To facilitate mutation screening in patients, a custom resequencing chip has been developed to detect sequence alterations of 267,550 bases of both sense and antisense sequences in 1,470 exons spanning 93 genes involved in inherited retinal dystrophy.
Abstract
Purpose.
Retinal dystrophy (RD) is a broad group of hereditary disorders with heterogeneous genotypes and phenotypes. Current available genetic testing for these diseases is complicated, time consuming, and expensive. This study was conducted to develop and apply a microarray-based, high-throughput resequencing system to detect sequence alterations in genes related to inherited RD.
Methods.
A customized 300-kb resequencing chip, Retina-Array, was developed to detect sequence alterations of 267,550 bases of both sense and antisense sequence in 1470 exons spanning 93 genes involved in inherited RD. Retina-Array was evaluated in 19 patient samples with inherited RD provided by the eyeGENE repository and four Centre d'Etudes du Polymorphisme Humaine reference samples through a high-throughput experimental approach that included an automated PCR assay setup and quantification, efficient post-quantification data processing, optimized pooling and fragmentation, and standardized chip processing.
Results.
The performance of the chips demonstrated that the average base pair call rate and accuracy were 93.56% and 99.86%, respectively. In total, 304 candidate variations were identified using a series of customized screening filters. Among 174 selected variations, 123 (70.7%) were further confirmed by dideoxy sequencing. Analysis of patient samples using Retina-Array resulted in the identification of 10 known mutations and 12 novel variations with high probability of deleterious effects.
Conclusions.
This study suggests that Retina-Array might be a valuable tool for the detection of disease-causing mutations and disease severity modifiers in a single experiment. Retinal-Array may provide a powerful and feasible approach through which to study genetic heterogeneity in retinal diseases.
Retinal dystrophy (RD), a broad group of hereditary disorders, is one of the major causes of incurable blindness in the western world. Inherited RD is genetically and phenotypically heterogeneous. Clinically, RD may present as a variety of phenotypes, including retinitis pigmentosa (RP), cone-rod dystrophy (CRD), Leber congenital amaurosis (LCA), Usher syndrome, and others. Molecular genetic analysis reveals that RD may result from mutations in a variety of genes and may show different inheritance patterns, including autosomal dominant, autosomal recessive, X-linked, and mitochondrial inheritance.1,2 Mutations within the same gene may be associated with different phenotypes. For example, mutations in ABCA4 have been identified in RP, CRD, and Stargardt disease (STGD).3,4 RD may be nonsyndromic, such as RP, LCA, and CRD, or syndromic, such as Usher syndrome and Bardet Biedl syndrome (BBS).2,5 Although monogenic forms have been reported in most families, some digenic forms have also been identified.1,3 During the past 20 years, >170 genes (plus 42 loci without known genes) have been identified for different retinal diseases by a variety of methods (http://www.sph.uth.tmc.edu/retnet/sum-dis.htm). Significant progress has been made in understanding the pathogenesis of these diseases, but many questions remain to be answered.
Genetic testing can identify causative mutations in specific genes and thus has an important impact on clinical diagnosis and genetic counseling. Finding a causative RD mutation often requires sequencing many individual candidate genes. Although efficient test strategies are being developed, current genetic testing used in regular diagnostic laboratories is based on Sanger sequencing and can be complicated, time consuming, and expensive.6,7 A high-throughput screening tool that can identify both known and novel mutations in multiple genes, in a fast and cost-effective manner, would be of great interest to both physicians and researchers. Next-generation sequencing technology has been intensively used in recent studies8,9 of human disease and has the potential for routine testing in a clinical setting; however, data handling and data mining associated with this technology remain a significant challenge. Microarray-based resequencing is able to detect sequence variations in multiple genes simultaneously and rapidly with high accuracy and reproducibility. This technology has been successfully used to sequence mitochondrial genomes or selected groups of genes in the nuclear genome for specific human diseases, such as RP, childhood hearing loss, amyotrophic lateral sclerosis, and hypertrophic cardiomyopathy.10–18 In contrast to massive parallel whole genome or exome sequencing, microarray-based resequencing may provide a better platform for analyzing a moderate amount of sequence (50–300 kb) in a repetitive manner.
A custom-designed, microarray-based, high-throughput resequencing system, Retina-Array (Affymetrix, Santa Clara, CA), was developed and evaluated in this study to identify the genetic causes of RD in a group of patients and to explore the potential clinical usefulness of high-throughput genomic DNA sequence screening for mutations in candidate genes.
Materials and Methods
Patient and DNA Samples
A set of 23 genomic DNA samples was included in this study. These included four Centre d'Etudes du Polymorphisme Humaine (CEPH) individual control samples (ND00001, ND00052, ND00068B, ND00268) from the Coriell Institute for Medical Research and 19 isolated samples from patients with diagnoses of RP (five patients), Usher syndrome (five patients), CRD (five patients), STGD (three patients), and choroideremia (CHM, one patient) harboring 18 known sequence changes from the eyeGENE repository (National Ophthalmic Disease Genotyping Network; http://www.nei.nih.gov/resources/eyegene.asp). This study was approved by the CNS Institutional Review Board of the National Institutes of Health, and informed consent was obtained from each participant. Genomic DNA was isolated from patient peripheral blood using a kit (Gentra Puregene Blood Kit; Qiagen Inc., Valencia, CA) in accordance with the manufacturer's protocol.
Affymetrix Custom Resequencing Chip Design
A 49-format (300-kb) Affymetrix resequencing array platform was used to design and construct a custom resequencing chip, Retina-Array. According to the design guide (GeneChip CustomSeq Custom Resequencing Array Design Guide, P/N 701263, revision 4; Affymetrix), the sequences of interest were identified and downloaded from the human genome database, converted to FASTA format, quality checked, and then used as the reference sequences for probe and primer selection. The sequences consisted of all coding exons plus 12 bp of flanking intronic sequences on either side of the exons allowing splice-site variations to be identified. Repetitive elements and internal duplications leading to cross-hybridization were identified using RepeatMasker shareware (http://www.repeatmasker.org/χ-bin/webrepeatmasker) or Micropeats (http://www.littlest.co.uk/software/bioinf/index.html) and were removed. Highly homologous sequences were also identified by running a homology check on both the amplified and the tiled sequences and were excluded. The resequencing array consisted of a number of probe cells, each of which contained many copies of a unique 25-base oligonucleotide probe of defined sequence. Eight probe cells queried a specific site in a known reference sequence (four interrogated the sense strand, and the other four interrogated the antisense strand, containing probes that were identical except for the central base [A, C, G, or T]) (GeneChip Sequence Analysis Software User's Guide, version 4.1; P/N 701930, revision 2; Affymetrix). As a positive control, the 814-bp sequence of plasmid (TAG IQ-EX; Affymetrix) was also tiled onto the chips. The Retina-Array chips were manufactured by Affymetrix.
High-Throughput PCR Assay Setup
All primer sequences along with amplicon sizes and PCR conditions are included in Supplementary Table S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7978/-/DCSupplemental. PCR assays for each sample were set up in 96-well PCR plates in a high-throughput fashion using either an automated pipetting system (epMotion 5070; Eppendorf, Hauppauge, NY) or a liquid handling automation system (Freedom EVO; Tecan US Inc., Durham, NC). Three PCR conditions were adopted as follows: short-range PCR was carried out in 25-μL reaction volumes using 50 ng DNA, 0.6 μM each primer (Sigma, St. Louis, MO), 0.25 mM dNTP, 2.5 mM MgCl2, and 1.25 U Taq Gold DNA polymerase in 1× Taq Gold PCR buffer (Applied Biosystems, Foster City, CA) as follows: 95°C for 12 minutes; 32 cycles at 94°C for 40 seconds, 63°C for 30 seconds, and 72°C for 1 minute; final 10-minute extension at 72°C. Long-range or GC-rich PCR was carried out in 25-μL reaction volumes using 50 ng DNA, 0.4 μM each primer (Sigma), 0.4 mM dNTP, 2.5 mM MgCl2, and 1.25 U TaKaRa LA Taq DNA polymerase in 1× LA or GC PCR buffer (TaKaRa Bio, Madison, WI) as follows: 94°C for 10 minutes; 32 cycles at 94°C for 40 seconds, 63°C for 40 seconds, and 72°C for 7 minutes; final 15-minute extension at 72°C. Besides the IQ-EX positive controls (1.0- and 7.5-kb PCR products) recommended by Affymetrix, custom positive/negative controls (with/without VMD2 DNA template) were also included for each sample.
High-Throughput PCR Product Quantification
PCR products were quantified in a high-throughput fashion (LabChip 90 system; Caliper Life Sciences, Hopkinton, MA). Briefly, the high-throughput system (LabChip 90; Caliper Life Sciences) automatically sampled approximately 150 nL PCR product per well directly from 96-well PCR plates, separated sample analytes electrophoretically on a small, microfluidic chip, calculated the size and concentration of each DNA fragment using both a DNA ladder and internal markers (HT DNA LabChip 5K or 12K Kit), and generated digital results that were easily imported into software (Excel; Microsoft, Redmond, WA) for the post-PCR reading data processing (Supplementary Fig. S1A, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7978/-/DCSupplemental).
Pooling, Purification, and Fragmentation of PCR Products
Equimolar amounts of 605 PCR products from each individual sample were initially pooled at a concentration of 250 picomolar per PCR amplicon to the array for the first seven samples in accordance with a resequencing array protocol (GeneChip CustomSeq, version 2.1; P/N 701231, revision 5; Affymetrix). Alternative pooling strategies, which pooled all amplicons with similar quantities in less than 10-fold difference, were also performed for the next 16 samples and compared with the Affymetrix protocol. Pooled PCR products were concentrated using centrifugal filters (Centricon Plus-70; Millipore Corp., Billerica, MA) and purified using PCR purification kits (QIAquick; Qiagen Inc., Valencia, CA). The samples were then subjected to DNA fragmentation using the fragmentation reagents from the resequencing assay kit (GeneChip; Affymetrix). The fragmentation reaction was conducted in a reaction volume of 46.6 μL at 37°C for 35 minutes with 0.015 U fragmentation reagent per microgram DNA and was inactivated at 95°C for 15 minutes. The efficacy of fragmentation was checked on a 20% TBE PAGE gel followed by staining with SYBR Gold nucleic acid gel stain (1:10,000; Invitrogen, Corp., Carlsbad, CA) with fragmentation adjusted so that DNA sizes ranged from 20 to 200 bp. Underfragmented DNAs were refragmented under the conditions specifically modified for the sample.
DNA Labeling and Hybridization to Retina-Array
Fragmented DNAs were terminally labeled using DNA-labeling reagents from the resequencing assay kit (GeneChip; Affymetrix) following the array protocol (GeneChip CustomSeq, version 2.1; P/N 701231, revision 5; Affymetrix). The oligonucleotide control reagent included in the kit contains a gridding control that serves as the hybridization control. Chip prehybridization, hybridization, washing, staining, and scanning were carried out following the manufacturer's protocols (Hybridization Oven 645, Fluidics Station 450, and Scanner 3000, respectively; GeneChip; Affymetrix), and instrument control software (GeneChip Command Console [AGCC], version 1.0; Affymetrix) was used for the microarray image data acquisition.
Microarray Data Analysis
Sequence analysis software (GSEQ, version 4.1; GeneChip; Affymetrix) was used for the initial sequence data analysis. Cell intensity files (*.cel) generated by AGCC were analyzed in GSEQ in a batch mode (a minimum of 15 independent samples recommended by Affymetrix; 23 individual samples used in this study) to generate the analysis result files (*.chp) using the resequencing algorithm version 2 under the default settings. The resequencing calling algorithm is based on the Adaptive Background Genotype Calling Scheme (ABACUS) developed by Culter et al.19 This algorithm specifies models for the presence or absence of 11 genotypes for diploid data: A, C, G, T, AC, AG, AT, CG, CT, GT, and no call. The call rate and accuracy of each fragment in an individual patient sample were automatically generated by the GSEQ software in a report file under either the default or custom-defined setting and were used to calculate the call rate and accuracy in that patient sample. Different settings of quality score threshold (QST; 0, 1, 2, 3, 6, 12) and base reliability threshold (BRT; 0, 0.5) within this base calling were tested to check their effects on the call rates and accuracies of the base calls. The user-defined criteria, which included both the PCR assay filter (filtering out those with consistently failed PCR assays for at least eight samples) and the call rate filter (filtering out those with call rates <80% at least for four samples), were also used to assess the performance of Retina-Array chips in this study. A bioinformatic pipeline including a series of custom screening filters was further developed for the post-GSEQ data analysis. The custom screening filters included a series of quality filters that were used to filter out those with variations also detected in CEPH cells (CEPH reference sequence filter) or those with >2 N (10%) or with >5 (25%) variation calls across the board (quality filters 1 and 2) implemented with either spreadsheet software (Excel; Microsoft) or technical computing software (MatLab 7; The MathWorks, Inc., Natick, MA) and one footprint effect filter that was used to filter out variation locations with nearby positions (within nine bases) that were rich in N call and variation calls after a manual visual check, as suggested by the Affymetrix Technical Notes (http://media.affymetrix.com/support/technical/technotes/customseq_arraybase_technote.pdf) and used in previous studies.20,21 All the chip data were also analyzed in a batch mode in software (Sequence Pilot module SeqC, version 3.3; JSI Medical Systems Corp., Costa Mesa, CA) under the default settings as an alternative validation. An internal database was created by downloading the reference sequences of all genes from Ensembl (http://uswest.ensembl.org/index.html). All SNP IDs were retrieved from dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) using SeqC.
Dideoxy Sequencing Analysis
To examine the reliability of the Retina-Array data, one panel of candidate variations identified by the Retina-Array chips was selected for validation by dideoxy sequencing. More specifically, a set of new primers was designed using Primer3 (http://frodo.wi.mit.edu/primer3/) if the variation was identified by long-range PCR; otherwise the same primers as those used in the chip experiment were used. The target DNA fragments were independently amplified from the same DNA sample. PCR reactions were separated by gel electrophoresis (1% SeaKem Gold Agarose; Lonza, Rockland, ME), and DNA bands were cut and extracted (QIAcube and QIAquick Gel Extraction kit; Qiagen Inc., Valencia, CA). Sequencing was performed using cycle sequencing kits (BigDye Terminator, version 3.1; Applied Biosystems, Foster City, CA) in both directions. Products were purified (Perform DTR V3 96-Well Short Plates Kits; Edge BioSystems, Gaithersburg, MD) and electrophoresed (ABI 3130xl Genetic Analyzer; Applied Biosystems), and sequencing data were analyzed (Sequencher version 4.8 software; Gene Codes Corp., Ann Arbor, MI).
Variation Effect Analysis
All validated variations identified in patient samples were checked to determine whether there was a previously reported mutation based on the available information in HGMD (https://portal.biobase-international.com/cgi-bin/portal/login.cgi) and UMD-USH2A (http://www.umd.be/USH2A/) mutation databases. For all potential novel pathogenic variations, the PolyPhen-2 (Polymorphism Phenotyping, version 2) Web-based service and SIFT Human Protein DB were used to predict the possible impact of amino acid substitutions on the structure and function of human proteins. The HumVar modeling of Polyphen-2 was used in these computational predictions (http://genetics.bwh.harvard.edu/cgi-bin/ggi/ggi2.cgi). This is a preferred model for the diagnosis of Mendelian diseases and distinguishes mutations with drastic effects from remaining human variations, including abundant mildly deleterious alleles.
Results
Design of the Retina-Array
A custom resequencing chip, Retina-Array, was designed on the 300-kb resequencing platform, which is the highest density format available from Affymetrix. As shown in Table 1 and Supplementary Table S1 (http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7978/-/DCSupplemental), a total of 93 genes associated with inherited retinal dystrophy, including RP (44), macular degeneration (20), CRD (18), Usher syndrome (8), BBS (13), LCA (13), congenital stationary night blindness (10) and other retinopathies (18), were included on the Retina-Array. As described in Materials and Methods (Supplementary Table S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7978/-/DCSupplemental), the Retina-Array was finally constructed to interrogate 1470 exons from 93 genes with the capability of resequencing both strands of 267,550 bases in a single experiment.
Table 1.
Clinical Classification of Candidate Genes on the Retina-Array
| Disease Category* | Genes (n) | Genes on the Array |
|---|---|---|
| Bardet-Biedl syndrome, AR | 13 | ARL6, BBS1, BBS2, BBS4, BBS5, BBS7, BBS9, BBS10, BBS12, CEP290,†MKKS, TRIM32, TTC8 |
| Chorioretinal atrophy or degeneration, AD | 1 | RGR |
| Cone or cone-rod dystrophy, AD | 9 | AIPL1, CRX, GUCA1A, GUCY2D, PROM1, PRPH2, RIMS1, SEMA4A, UNC119 |
| Cone or cone-rod dystrophy, AR | 7 | ABCA4, CACNA2D4, CERKL, CNGB3, KCNV2, RDH5, RPGRIP1 |
| Cone or cone-rod dystrophy, XL | 2 | CACNA1F, RPGR |
| Congenital stationary night blindness, AD | 3 | GNAT1, PDE6B, RHO |
| Congenital stationary night blindness, AR | 5 | CABP4, GRK1, GRM6, RDH5, SAG |
| Congenital stationary night blindness, XL | 2 | CACNA1F, NYX |
| Deafness alone or syndromic, AD | 1 | MYO7A† |
| Deafness alone or syndromic, AR | 4 | CDH23, MYO7A, PCDH15, USH1C |
| Leber congenital amaurosis, AD | 2 | CRX, IMPDH1 |
| Leber congenital amaurosis, AR | 11 | AIPL1, CEP290, CRB1, CRX, GUCY2D, LRAT, RD3, RDH12, RPE65, RPGRIP1, TULP1 |
| Macular degeneration, AD | 10 | BEST1, C1QTNF5, EFEMP1, ELOVL4, FSCN2, GUCA1B, HMCN1, PROM1, PRPH2, TIMP3 |
| Macular degeneration, AR | 2 | ABCA4, CFH |
| Macular degeneration, XL | 1 | RPGR |
| Macular degeneration, age related | 7 | ABCA4,†ARMS2,†CFH,†FBLN5,†HMCN1,†HTRA1,†TLR4† |
| Retinitis pigmentosa, AD | 20 | ABCA4,†BEST1, CA4, CRX, FSCN2, GUCA1B, IMPDH1, NR2E3, NRL, PRPF3, PRPF8, PRPF31, PRPH2, RDH12, RGR,†RHO, ROM1, RP1, RP9, SEMA4A |
| Retinitis pigmentosa, AR | 22 | ABCA4, CERKL, CNGA1, CNGB1, CRB1, LRAT, MERTK, NR2E3, NRL, PDE6A, PDE6B, PROM1, RGR, RHO, RLBP1, RP1, RPE65, SAG, SEMA4A,†TTC8, TULP1, USH2A |
| Retinitis pigmentosa, XL | 2 | RP2, RPGR |
| Syndromic/systemic diseases with retinopathy, AR | 2 | CEP290, LRP5 |
| Usher syndrome, AR | 8 | CDH23, CLRN1, GPR98, MYO7A, PCDH15, USH1C, USH1G, USH2A |
| Other retinopathy, AD | 4 | BEST1, CRB1, FZD4, LRP5 |
| Other retinopathy, AR | 10 | BEST1, CDH3, CNGB3, CYP4V2, LRP5, NR2E3, OAT, PROM1, RBP4, RLBP1 |
| Other retinopathy, XL | 4 | CACNA1F, CHM, NDP, RS1 |
Genes in bold are associated with at least two phenotypes. AD, autosomal dominant; AR, autosomal recessive; XL, X-linked.
GeneCards: http://www.genecards.org/index.shtml.
Development of a High-Throughput Resequencing System
A high-throughput experimental approach using the Retina-Array has been established in this study. The protocol includes an automated PCR assay setup using robots, automated PCR product quantification using a high-throughput system (LabChip 90; Caliper Life Sciences), efficient post-reading data processing (Excel; Microsoft), optimized pooling and fragmentation strategies, and standardized chip processing procedures. A total of 605 PCR assays, including 235 short-range (≤1500 bp), 355 long-range (>1500–6000 bp), and 15 GC-rich (variable sizes) PCR assays were designed (Supplementary Table S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7978/-/DCSupplemental). Each pair of primers was individually tested, and each amplicon was verified by dideoxy sequencing. A high-throughput PCR setup using a 96-well plate format was developed and optimized. All PCR assay results for each individual sample were documented by the high-throughput system (LabChip 90; Caliper Life Sciences) (Supplementary Figs. S1B, S1C, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7978/-/DCSupplemental). The size and concentration of each amplicon were imported into data processing (Excel; Microsoft) files for post-quantification data processing and afterward post-GSEQ data analysis. In this pilot study, approximately 22.8% of the 605 PCR reactions had no detectable products, at least for two samples; most of these were long-range and GC-rich PCR assays. Our experimental data indicated that the strategy of pooling and fragmentation used in this study works reliably and is more efficient than the standard Affymetrix resequencing array protocol (GeneChip CustomSeq, version 2.1; P/N 701231 revision 5), especially with the use of the high-density chip format and several hundred PCR assays performed (data not shown).
Evaluation of the Retina-Array
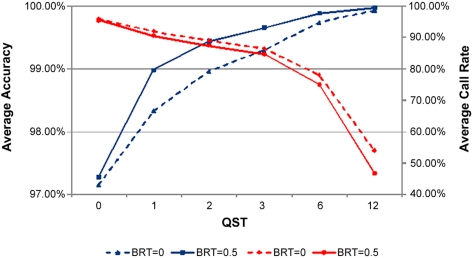
The performance of the Retina-Array chip was evaluated in several ways. First, the average call rate and accuracy of base calling across the board were assessed under several conditions. The default settings of QST at 3 and BRT at 0.5 optimized the combined call rate and accuracy, giving an average call rate of 84.65% and an average accuracy of 99.66% (Fig. 1). These were therefore used in the subsequent post-GSEQ data analysis. For the IQ-EX positive control, an average call rate of 92.35% and an average accuracy of 99.95% were observed under the default settings.
Figure 1.
Average call rates and average accuracies with different GSEQ parameter settings. Average call rate (red) represents total bases called (excluding no calls)/total bases interrogated by array across the board; average accuracy (blue) represents correct base calls/total bases called (excluding no calls) across the board. QST, quality score threshold; BRT, base reliability threshold across samples. Different settings of QST and BRT in the GSEQ data analysis affected the average base call rate and average accuracy. The average base call rate and average accuracy of the Retina-Array ranged from 95.79% and 97.15% under the settings of QST at 0 and BRT at 0 to 46.77% and 99.97% under the settings of QST at 12 and BRT at 0.5. The default settings of QST at 3 and BRT at 0.5 gave an average call rate of 84.65%, and an average accuracy of 99.66% was used in the subsequent post-GSEQ data analysis.
Second, two user-defined criteria, the PCR assay filter and the call rate filter, were used to further assess the actual performance of the chips in this study. Under the user-defined criteria, 181,897 bp (1050 exon fragments, ∼70% of the tiled exon fragments) per array were efficiently sequenced, giving an average call rate of 93.56% (84.27% ∼ 96.54%) with an average accuracy of 99.86% (99.70% ∼ 99.93%). In contrast, without filtering by user-defined criteria, 267,550 bp (1508 exon fragments) per array were originally sequenced, giving an average call rate of 84.79% (75.55% ∼ 89.39%) and an average accuracy of 99.69% (99.30% ∼ 99.84%) (Supplementary Table S2, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7978/-/DCSupplemental).
Third, to check the pick sensitivity of the chips for the detection of previously reported sequence alterations in this group of patient samples, supervised data analysis of both automatic GSEQ base calling and manual visual checking was performed. Of the 18 previously reported sequence alterations in ABCA4 for STGD and CHM for CHM patient samples, six presumably nonpathogenic polymorphisms were located in intronic sequences and thus fall outside the designed detection scope of the array. Among the remaining 12 sequence alterations in the target region, nine (seven heterozygous, one homozygous, and one hemizygous) were detected correctly, whereas three gave an N call, suggesting that approximately 75% of previously reported sequence alterations were correctly detected by the Retina-Array chips (Supplementary Table S3, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7978/-/DCSupplemental). The remaining three were identified as potential problems, but no specific base change was identified.
Performance of Bioinformatic Filters
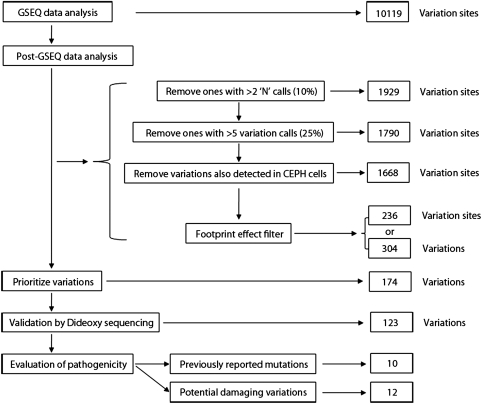
To efficiently screen the candidate variations and identify the most reliable sequence changes, a bioinformatic pipeline, including a series of custom-screening filters, was developed and assessed for the post-GSEQ data analysis (Fig. 2). The first filter applied, referred to as the overall quality filter, eliminated unreliable variations that could be attributed to PCR failures, poor hybridization, nonspecific hybridization, or system errors resulting from chip fabrication. This filtering step led to a significant reduction in the candidate variation sites from the 10,119 originally derived from GSEQ data analysis to 1668. The second filter, referred to as the footprint effect filter or nearby SNP effect filter, assumed that a true variation is most likely to induce false-variation calls at locations within nine bases on either side of the true variation. This filtering step further decreased the number of candidate variation sites to 236. A number of identical variations were detected in multiple DNA samples in our study (Table 2). A total of 13,861 variations were initially identified by the Retina-Array chips using the GSEQ under the default settings, and the average number of variations detected in each individual sample was 730. After the post-GSEQ data analysis using custom screening filtering, a total of 304 candidate variations, averaging 16 in each patient sample, were finally accepted from the initial list (Fig. 2, Table 2).
Figure 2.
Flow chart of the variation analysis.
Table 2.
Variations Identified by Retina-Array before and after Validation by Dideoxy Sequencing
| Variations | Missense | Silent | Nonsense | Splice Site | 5′ UTR | Total (%) |
|---|---|---|---|---|---|---|
| All | ||||||
| Known SNP | 50 | 69 | 0 | 11 | 1 | 131 (42.1) |
| Novel variation | 104 | 55 | 4 | 10 | 0 | 173 (56.9) |
| Total (%) | 154* (50.7) | 124* (40.8) | 4 (1.3) | 21* (6.9) | 1 (0.3) | 304* |
| Unique | ||||||
| Known SNP | 37 | 36 | 0 | 6 | 1 | 80 (33.9) |
| Novel variation | 91 | 51 | 4 | 10 | 0 | 156 (66.1) |
| Total (%) | 128 (54.2) | 87 (36.9) | 4 (1.7) | 16 (6.8) | 1 (0.4) | 236 |
| Selected for validation (confirmed/ not confirmed) | ||||||
| Known SNP | 45/0 | 7/0 | 0/0 | 4/0 | 1/0 | 57/0 |
| Novel variation | 47/43 | 14/0 | 2/2 | 3/6 | 0/0 | 66/51 |
| Total (%) | 135* (77.6) | 21* (12.1) | 4 (2.3) | 13 (7.5) | 1 (0.6) | 174 |
Some variations were detected in multiple DNA samples.
Identification and Validation of Known and Novel Variations
Among 304 variants or 236 unique variations identified by the Retina-Array chips, there are 80 (33.9%) known SNPs and 156 (66.1%) novel variations, respectively. These include 128 (54.2%) missense, 4 (1.7%) nonsense, 87 (36.9%) silent, 16 (6.8%) splice-site, and 1 (0.4%) 5′ untranslated region variations (Table 2).
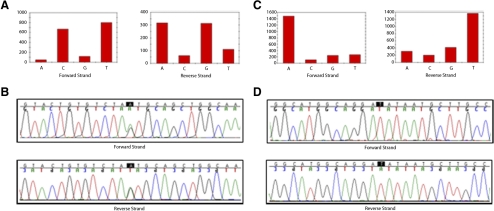
The variations detected by the Retina-Array chips were independently validated by dideoxy sequencing using the same batch of DNA samples (Table 2). Variations selected for validation were reported mutations, expected deleterious variations, missense variations, and some synonymous or benign variations. Of 174 variations selected for validation, including 135 (77.6%) missense, 4 (2.3%) nonsense, 21 (12.1%) silent, 13 (7.5%) splice-site, and 1 (0.6%) 5′ untranslated region variation, 123 (70.7%) were confirmed, including 92 missense, 2 nonsense, 21 silent, 7 splice-site, and 1 5′ untranslated region variations (Table 2). This number included all 57 known SNPs (57/57, 100% confirmed) and 66 novel variations (66/117, 56.4% confirmed; Table 2). Fifty-one novel variations could not be confirmed by sequencing, consistent with the much lower a priori odds that they would be real. This number included 120 heterozygous (114/120, 95% correctly detected by the chips; 6/120, 5% reported in the homozygous state by the chips), 2 homozygous (2/2, 100% correctly detected by the chips), and 1 hemizygous variation (1/1, 100% correctly detected by the chips). Complete information about the 123 variations validated by dideoxy sequencing—including gene name, reference sequence ID, nucleotide change, amino acid change, genotype, variation category, SNP ID, a reference to the previously reported mutation, PolyPhen-2 score, and SIFT score—is shown in Supplementary Table S4, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7978/-/DCSupplemental. Two examples of typical variations detected by the Retina-Array and validated by dideoxy sequencing, including one known heterozygous variation and one novel hemizygous variation, are shown in Figures 3A to 3D.
Figure 3.
Validation of Retina-Array data by dideoxy sequencing. Two examples of variations identified by the Retina-Array and validated by dideoxy sequencing. (A, B) Known variations (rs41317471, c.4586A>G, p.Asn1529Ser) identified in the HMCN1gene (NM_031935.2) in the heterozygous state in patient 17. (C, D) Novel variations (c.4161G>T, p.Glu1387Asp) identified in the CACNA1F gene (NM_005183.2) on the X chromosome in a hemizygous state in patient 6. (A, C) Resequencing chip data. (B, D) Dideoxy sequencing data. The highlighted base indicates the variation location.
Evaluation of Potential Pathogenic Variations
A series of computational analyses was adopted to evaluate the potential pathogenic effects of the 123 validated variations (Supplementary Table S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7978/-/DCSupplemental). Some of the previously reported mutations, such as rs696723, rs4986791, and rs41281314, are likely to be benign SNPs because of the minor allele frequency of 5%. Unfortunately, because of the manner of recruitment, we could not examine the inheritance of the sequence changes in parents and siblings of the patients. Ten previously reported mutations were summarized in Table 3. Among the potential novel pathogenic variations, there are two nonsense variants (PROM1, c.1557C>A, p.Tyr519Ter, patient 10, CRD; ABCA4, c.3595C>T, p.Gln1199Ter, patient 13, STGD), one splice-site variant (USH2A, c.14,792–2A>G, patient 2, RP), and one 5′ untranslated region variant (KCNV2, c.-2C>T, patient 9, CRD). The genes in which these identified variants occur have an established relationship with the clinical presentations of the patients; hence, these variations are very likely pathogenic mutations (Table 3). Five variations were consistently computationally predicted as damaging using both PolyPhen-2 and SIFT. In addition, three variations were computationally predicted as probably damaging using either Polyphen-2 or SIFT only (Table 3; Supplementary Table S4, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7978/-/DCSupplemental). In addition, we observed a high frequency of USH2A gene rare variations (Supplementary Table S4, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7978/-/DCSupplemental). There were 13 USH2A alleles in 9 patients from four disease categories. Some of these variants have been reported in linkage disequilibrium with USH2A deletions, which would not be detectable by this method. It is still not clear whether some of these variants have pathogenic effects.22
Table 3.
Ten Previously Reported Mutations and 12 Potential Novel Pathogenic Variations
| Patient ID | Sex | Diagnosis | Gene | Nucleotide Change | Amino Acid Change | HGMD No. | PolyPhen-2 | SIFT | SNP ID |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Male | RP | TLR4 | c.842G>A | p.Cys281Tyr | CM035921 | Poss/0.900 | Tolerated/0.25 | |
| 2 | Female | RP | USH2A | c.14792–2A>G | Novel | ||||
| 3 | Female | RP | CNGB1 | c.2747G>A | p.Arg916His | Novel | Prob/0.994 | Damaging/0.00 | |
| RPGR | c.223A>G | p.Ile75Val | CM011007 | Benign/0.015 | Tolerated/1.00 | ||||
| 19 | Male | RP | BEST1 | c.727G>A | p.Ala243Thr | CM004434 | Prob/0.946 | Damaging/0.01 | |
| 5 | Female | Usher syndrome | GUCY2D | c.1720C>T | p.Arg574Cys | Novel | Prob/0.933 | Damaging/0.02 | |
| LRAT | c.74T>A | p.Phe25Tyr | Novel | Prob/0.98 | Tolerated/0.11 | rs75368761 | |||
| GUCA1B | c.253G>A | p.Val85Met | CM045052 | Benign/0.023 | Damaging/0.02 | ||||
| ABCA4 | c.6320G>A | p.Arg2107His | CM990074 | Prob/0.996 | Damaging/0.00 | rs62642564 | |||
| 6 | Male | Usher syndrome | CACNA1F | c.4161G>T | p.Glu1387Asp | Novel | Benign/0.266 | Damaging/0.00 | |
| TRIM32 | c.1222C>T | p.Arg408Cys | Novel | Poss/0.795 | Damaging/0.04 | rs3747835 | |||
| 7 | Female | Usher syndrome | USH2A | c.4714C>T | p.Leu1572Phe | Novel | Poss/0.673 | Damaging/0.01 | |
| 8 | Male | Usher syndrome | RPGRIP1 | c.3358A>G | p.Ile1120Val | CM076486 | Poss/0.833 | Tolerated/0.09 | |
| 9 | Female | CRD | KCNV2 | c.−2C>T | Novel | rs75316505 | |||
| BBS12 | c.1381A>C | p.Asn461His | Novel | Poss/0.706 | Damaging/0.01 | rs10027479 | |||
| 10 | Male | CRD | PROM1 | c.1557C>A | p.Tyr519Ter | Novel | |||
| ABCA4 | c.4297G>A | p.Val1433Ile | CM990050 | Benign/0.112 | Tolerated/0.09 | rs56357060 | |||
| 18 | Male | CRD | ABCA4 | c.4793C>A | p.Ala1598Asp | CM003386 | Poss/0.638 | Damaging/0.01 | rs61750155 |
| 13 | Male | STGD | ABCA4 | c.3595C>T | p.Gln1199Ter | Novel | |||
| 16 | Female | STGD | ABCA4 | c.5882G>A | p.Gly1961Glu | CM970016 | Prob/1.000 | Damaging/0.00 | rs1800553 |
| HMCN1 | c.5482A>G | p.Ile1828Val | Novel | Prob/0.896 | Tolerated/0.4 | ||||
| 14 | Male | CHM | BBS5 | c.551A>G | p.Asn184Ser | CM044580 | Prob/0.986 | Damaging/0.00 |
All mutations/variations were identified in their respective genes in the heterozygous state except one variation on CACNA1F in the hemizygous state. Genes in bold represent the known genes involved in their respective diseases. Patients 4, 11, 12, 15, and 17 were not included because no variations were listed in the table.
Overall, this study identified informative sequence changes in at least 15 of 19 patients (79%), including patients 1 to 3, 5 to 10, 13 to 16, 18, and 19. Based on the established pattern of inheritance, the significant nature of the sequence changes, and an agreement between the gene and the corresponding clinical disease, mutations found in patients 5, 10, 14, 15, and 19 are likely to provide a valid clinical molecular diagnosis. Mutations found in patients 2, 3, 9, 13, 16, and 18 revealed strong support for additional analysis of the genes in which they occur. Mutation linkage disequilibrium information strongly suggested a potential target gene for further genetic analysis in patient 7.
Discussion
Many RD patients do not have sufficient family history to reveal a definitive inheritance pattern. Therefore, it is often difficult or impossible to identify a specific target gene or a gene panel for genetic analysis. Retina-Array makes it feasible to rapidly screen DNA samples for both known and novel mutations causing the disease and for the disease severity modifiers in the other genes in a single experiment. We randomly selected 19 patients from the eyeGENE registry for this study. These patients represented five categories of clinical presentations. Although four patients had a clinical diagnosis of either STGD or CHM, the remaining 15 patients did not have sufficient information to suggest a target gene or a gene panel for analysis. Using the Retina-Array, this study identified at least 10 previously reported mutations and at least 12 potential novel variations with a high probability of deleterious effects in 15 of the 19 patients. Thus, this approach shows promise as a screening tool when followed by confirmatory dideoxy sequencing for the final clinical diagnosis and brings in challenges for molecular diagnosis.
The advantages of the chip-based assay include its simplicity, accuracy, efficiency, and cost-effectiveness when compared with other large-scale sequencing platforms, although the detection of insertions, deletions, and inversions remains a challenge for this approach. Next-generation sequencing technology has been intensively used in recent studies of human disease and has the potential for routine testing in a clinical setting; however, data handling and data mining associated with this technology remain significant challenges.8,9 Although chip-based assays are less daunting than next-generation sequencing in terms of their bioinformatic challenges, they remain limited to the detection of mutations in genes identified as causes of retinal degeneration. The chip-based assay approach is especially useful for medium-sized applications of targeted disease sequencing. Retina-Array provides the highest density among resequencing chips available for the detection of sequence changes in multiple genes related to inherited RD.13,23,24
The performance of the chips with the average base pair call rate of 93.56% and the average accuracy of 99.86% is similar to that obtained with other customized resequencing chips.10,21,25 Although chip design and fabrication were critical for mutation detection, efficient PCR amplification, PCR quantification, practical PCR product pooling, and proper DNA fragmentation all were important components of this approach. Approximately 22.8% of PCR assays were inefficient or failed in this pilot study. This contributed significantly to N calls and false-positive calls, emphasizing that further optimization of the PCR assays could provide better coverage of sequence analysis. Although some of the failures can be fixed by redesigning the PCR primers and reactions, there will be a number for which the genomic context itself makes amplification difficult, and these will be resistant to improvement. With further development, we are confident that the amplification rate can be improved. The high-throughput system (LabChip 90; Caliper Life Sciences) efficiently identified and documented these results and assisted not only in pooling the PCR products but also in the post-GSEQ data analysis. All calls that originated from a failed PCR amplification, which could be easily identified either by checking the system reading data or examining call rate as a function of the PCR amplicon in a specific sample, were removed to ensure reliable and specific identification of variations. Based on the results of this study, the current cost of $1500 per sample, which includes chip, primers, reagents, and consumable materials, could be reduced to less than $1000 per sample as the PCR reaction volume is scaled down.
Although improved resequencing array base-calling algorithms greatly minimize false-negative calls, bioinformatic tools are essential to decrease false-positive calls and to increase the overall accuracy of this platform. Several major factors, including PCR failures, nearby SNP effects, cross-hybridization, low-sequence complexity, and non-biallelic calls, have been shown to be responsible for the majority of false-positive calls (http://media.affymetrix.com/support/technical/technotes/customseq_arraybase_technote.pdf). Additional computational methods and bioinformatic tools focusing on different aspects of this problem have been developed recently.10,20,21 In this study, a bioinformatic pipeline, including a series of custom screening filters, was developed to systematically remove false-positive calls and to ensure the most reliable identification of sequence variations in patient samples. Through this strategy, a total of 304 candidate variations with an average of 16 were identified in each patient sample. However, unsupervised data analysis did not reveal complete mutation information in patients 14, 15, and 16. Given that these patients were previously analyzed for their target genes and that our supervised data analysis confirmed the mutation status except for a single ambiguous call at ABCA4 c.1A>G, further study with a larger cohort analyzing additional filtering strategies and experimental procedures may help to evaluate the false-negative rate of this approach.
In conclusion, in this study a custom-designed, microarray-based, resequencing system has been developed and validated for the detection of sequence changes in 93 genes involved in inherited RD. This array was demonstrated to be a valuable screening tool for the high-content detection of both known and novel mutations in a single experiment. We believe that Retina-Array could also prove a valuable resource to explore phenotype-genotype relationships and gene-gene and gene-environment interactions. Further evaluation using a larger number of patients to improve the procedure and analysis strategy is warranted.
Supplementary Material
Acknowledgments
The authors thank the staff at the National Institutes of Health/National Eye Institute eyeGENE Coordinating Center and Kerry Goetz, Vida Ndifor, Matthew Brooks, Christian Antolik, and Anand Swaroop for their technical and scientific help during the course of this study.
Footnotes
Supported by the Department of Health and Human Services/National Institutes of Health/National Eye Institute extramural, clinical, and intramural programs.
Disclosure: J. Song, None; N. Smaoui, None; R. Ayyagari, None; D. Stiles, None; S. Benhamed, None; J.I.M. MacDonald, None; S.P. Daiger, None; S.J. Tumminia, None; F. Hejtmancik, None; X. Wang, None
References
- 1. Rivolta C, Sharon D, DeAngelis MM, et al. Retinitis pigmentosa and allied diseases: numerous diseases, genes, and inheritance patterns. Hum Mol Genet. 2002;11:1219–1227 [DOI] [PubMed] [Google Scholar]
- 2. Hamel CP. Cone rod dystrophies. Orphanet J Rare Dis. 2007;2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Daiger SP. Identifying retinal disease genes: how far have we come, how far do we have to go? Novartis Found Symp. 2004;255: 17–27; discussion 27–36, 177–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. MacDonald IM, Tran M, Musarella MA. Ocular genetics: current understanding. Surv Ophthalmol. 2004;49:159–196 [DOI] [PubMed] [Google Scholar]
- 5. Saihan Z, Webster AR, Luxon L, et al. Update on Usher syndrome. Curr Opin Neurol. 2009;22:19–27 [DOI] [PubMed] [Google Scholar]
- 6. Downs K, Zacks DN, Caruso R, et al. Molecular testing for hereditary retinal disease as part of clinical care. Arch Ophthalmol. 2007;125:252–258 [DOI] [PubMed] [Google Scholar]
- 7. Brooks BP, Macdonald IM, Tumminia SJ, et al. Genomics in the era of molecular ophthalmology: reflections on the National Ophthalmic Disease Genotyping Network (eyeGENE). Arch Ophthalmol. 2008;126:424–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bowne SJ, Sullivan LS, Koboldt DC, et al. Identification of disease-causing mutations in autosomal dominant retinitis pigmentosa (adRP) using next-generation DNA sequencing. Invest Ophthalmol Vis Sci. 2011;52:494–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145 [DOI] [PubMed] [Google Scholar]
- 10. Kothiyal P, Cox S, Ebert J, et al. High-throughput detection of mutations responsible for childhood hearing loss using resequencing microarrays. BMC Biotechnol. 2010;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hacia JG. Resequencing and mutational analysis using oligonucleotide microarrays. Nat Genet. 1999;21:42–47 [DOI] [PubMed] [Google Scholar]
- 12. Maitra A, Cohen Y, Gillespie SE, et al. The Human MitoChip: a high-throughput sequencing microarray for mitochondrial mutation detection. Genome Res. 2004;14:812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mandal MN, Heckenlively JR, Burch T, et al. Sequencing arrays for screening multiple genes associated with early-onset human retinal degenerations on a high-throughput platform. Invest Ophthalmol Vis Sci. 2005;46:3355–3362 [DOI] [PubMed] [Google Scholar]
- 14. Lin B, Wang Z, Vora GJ, et al. Broad-spectrum respiratory tract pathogen identification using resequencing DNA microarrays. Genome Res. 2006;16:527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takahashi Y, Seki N, Ishiura H, et al. Development of a high-throughput microarray-based resequencing system for neurological disorders and its application to molecular genetics of amyotrophic lateral sclerosis. Arch Neurol. 2008;65:1326–1332 [DOI] [PubMed] [Google Scholar]
- 16. Waldmuller S, Muller M, Rackebrandt K, et al. Array-based resequencing assay for mutations causing hypertrophic cardiomyopathy. Clin Chem. 2008;54:682–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hartmann A, Thieme M, Nanduri LK, et al. Validation of microarray-based resequencing of 93 worldwide mitochondrial genomes. Hum Mutat. 2009;30:115–122 [DOI] [PubMed] [Google Scholar]
- 18. Leski TA, Lin B, Malanoski AP, et al. Testing and validation of high density resequencing microarray for broad range biothreat agents detection. PLoS One. 2009;4:e6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cutler DJ, Zwick ME, Carrasquillo MM, et al. High-throughput variation detection and genotyping using microarrays. Genome Res. 2001;11:1913–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pandya GA, Holmes MH, Sunkara S, et al. A bioinformatic filter for improved base-call accuracy and polymorphism detection using the Affymetrix GeneChip whole-genome resequencing platform. Nucleic Acids Res. 2007;35:e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang HY, Gopalan V, Aksentijevich I, et al. A custom 148 gene-based resequencing chip and the SNP explorer software: new tools to study antibody deficiency. Hum Mutat. 2010;31:1080–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McGee TL, Seyedahmadi BJ, Sweeney MO, et al. Novel mutations in the long isoform of the USH2A gene in patients with Usher syndrome type II or non-syndromic retinitis pigmentosa. J Med Genet. 2010;47:499–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Clark GR, Crowe P, Muszynska D, et al. Development of a diagnostic genetic test for simplex and autosomal recessive retinitis pigmentosa. Ophthalmology. 2010;117:2169–2177, e2163 [DOI] [PubMed] [Google Scholar]
- 24. Booij JC, Bakker A, Kulumbetova J, et al. Simultaneous mutation detection in 90 retinal disease genes in multiple patients using a custom-designed 300-kb retinal resequencing chip. Ophthalmology. 2011;118:160–167, e163 [DOI] [PubMed] [Google Scholar]
- 25. Bruce CK, Smith M, Rahman F, et al. Design and validation of a metabolic disorder resequencing microarray (BRUM1). Hum Mutat. 2010;31:858–865 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.