells were lysed in 200 μl of ice-cold 1× passive lysis buffer by shaking for 10 min. The cell lysates were centrifuged for 5 min at 10,000 rpm at 4 C to remove cell debris. To determine hRluc activity, 20 μl of supernatant were assayed by addition of 1.0 μg of coelenterazine in 100 μl of 0.05 m sodium PBS at pH 7.0, followed by photon counting in the luminometer (model T 20/20, Turner Designs, Sunnyvale, CA) for 10 sec. The Fluc activity was determined as described for hRluc activity, except for the use of 100 μl of LARII substrate from Promega instead of coelenterazine. Protein concentrations from the cell lysates were determined by using the Bradford assay reagent (Bio-Rad Laboratories, Inc., Hercules, CA). Rluc activities were normalized for protein content and for transfection efficiency using Fluc activity, and they were expressed as relative light units/μg protein · min of counting.
To create stable HEK293T cells coexpressing the ERα/β heterodimerization system, the vectors expressing NFluc-ERα-LBD and ERβ-LBD-CFluc were cotransfected and selected by using puromycin. The cells were plated in 96-well plates, and single clones of cells coexpressing both the fusion proteins at approximately equal levels were expanded and used for different in vitro and in vivo experiments.
Western blot analysis
To determine the expression level of different fusion proteins containing the split-Fluc fragments with ERα or ERβ, cell lysates in triplicate wells assayed for luciferase activity by luminometry were then combined, and protein concentrations were determined using the Bradford assay reagent. A 30-μg portion of total protein in 1×Lamelli loading buffer with β-mercaptoethanol (Bio-Rad) was heated at 95 C for 5 min, resolved using a 4–12% SDS-PAGE gradient gel (Invitrogen), and electroblotted onto a nitrocellulose membrane (Schleicher & Schuell, Keene, NH) of 0.2-μm pore size. The membrane was blocked with 5% nonfat dry milk in Tris-buffered saline containing 0.01% Tween 20 (pH 7.6) for 1 h and probed overnight at 4 C on a rotating platform with the appropriate antibodies as follows: goat polyclonal antibody against firefly luciferase (1:1000 dilution, Abcam, Cambridge, MA); secondary antibodies were peroxidase-conjugated donkey antigoat IgG (1:3000 dilution for both, Abcam). Mouse monoclonal antibody against Renilla luciferase (1:1000 dilution, Chemicon, Temecula, CA); secondary antibodies were peroxidase conjugated donkey antimouse IgG (1:5000 dilution for both; Rockland Immunochemicals, Philadelphia, PA). All Western blot membranes were reprobed with a mouse monoclonal antibody against human α-tubulin (1:5000, clone B-5-1-2; Sigma) to control for protein loading. Immunoblots were developed using the LumiGlo enhanced chemiluminescence method (Cell Signaling Technology, Danvers, MA), following manufacturer's instructions using blue films (Midwest Scientific, St. Louis, MO).
Bioluminescence imaging in living mice
All the animal handling was performed in accordance with Stanford University Animal Research Committee guidelines. Mice were gas anesthetized using isoflurane (2% isoflurane in 100% oxygen, 1 liter/min) during all injection and imaging procedures and were imaged using a cooled charge-coupled device camera (Xenogen IVIS-29; Xenogen Corp., Alameda, CA). An aliquot of 5 × 106 HEK293T control cells or HEK293T cells stably coexpressing ERα-LBD-CFluc and NFluc-ERβ-LBD were implanted sc in the bottom left and right flanks, respectively, of 7-wk-old female nude mice (n = 5) (nu/nu, Charles River Laboratories, Inc., Wilminton, MA). The animals were imaged on 1d (before injecting ligand) and daily for 5 d with the injection of 50 μg ligand MPP every day. Similarly, another group of animals (n = 5) was imaged every day, with the injection of ligand MPP for the first 2 d and then discontinuing any further ligand injection for an additional period of 8 d (to see drug withdrawal effect). d-Luciferin (3 mg in PBS) was injected ip, and animals were imaged with the IVIS imaging system with an acquisition time of 1 min each until the signal reached the peak. Images were obtained using Living Image Software (Xenogen Corp.) and analyzed using Igor Image Analysis Software (Wavemetrics, Seattle, WA). To quantify the measured light, regions of interest were drawn over the area of cell implants, and the maximum photons/sec/cm2/steradian (sr) was obtained, as validated previously.
Data analysis
Each experiment was repeated at least three times, and results are expressed as mean ± sd or sem. Statistical differences were determined by Student's t test using P < 0.05 as cut-off point.
Supplementary Material
Acknowledgments
We thank Dr. Benita S. Katzenellenbogen for helpful suggestions.
This work was supported by National Institutes of Health Grants: NC ICMIC P50 CA114747, NCI SAIRP R24 CA92865, and R01 CA82214 (to S.S.G.) and R37 DK015556 and P01 AG024387 (to J.A.K.). T.F.M. received partial salary support from the NIHR Cambridge Biomedical Research Centre. R.P. was funded by the department of Radiology, Stanford University.
Disclosure Summary: The authors have nothing to disclose.
NURSA Molecule Pages†:
Nuclear Receptors: ER-α | ER-β;
Ligands: 17β-estradiol | Genistein | 4-Hydroxytamoxifen | Raloxifene | Estriol | Estrone | Diarylpropionitrile | Diethylstilbestrol.
Annotations provided by Nuclear Receptor Signaling Atlas (NURSA) Bioinformatics Resource. Molecule Pages can be accessed on the NURSA website at www.nursa.org.
- E2
- Estradiol
- ER
- estrogen receptor
- DES
- diethylstilbestrol
- DPN
- diarylpropionitrile
- Fluc/fluc
- firefly luciferase gene/protein
- Gen
- genistein
- HEK
- human embryonic kidney
- LBD
- ligand-binding domain
- MPP
- methyl piperidino pyrazole
- OHT
- hydroxytamoxifen
- PPT
- propyl pyrazole triol
- Ral
- raloxifene
- SERM
- selective ER modulator
- THC
- tetrahydrochrysene.
References
- 1. Kuiper GG, Carlsson B, Grandien K, Enmark E, Häggblad J, Nilsson S, Gustafsson JA. 1997. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors α and β. Endocrinology 138:863–870 [DOI] [PubMed] [Google Scholar]
- 2. Mosselman S, Polman J, Dijkema R. 1996. ERβ: identification and characterization of a novel human estrogen receptor. FEBS Lett 392:49–53 [DOI] [PubMed] [Google Scholar]
- 3. Koehler KF, Helguero LA, Haldosén LA, Warner M, Gustafsson JA. 2005. Reflections on the discovery and significance of estrogen receptor β. Endocr Rev 26:465–478 [DOI] [PubMed] [Google Scholar]
- 4. Harris HA. 2007. Estrogen receptor-β: recent lessons from in vivo studies. Mol Endocrinol 21:1–13 [DOI] [PubMed] [Google Scholar]
- 5. Minutolo F, Macchia M, Katzenellenbogen BS, Katzenellenbogen JA. 2011. Estrogen receptor β ligands: Recent advances and biomedical applications. Med Res Rev 31:364–442 [DOI] [PubMed] [Google Scholar]
- 6. Shanle EK, Xu W. 2010. Selectively targeting estrogen receptors for cancer treatment. Adv Drug Deliv Rev 62:1265–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pettersson K, Gustafsson JA. 2001. Role of estrogen receptor β in estrogen action. Annu Rev Physiol 63:165–192 [DOI] [PubMed] [Google Scholar]
- 8. Charn TH, Liu ET, Chang EC, Lee YK, Katzenellenbogen JA, Katzenellenbogen BS. 2010. Genome-wide dynamics of chromatin binding of estrogen receptors α and β: mutual restriction and competitive site selection. Mol Endocrinol 24:47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cowley SM, Hoare S, Mosselman S, Parker MG. 1997. Estrogen receptors α and β form heterodimers on DNA. J Biol Chem 272:19858–19862 [DOI] [PubMed] [Google Scholar]
- 10. Pettersson K, Grandien K, Kuiper GG, Gustafsson JA. 1997. Mouse estrogen receptor β forms estrogen response element-binding heterodimers with estrogen receptor α. Mol Endocrinol 11:1486–1496 [DOI] [PubMed] [Google Scholar]
- 11. Schultz JR, Loven MA, Melvin VM, Edwards DP, Nardulli AM. 2002. Differential modulation of DNA conformation by estrogen receptors α and β. J Biol Chem 277:8702–8707 [DOI] [PubMed] [Google Scholar]
- 12. Zhao C, Dahlman-Wright K, Gustafsson JA. 2008. Estrogen receptor β: an overview and update. Nucl Recept Signal 6:e003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chang EC, Charn TH, Park SH, Helferich WG, Komm B, Katzenellenbogen JA, Katzenellenbogen BS. 2008. Estrogen receptors α and β as determinants of gene expression: influence of ligand, dose, and chromatin binding. Mol Endocrinol 22:1032–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lindberg MK, Movérare S, Skrtic S, Gao H, Dahlman-Wright K, Gustafsson JA, Ohlsson C. 2003. Estrogen receptor (ER)-β reduces ERα-regulated gene transcription, supporting a “ying yang” relationship between ERα and ERβ in mice. Mol Endocrinol 17:203–208 [DOI] [PubMed] [Google Scholar]
- 15. Paulmurugan R, Gambhir SS. 2003. Monitoring protein-protein interactions using split synthetic renilla luciferase protein-fragment-assisted complementation. Anal Chem 75:1584–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paulmurugan R, Gambhir SS. 2005. Firefly luciferase enzyme fragment complementation for imaging in cells and living animals. Anal Chem 77:1295–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paulmurugan R, Gambhir SS. 2006. An intramolecular folding sensor for imaging estrogen receptor-ligand interactions. Proc Natl Acad Sci USA 103:15883–15888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paulmurugan R, Tamrazi A, Katzenellenbogen JA, Katzenellenbogen BS, Gambhir SS. 2008. A human estrogen receptor (ER)α mutation with differential responsiveness to nonsteroidal ligands: novel approaches for studying mechanism of ER action. Mol Endocrinol 22:1552–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tamrazi A, Carlson KE, Daniels JR, Hurth KM, Katzenellenbogen JA. 2002. Estrogen receptor dimerization: ligand binding regulates dimer affinity and dimer dissociation rate. Mol Endocrinol 16:2706–2719 [DOI] [PubMed] [Google Scholar]
- 20. Stauffer SR, Coletta CJ, Tedesco R, Nishiguchi G, Carlson K, Sun J, Katzenellenbogen BS, Katzenellenbogen JA. 2000. Pyrazole ligands: structure-affinity/activity relationships and estrogen receptor-α-selective agonists. J Med Chem 43:4934–4947 [DOI] [PubMed] [Google Scholar]
- 21. Sun J, Huang YR, Harrington WR, Sheng S, Katzenellenbogen JA, Katzenellenbogen BS. 2002. Antagonists selective for estrogen receptor α. Endocrinology 143:941–947 [DOI] [PubMed] [Google Scholar]
- 22. Meyers MJ, Sun J, Carlson KE, Marriner GA, Katzenellenbogen BS, Katzenellenbogen JA. 2001. Estrogen receptor-β potency-selective ligands: structure-activity relationship studies of diarylpropionitriles and their acetylene and polar analogues. J Med Chem 44:4230–4251 [DOI] [PubMed] [Google Scholar]
- 23. Muthyala RS, Ju YH, Sheng S, Williams LD, Doerge DR, Katzenellenbogen BS, Helferich WG, Katzenellenbogen JA. 2004. Equol, a natural estrogenic metabolite from soy isoflavones: convenient preparation and resolution of R- and S-equols and their differing binding and biological activity through estrogen receptors α and β. Bioorg Med Chem 12:1559–1567 [DOI] [PubMed] [Google Scholar]
- 24. Meyers MJ, Sun J, Carlson KE, Katzenellenbogen BS, Katzenellenbogen JA. 1999. Estrogen receptor subtype-selective ligands: asymmetric synthesis and biological evaluation of cis- and trans-5,11-dialkyl- 5,6,11, 12-tetrahydrochrysenes. J Med Chem 42:2456–2468 [DOI] [PubMed] [Google Scholar]
- 25. Massoud TF, Paulmurugan R, Gambhir SS. 2010. A molecularly engineered split reporter for imaging protein-protein interactions with positron emission tomography. Nat Med 16:921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Couse JF, Korach KS. 1999. Estrogen receptor null mice: what have we learned and where will they lead us? Endocr Rev 20:358–417 [DOI] [PubMed] [Google Scholar]
- 27. Paulmurugan R, Umezawa Y, Gambhir SS. 2002. Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc Natl Acad Sci USA 99:15608–15613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eckert RL, Mullick A, Rorke EA, Katzenellenbogen BS. 1984. Estrogen receptor synthesis and turnover in MCF-7 breast cancer cells measured by a density shift technique. Endocrinology 114:629–637 [DOI] [PubMed] [Google Scholar]
- 29. Shiau AK, Barstad D, Radek JT, Meyers MJ, Nettles KW, Katzenellenbogen BS, Katzenellenbogen JA, Agard DA, Greene GL. 2002. Structural characterization of a subtype-selective ligand reveals a novel mode of estrogen receptor antagonism. Nat Struct Biol 9:359–364 [DOI] [PubMed] [Google Scholar]
- 30. Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hilal T, Puetter V, Otto C, Parczyk K, Bader B. 2010. A dual estrogen receptor TR-FRET assay for simultaneous measurement of steroid site binding and coactivator recruitment. J Biomol Screen 15:268–278 [DOI] [PubMed] [Google Scholar]
- 32. Tamrazi A, Carlson KE, Katzenellenbogen JA. 2003. Molecular sensors of estrogen receptor conformations and dynamics. Mol Endocrinol 17:2593–2602 [DOI] [PubMed] [Google Scholar]
- 33. Tamrazi A, Carlson KE, Rodriguez AL, Katzenellenbogen JA. 2005. Coactivator proteins as determinants of estrogen receptor structure and function: spectroscopic evidence for a novel coactivator-stabilized receptor conformation. Mol Endocrinol 19:1516–1528 [DOI] [PubMed] [Google Scholar]
- 34. Koterba KL, Rowan BG. 2006. Measuring ligand-dependent and ligand-independent interactions between nuclear receptors and associated proteins using bioluminescence resonance energy transfer (BRET). Nucl Recept Signal 4:e021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lupien M, Jeyakumar M, Hébert E, Hilmi K, Cotnoir-White D, Loch C, Auger A, Dayan G, Pinard GA, Wurtz JM, Moras D, Katzenellenbogen J, Mader S. 2007. Raloxifene and ICI182,780 increase estrogen receptor-α association with a nuclear compartment via overlapping sets of hydrophobic amino acids in activation function 2 helix 12. Mol Endocrinol 21:797–816 [DOI] [PubMed] [Google Scholar]
- 36. Powell E, Xu W. 2008. Intermolecular interactions identify ligand-selective activity of estrogen receptor α/β dimers. Proc Natl Acad Sci U S A 105:19012–19017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Paulmurugan R, Gambhir SS. 2007. Combinatorial library screening for developing an improved split-firefly luciferase fragment-assisted complementation system for studying protein-protein interactions. Anal Chem 79:2346–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Paulmurugan R, Massoud TF, Huang J, Gambhir SS. 2004. Molecular imaging of drug-modulated protein-protein interactions in living subjects. Cancer Res 64:2113–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.