The partial genomes of seven ocular Herpes simplex type-1 virus isolates were sequenced in a multiplex format. The data will be useful for studies of virulence and population structure and for the design of mutagenesis strategies for structure–function studies of each of the proteins.
Abstract
Purpose.
Little is known about the role of sequence variation in the pathology of HSV-1 keratitis virus. The goal was to show that a multiplex, high-throughput genome-sequencing approach is feasible for simultaneously sequencing seven HSV-1 ocular strains.
Methods.
A genome sequencer was used to sequence the HSV-1 ocular isolates TFT401, 134, CJ311, CJ360, CJ394, CJ970, and OD4, in a single lane. Reads were mapped to the HSV-1 strain 17 reference genome by high-speed sequencing. ClustalW was used for alignment, and the Mega 4 package was used for phylogenetic analysis (www.megasoftware.net). Simplot was used to compare genetic variability and high-speed sequencing was used to identify SNPs (developed by Stuart Ray, Johns Hopkins University School of Medicine, Baltimore, MD, http://sray.med.som.jhml.edu/SCRoftware/simplot).
Results.
Approximately 95% to 99% of the seven genomes were sequenced in a single lane with average coverage ranging from 224 to 1345. Phylogenetic analysis of the sequenced genome regions revealed at least three clades. Each strain had approximately 200 coding SNPs compared to strain 17, and these were evenly spaced along the genomes. Four genes were highly conserved, and six were more variable. Reduced coverage was obtained in the highly GC-rich terminal repeat regions.
Conclusions.
Multiplex sequencing is a cost-effective way to obtain the genomic sequences of ocular HSV-1 isolates with sufficient coverage of the unique regions for genomic analysis. The number of SNPs and their distribution will be useful for analyzing the genetics of virulence, and the sequence data will be useful for studying HSV-1 evolution and for the design of structure–function studies.
Herpes simplex virus (HSV)-1 is a significant human pathogen causing diseases such as mucocutaneous ulcers, keratitis, and encephalitis. In the United States, HSV-1 keratitis is the leading cause of blindness due to infections and the leading cause of sporadic encephalitis.1,2 Studies in animal models have shown that the severity of an HSV-1 infection depends on three factors. The first is the innate resistance of the host. Strains of mice vary widely in their susceptibility, and some host genes involved in this innate resistance have been identified.3–11 The second factor is the host immune response. Animals with various defects in innate and acquired immunity have difficulty in controlling the virus, resulting in lethal infections.12,13 The host immune response is important in blinding keratitis, as corneal damage is due to an immunopathologic response.14–16
The third factor is the genetic makeup of the virus. Strains of HSV-1 display virulence patterns in mice ranging from no disease to lethal encephalitis.17,18 The severity of keratitis also varies widely between strains, but the genetic basis for these differences is poorly understood. Deletion of an entire gene from the virus can have significant effects on virulence in animal models, but in nature, it is more likely that virulence differences are due to effects of multiple genes and the combination of alleles carried by a given strain of virus. Studies on the genetic basis of virulence would be facilitated if additional genomic sequence data were available to enhance targeted mutagenesis strategies for studying the structure and function of viral genes.
Although the sequence of one complete HSV-1 genome has been available for some time19–22 and two more genomes were recently sequenced,23 little is known about the total sequence divergence of HSV-1. The genome of HSV-1 is approximately 152,000 base pairs, with a GC content of 68%. The genome is divided into unique long and unique short regions, each of which is flanked by long inverted repeats. Seventy-seven protein-coding open reading frames have been annotated to date. Variability in the length of the genome of individual strains is due to the presence of shorter repeated elements, including microsatellite repeats up to 100 bases long and tandemly reiterated sequences up to 500 bases long. These longer repeats are denoted as variable-number tandem repeats (VNTRs), and the number in any given strain of virus can vary. The advent of high-throughput sequencing platforms has made it feasible to sequence larger numbers of HSV-1 genomes to get a more complete picture of the sequence diversity and population structure of the virus.
We previously described the virulence properties of several ocular isolates of HSV-124 and demonstrated that recombinants between three of these strains, OD4, CJ394, and 994, generated viruses with a wide range of virulence phenotypes.25,26 In addition we isolated and characterized several OD4/CJ394 recombinants and showed that the transfer of different combinations of genes from strain CJ394 to OD4 resulted in either increased ocular virulence or increased ocular and neurovirulence.25,27 Sequencing of these recombinant genomes26 has the potential to quickly identify virulence determinants.
We report the partial genomic sequences of seven ocular isolates of HSV-1. All the strains were sequenced in a single lane of a genome sequencer (GAIIx flowcell; Illumina), and the results showed that this multiplexing strategy results in high coverage of the unique regions of the genome. Analysis of the data revealed that HSV-1 separated into at least three clades, supporting results in previous studies, and that certain genes are highly conserved while others have higher variability. There were approximately 200 coding single-nucleotide polymorphisms (SNPs) in each genome compared to strain 17 and they are evenly spaced across the genomes. We also show that variability was distributed across some genes, while in other genes it was localized to specific regions. Coverage was low in the repeat sequences as a result of high GC content and the repetitive nature of the sequences, but useful data were obtained from these regions.
Materials and Methods
Cell Culture
Vero cells were grown in Dulbecco's modified Eagle's medium (DMEM) with 5% serum and antibiotics, as described previously.24 For genomic DNA isolation, infections were performed in DMEM supplemented with 2% serum and antibiotics.
HSV-1 Viral Strains
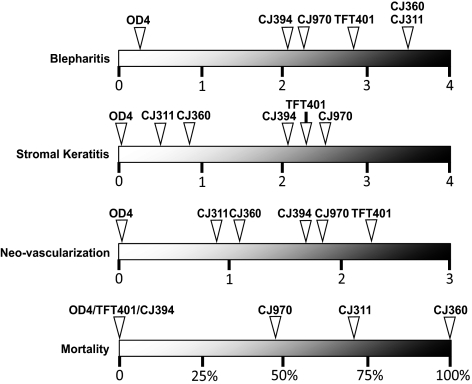
The viral strains used for this study were derived from plaque purified ocular clinical isolates, originally collected in Seattle, Washington. The ocular disease phenotypes of HSV-1 strains TFT401, CJ311, CJ360, CJ394, CJ970, 134, and OD4 have been reported,24,25 and a visual summary is found in Figure 1. Briefly, HSV-1 strain TFT401 causes stromal keratitis in mice. Viral strains CJ311 and CJ360 are highly neurovirulent in mice with 70% and 100% mortality respectively, due to encephalitis. Mice infected with strain CJ394 exhibit moderate keratitis and vascularization. Strain CJ970 infection results in severe stromal keratitis and corneal neovascularization with 50% mortality. Strain OD4 contains multiple attenuating mutations and is avirulent, even in nude mice24,27 (CRB, unpublished data, 1996). The virulence phenotype of strain 134 has not yet been evaluated in animals.
Figure 1.
Schematic summarizing the mean peak ocular disease scores for blepharitis, stromal keratitis, neovascularization, and percent mortality of 4- to 6-week-old Balb/c mice infected with viral strains TFT401, CJ311, CJ360, CJ394, CJ970, and OD4. The scoring system and virulence data were presented in a previous publication.24 The virulence characteristics of 134 have not yet been determined.
Viral DNA Preparation
For high-throughput genome sequencing, viral DNA was prepared by using a modification of a previously described DNA isolation protocol.28 Briefly, 20 confluent 10-cm plates of Vero cells were infected at a multiplicity of infection (MOI) of 0.1. Twenty-four hours after the cells reached 100% cytopathic effect (CPE), they were scraped and then centrifuged at 2000g for 10 minutes at 4°C. The cell pellet was resuspended in 5 mL of medium, subjected to three freeze–thaw cycles (−80°C/37°C), and then centrifuged at 2000g to remove debris. The supernatants were then combined, layered onto a 36% sucrose cushion in PBS and then centrifuged for 80 minutes at 24,451g in a rotor (SW28; Beckman Instruments, Fullerton, CA). The resulting pellet was resuspended in 5 mL of PBS, applied to another 36% sucrose cushion in PBS and then centrifuged 80 minutes at 26,295g in another rotor (SW41; Beckman). The viral pellet was resuspended in 5 mL of TE buffer (10 mM Tris [pH 7.4], 1 mM EDTA) with 0.15 M sodium acetate and 50 μg/mL RNAase A, and incubated for 30 minutes at 37°C. Proteinase K and SDS (50 μg/mL and 0.1%, respectively) were then added, and the solution was incubated for 30 minutes at 37°C. The viral DNA was then purified by phenol:chloroform extraction and ethanol precipitation, resuspended in deionized water, and stored at −20°C.
Construction and Sequencing of Gene Libraries
A total of 5 μg of high-quality genomic DNA for each strain was submitted to the University of Wisconsin-Madison DNA Sequencing Facility for paired-end library preparation. The DNA was divided in two for duplicate library preparation and each library was generated using a paired-end sample preparation kit (Illumina Inc., San Diego, CA) with the following modifications: Paired-end adapters and primers were replaced with adapters, primers, and indexing primers (Multiplexing Sample Preparation Oligo Kit; Illumina, Inc.). Products of the ligation reaction were purified by gel electrophoresis, using 2% agarose gels (SizeSelect; Invitrogen, Carlsbad, CA) targeting 325-bp fragments. The quality and quantity of the DNA was assessed with a chip assay (DNA 1000 series; Agilent, Palo Alto, CA) and a dsDNA kit (QuantIT PicoGreen dsDNA Kit; Invitrogen), respectively, and libraries were standardized to 10 nM. Cluster generation was performed (Paired-End Cluster Generation Kit [ver. 4] and the Cluster Station; Illumina, Inc.). All seven HSV-1 genomic samples were run in a single lane of a standard flowcell (Illumina). A paired-end, 2 × 75-bp run was performed, using standard 36-bp cluster sequencing kits (SBS, ver. 4 and SCS 2.6 software), on a high throughput sequencer (GAIIx, Illumina). This generated two 76-bp sequences from the ends of each fragment. The images were then analyzed (Pipeline, ver. 1.6; Illumina).
Sequence Read Alignment
For each strain, the paired-end reads were aligned to the strain 17 HSV-1 genome (RefSeq Accession number NC_001806; National Center for Biotechnology Information, Bethesda MD, available at www.ncbi.nlm.nih.gov/locuslink/refseq) using the high-speed sequencing system (Genomics Workbench, ver 4.0; CLC-Bio Cambridge, MA). The system maps reads in a two-step process. In the first step, individual reads were mapped to the reference genome with a local alignment scheme. The alignments were performed with a mismatch cost of two and insertion–deletion costs of three. In the second step, putative alignments of individual reads to the reference were removed when there was no window of at least 50% of the read length that had at least 80% similarity to the reference genome. In cases where reads had bases outside of the window of local alignment, the read was not necessarily excluded from the final reference alignment, but those bases were excluded from any estimates of coverage or polymorphism. ClustalW29 from the Mega 4 package30 was used to create whole-genome multiple alignments of the genomic sequence of HSV-1 strain 17 and the ungapped consensus sequences from the reference assemblies of TFT401, 134, CJ311, CJ360, CJ394, CJ970, and OD4 (www.megasoftware.net).
SNP and INDEL Identification
High speed sequencing was used to detect single-nucleotide polymorphisms (SNPs) using the neighborhood quality standard algorithm.31,32 In order for a variable site to be declared an SNP, there had to be a minimum coverage of four reads spanning the position in the alignment of the reads to the reference genome. In addition, 35% of the reads had to differ from the reference, or there had to be a minimum of five reads with a variant base. However, if the particular variant base was detected in at least five reads, it was also regarded as an SNP candidate pending validation via visual inspection of the alignment. Potential SNPs were filtered on the basis of base-calling quality scores at the variant position and the five neighboring bases on either side of the position. To pass this step, the average quality at the polymorphic site within a read had to have a minimum Phred quality score of 20, and the 10 adjacent bases had to have an average quality score of 15.
Polymorphisms of INDELS of up to 10 bases were detected (DIP tool; Genomics Workbench). The procedure used is similar to that used for SNP detection, except that quality scores in the vicinity of INDELS are not taken into account, and there is no requirement for the minimum number of variants. As with the SNP detection, at least 35% of the reads had to differ from the reference, and there had to be a minimum coverage of four reads for a site to be regarded as polymorphic.
Correlation between SNPs and Disease Phenotype
To determine whether the observed SNPs correlate with the phenotypes of interest, we clustered the SNPs by gene and counted the number of mutations in a given gene for each strain. We ordered the strains by a chosen phenotype and determined whether such an ordering is consistent with the ordering induced by the SNP counts (i.e., when we order strains by phenotype, the associated mutation counts are consistent if they are monotonically nonincreasing or nondecreasing). Under the null hypothesis that each possible ordering of the strains is equally likely, we used an exact permutation test to calculate how probable it is that we would get an ordering of mutation counts that is consistent with the phenotype ordering by chance. We adjusted for multiple comparisons using the Bonferroni correction.
Phylogenetic Analysis
The genomic sequences used for analysis were obtained from this work and from the NCBI Reference Database (www.NCBI.nlm.nih.gov; Bethesda, MD). The genomes of HSV-2 HG52, HSV-1 strain 17, H129, TFT401, 134, CJ311, CJ360, CJ394, CJ970, and OD4 were aligned with ClustalW. The Mega 4 package was used to construct a consensus bootstrapped (1000 replicates33) neighbor-joining tree.34 Branches corresponding to partitions reproduced in less than 50% bootstrap replicates were collapsed. The Tamura-Nei method35 was used to calculate evolutionary distance. A gamma distribution with a shape parameter of 1 was used to model rate variation among sites. All positions containing gaps and missing data were eliminated from the dataset. Phylogenetic trees inferred using the maximum composite likelihood and maximum parsimony methods produced similar results. For phylogenetic analysis to detect possible recombination, the UL1, UL19, UL29, UL42, UL53, and US12 genes from HSV-2 HG52 as well as the HSV-1 strains 17, F, H129, TFT401, 134, CJ311, CJ360, CJ394, CJ970, and OD4 were translated into their inferred amino acid sequences and aligned with ClustalW, and the phylogenetic analysis was performed as described for the entire genomes.
Genome Similarity
To examine genomic nucleotide variation, the online software package mVISTA Limited Area Global Alignment of Nucleotides (LAGAN) (http://genome.lbl.gov/vista/index.shtml) was used to scan the HSV-1 genomic sequences.36 Individual genomic sections were scanned for nucleotide variation using Simplot from the RDP3 software package (developed by Stuart Ray, Johns Hopkins University School of Medicine, Baltimore, MD, http://sray.med.som.jhml.edu/SCRoftware/simplot).37 The Kimura substitution model was used to perform the Simplot calculations.38
Results
Multiplexed High-Throughput Sequencing
Viral DNA from seven ocular HSV-1 isolates was purified using large-scale preparations. Genome libraries were generated, and each isolate was indexed for multiplexing. All seven samples were sequenced in a single lane of one run on the genome analyzer (paired-end, 75 bp length; GAIIx, Illumina). The number of total sequence reads for each genome ranged from 3,374,912 in strain 134 to 8,811,474 in strain OD4, with a total of 4.32 × 107 reads for the entire run (Table 1).
Table 1.
Multiplexed, High-Throughput Sequencing Statistics
| HSV-1 Strain | Total Sequence Reads | Reads Matching Strain 17 Reference n (%) | Reads Matching Human Genome n (%)* | Genome Assembly bp Size n (%)†‡ | Total Bases in ClustalW Alignment§‖ | Average¶ Coverage |
|---|---|---|---|---|---|---|
| TFT401 | 6,376,370 | 456,378 (7) | 5,129,056 (80) | 144,622 (95) | 152,315 | 224 |
| 134 | 3,374,912 | 838,330 (25) | 2,179,159 (65) | 149,697 (98) | 152,338 | 410 |
| CJ311 | 4,709,860 | 2,743,844 (58) | 1,653,018 (35) | 150,153 (98) | 152,352 | 1345 |
| CJ360 | 4,633,424 | 2,420,111 (52) | 1,901,988 (41) | 147,074 (96) | 152,317 | 1190 |
| CJ394 | 5,532,714 | 1,655,101 (30) | 3,316,019 (60) | 148,466 (97) | 152,331 | 813 |
| CJ970 | 5,557,308 | 1,260,259 (23) | 3,703,681 (67) | 149,127 (98) | 152,324 | 618 |
| OD4 | 8,811,474 | 1,131,373 (13) | 6,675,074 (76) | 150,381 (99) | 152,318 | 554 |
Total number of reads sequenced in run: 4.32 × 107.
The human genome (hg19; build 37) was used to map short base reads to determine host contamination.
The number of bases mapped to the HSV-1 strain 17 genome. Submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank; provided in the public domain by the National Center for Biotechnology Information, Bethesda, MD).
The approximate percentage of the HSV-1 genome sequenced for each strain.
The total number of bases in alignment with Strain 17 using ClustalW from the Mega 4 package (European Bioinformatics Institute, European Molecular Biology Laboratory, Heidelberg, Germany; available at http://www.ebi.ac.uk/clustalw/).
Sequencing gaps of greater than 20 base pairs were recorded for each strain sequenced (Supplementary Table S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11–7812/-/DCSupplemental), with most of the gaps mapping to the terminal or internal repeat regions. Visual mapping of the gaps is found in Figure 4.
Based on strain 17 genome size of 152,261 bp.
Genome Alignment
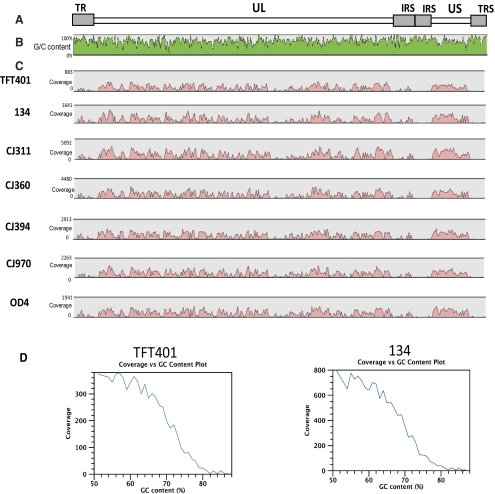
An ungapped consensus sequence for each HSV-1 isolate was generated by mapping, or aligning, the short-sequence reads to the strain 17 reference sequence. This alignment resulted in the average genome coverage ranging from 224x in strain TFT401 to 1345x in 311 (Fig. 2; Table 1). Patterns of relative genome coverage were remarkably consistent and lack of coverage in the terminal and internal repeat regions was mostly due to the high GC content in those areas of the genome (Fig. 2; Supplementary Fig. S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7812/-/DCSupplemental). Conversely, areas of high coverage tended to be areas of relatively low GC bias (Fig. 2, Supplementary Fig. S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7812/-/DCSupplemental). Each ungapped genome consensus sequence was then aligned to HSV-1 strain 17, with ClustalW used to produce a full genome alignment. The number of sequenced base pairs mapped in each ungapped consensus sequence and genome alignment is listed in Table 1. The ungapped consensus sequences varied in length from 144,622 bp for TFT401 to 150,381 bp for strain OD4. The ClustalW algorithm produced alignments ranging in size from 152,315 bp for TFT401 to 152,352 bp for CJ311. In addition, sequencing gaps of greater than 20 bp were recorded for each strain sequenced (Supplementary Table S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7812/-/DCSupplemental) with most of the gaps mapping to the terminal or internal repeat regions. To determine the amount of host genomic DNA contamination, the short sequence reads were aligned to the human genome (hg19; build 37). The amount of host DNA contamination ranged from 35% of the total sequence reads in CJ311 pool to 80% of the total sequence reads in the TFT401 pool (Table 2).
Figure 2.
Coverage of multiplexed, sequenced, and assembled HSV-1 genomes. strain 17 was used as the reference sequence to assemble each of the genomes. (A) HSV-1 genome structure. (B) The percentage of GC content for the strain 17 genome. (C) Depth of coverage for each of the HSV-1 genomes sequenced. Pink shading: depth of coverage in a given area of the genome. (D) Depth of coverage versus GC content percentage.
Table 2.
Genomes and Accession Numbers
De novo genome assembly was also performed as described by Szpara et al.23 in an attempt to increase coverage and to identify the length of VNTRs. The results from de novo assembly were not a substantial improvement from the reference assembly using strain 17; therefore, we present only the reference assembly sequence. In addition, we chose not to use a proxy-HSV-1 strain 17 sequence, as was done previously,23 because it has not been established that VNTR regions are phenotypically silent. The assembled partial genome consensus sequences were deposited in GenBank with protein annotations (Table 2) (GenBank; http://www.ncbi.nlm.nih.gov/Genbank; provided in the public domain by the National Center for Biotechnology Information, Bethesda, MD).
Genomic Phylogenetic Analysis
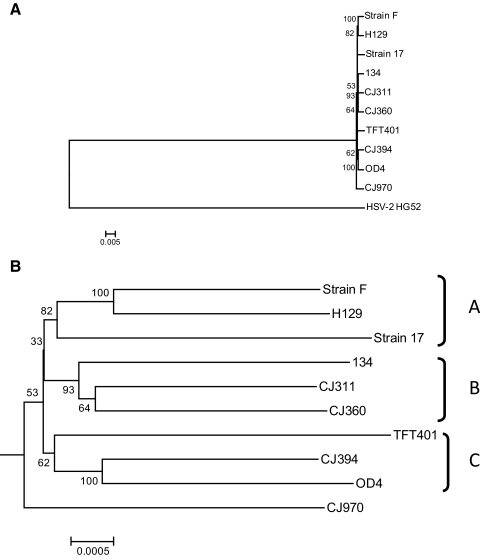
After genome sequencing of the HSV-1 isolates, two other HSV-1 strains (F and H129) and one HSV-2 genome were accessed from the reference database for phylogenetic analysis (Table 2). The nucleotide sequences of the genomes were aligned using ClustalW, and phylogenetic trees were constructed. Figure 3A shows a consensus boot-strapped, neighbor-joining tree using all genomes and illustrates that the HSV-1 viruses form a single robust group. Figure 3B shows an expansion of the HSV-1 specific node and demonstrates that the strains form three main clades denoted A, B, and C. Sequence comparison of the gE and gI genes from numerous viral strains also identified three clades.39 The relationship between the clades identified in this study and the previous study are not clear, as only two strains, 17 and F, were common and placed differently. Strain CJ970 fell outside of these clades and occupied a basal position in the tree, suggesting the existence of a fourth clade, but the bootstrap value and therefore confidence in the placement of strain CJ970 is low. Minimum evolution and maximum parsimony trees produced similar results (data not shown). The placement of strain TFT401 was somewhat unstable, as it sometimes grouped with strain CJ970 depending on the parameters used for analysis.
Figure 3.
Whole genome phylogenetic analysis of multiple HSV-1 viral strains. The HSV-1 genomes analyzed include the seven ocular isolates sequenced in this work, as well as the previously published genomes of strains 17, F, and H129. (A) Consensus bootstrap neighbor-joining tree using HSV-2 strain HG52 as an outgroup. (B) Expansion of the HSV-1 specific node from the neighbor-joining tree in (A). The genomes were aligned with ClustalW and then consensus bootstrap (1000 replicates) neighbor-joining trees, using the Tamura-Nei algorithm, were generated with the Mega4 package. The phylogenetic distance is located at the bottom of each tree.
Genomic Similarity Analysis
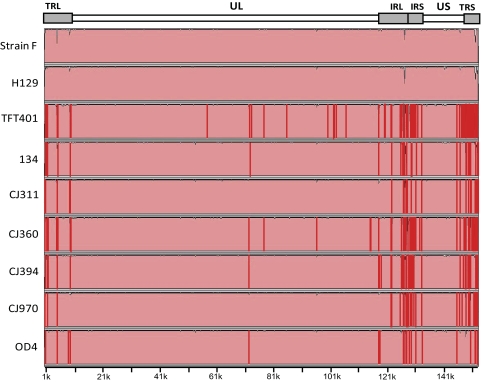
To identify genomic regions with significant variation, mVISTA LAGAN was used to scan the partial genomic sequences, again using strain 17 as the baseline. Figure 4 shows considerable variation in the terminal and internal repeat regions due to a combination of variation in the VNTR lengths as well as lower coverage of these regions. The unique long (UL) and unique short (US) coding regions appear highly similar between the strains, with only small areas of sequence diversity. In the UL coding region higher sequence diversity was seen near the UL1 gene and the second was in the UL reiteration sequence (nucleotides 71,604–71,814). The sequence dissimilarity in the GC-rich UL reiteration sequence was due to low coverage. The US reiteration sequence was also more variable due to low coverage and variability in VNTR length (Fig. 2). Variability was higher in strains TFT401, 134, CJ394, and OD4 in the US region, but due to the scale of Figure 4, this is difficult to discern. Sequence gaps in each respective genome were denoted by red bars in Figure 4.
Figure 4.
Global sequence comparison of multiple HSV-1 genomes compared with the reference strain 17. The comparison plots were generated in mVista LAGAN (http://genome.lbl.gov/vista/index.shtml). Top: HSV-1 genome map. The genomic nucleotide positions using HSV-1 strain 17 as a reference are located on the x-axis. Red lines: manually applied indicators of sequencing gaps greater than 20 bp, catalogued in Supplementary Table S1 (http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7812/-/DCSupplemental).
SNP Detection and Mapping
Future studies on the genetics of virulence will involve construction of recombinants between the strains described here and will depend on the identification of SNPs and their spacing across the genomes. Therefore, we cataloged individual nucleotide changes using strain 17 as the reference sequence with a threshold of 4× coverage and 30% alternate base frequency for detection. The total number of SNPs in each genome resulting from this analysis are shown in Table 3 and ranged from 628 in strain CJ311 to 762 in strain 134. The inferred amino acid coding changes varied from a minimum of 188 in strain CJ311 to a maximum of 232 in strain OD4. Thus, on average, there are approximately 200 coding SNPs per genome. Most of the protein coding SNPs contain a low level of allelic variability, suggesting that each strain is relatively pure population. Only 13 potential protein-coding complex SNPs were detected in all seven strains, and a list of these is found in Table 4.
Table 3.
SNPs and Amino Acid Coding Changes Detected in Each HSV-1 Strain Sequenced
| TFT401 | 134 | CJ311 | CJ360 | CJ394 | CJ970 | OD4 | |
|---|---|---|---|---|---|---|---|
| SNPs | 701 | 762 | 628 | 736 | 712 | 744 | 750 |
| SNP amino acid coding changes | 217 | 224 | 188 | 210 | 201 | 214 | 232 |
| INDELs | 37 | 61 | 54 | 58 | 49 | 55 | 53 |
| INDEL amino acid coding changes | 5 | 7 | 7 | 8 | 6 | 6 | 7 |
Data are the number of SNPs and Coding changes detedted.
Table 4.
Complex SNPs and INDELs in Protein Coding Regions
| Strain | Strain 17 Position | Reference Nucleotide | Allele Variation | Frequencies (%) | Base Call | Coverage | Protein | Amino Acid Change |
|---|---|---|---|---|---|---|---|---|
| TFT401 | 19403 | — | T/— | 50/50 | — | 4 | UL9 | — |
| TFT401 | 144830 | G | G/T | 57.1/42.9 | G | 354 | US10 | — |
| 134 | 79414 | — | —/A | 52.5/45.8 | — | 59 | UL36; VP1/2 | — |
| 134 | 79416 | G | G/— | 54.4/43.9 | G | 57 | UL36; VP1/2 | — |
| 134 | 99188 | CGT | —/CGT | 54.8/43.7 | — | 135 | UL46; VP11/12 | del589A |
| 134 | 141630 | A | A/G | 63.7/36.3 | A | 146 | US8; gE | — |
| CJ311 | 40230 | T | T/G | 64.9/35.1 | T | 1073 | UL19; VP5 | — |
| CJ311 | 79421 | A | A/C | 63.6/36.4 | A | 11 | UL36; VP1/2 | — |
| CJ360 | 40230 | T | T/G | 58.8/41.2 | T | 1127 | UL19; VP5 | — |
| CJ360 | 99188 | CGT | —/CGT | 56.9/39.2 | — | 153 | UL46; VP11/12 | del589A |
| CJ360 | 141799 | A | A/G | 64.4/35.6 | A | 427 | US8; gE | — |
| CJ394 | 141799 | A | A/G | 54.7/45.3 | A | 311 | US8; gE | — |
| CJ970 | 19176 | C | C/T | 57.1/42.9 | C | 18 | UL8 | — |
| CJ970 | 24282 | G | A/G | 58.5/41.5 | A | 172 | UL10 | R360H |
| CJ970 | 26636 | C | C/A | 63.6/36.2 | C | 434 | UL12 | — |
| CJ970 | 71812 | G | G/A | 57.1/42.9 | G | 7 | UL36; VP1/2 | — |
| CJ970 | 71818 | G | G/A | 55.6/44.4 | G | 9 | UL36; VP1/2 | — |
| CJ970 | 79625 | G | G/T | 62.5/37.5 | G | 8 | UL36; VP1/2 | — |
| CJ970 | 99510 | — | —/CGG | 50/50 | — | 4 | UL46; VP11/12 | — |
| CJ970 | 144838 | — | —/T | 62.1/37.9 | — | 29 | US10 | — |
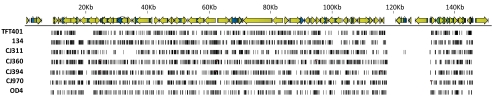
Figure 5 shows the fingerprint SNP pattern for each HSV-1 strain. The SNPs were generally distributed evenly across the genomes; however, some gaps were present. For example, OD4 contains an SNP gap incorporating parts of UL8 and UL9, and strain 311 has a gap between the UL11 and UL17 genes. Few SNPs were detected in the large repeat regions because of software limitations. For this reason, the open reading frames in the terminal and internal repeats were manually inspected, and the coding changes were catalogued. All the SNPs resulting in amino acid coding changes detected in the seven genomes sequenced for this work can be found in Supplementary Table S2, and the raw SNP data can be found in Supplementary Table S3 (both available at http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7812/-/DCSupplemental).
Figure 5.
SNP distribution across the genomes. The annotated HSV-1 genome is shown across the top and the SNPs for each strain (using strain 17 as the reference) are shown below with each tick mark indicating an SNP. The repeat regions were not plotted because of lower coverage.
INDEL Detection
In addition to SNP detection, INDEL detection was performed with parameters similar to those in the SNP detection. The total number of INDELS ranged from 37 in strain TFT401 to 61 in strain 134 (Table 3). The inferred amino acid changes resulting from the INDELS ranged from five in strain TFT401 to eight in CJ360. All the INDELs encoding amino acid changes are listed with the SNPs changes in Table 3. Most of the detected INDELs, like the SNPs, contained a low level of allelic variability, which implies a relatively homogenous viral population. Seven total potential protein coding complex INDELs were found, and these are listed in Table 4. Two frameshift mutations each were detected in the UL2 and UL17 genes; however, the most recent annotation of the HSV-1 strain 17 genomic sequences states that two frame shifts are likely to be present in each these genes and these INDELs were not considered further. There were three noteworthy frameshift mutations resulting in alternate translational stops. The first was a nucleotide deletion at amino acid 117 in the UL13 protein kinase in strains CJ311, CJ970, and OD4, with the second being a nucleotide deletion at amino acid 20 of the UL55 protein in strain 134. The third was a base deletion at the extreme 3′ end (Q478) of the UL42 gene in strain CJ360. Unlike the previous two frameshift mutations leading to premature stop codons, the UL42 mutation leads to an extra 16 amino acids being added to the protein sequence. The raw INDEL data can be found in Supplementary Table S4 (http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7812/-/DCSupplemental).
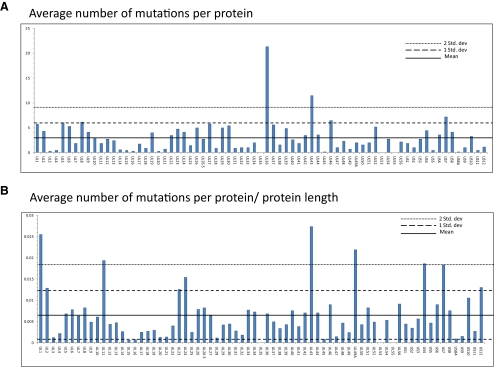
Protein Variability
To identify proteins with significantly different levels of amino acid variation compared with HSV-1 strain 17, we calculated both the average number of amino acid changes (Fig. 6A) and the average number of changes relative to protein length (Fig. 6B) for all the annotated genes of the seven sequenced strains. Six proteins fell beyond two standard deviations in the average number of amino acid changes normalized to protein length; UL1, UL11, UL43, UL49A, gG, and gI. Complete sequence conservation was found for UL35 and gK. Also of note, one nonsense mutation was identified, as the OD4 virus encoded a premature stop at position 49 in the virion host shutoff protein (UL41).
Figure 6.
Amino acid variation in proteins along the genome from multiple HSV-1 strains. (A) The average number of amino acid substitutions for each protein in HSV-1 strains TFT401, 134, CJ311, CJ360, CJ394, CJ970, and OD4 compared to strain 17. (B) The average number of amino acid changes normalized to protein length. The mean, one standard deviation, and two standard deviations for each graph have been plotted. The name of each protein is located on the x-axis. The γ134.5, ICP0 and ICP4 proteins were not included, because of low sequence coverage. The reported numbers do not reflect the frameshift mutations found in UL2, UL13, UL17, UL42, and UL55, so as not to skew the data.
The genomic regions containing the UL1, UL11, UL43, UL49A, gG, and gI proteins were scanned with Simplot to determine nucleotide similarity and see whether the variation in these genes and others is localized or distributed across the protein (Fig. 7). The similarity scans illustrated that both patterns were present. In the UL1, UL1, US4, and US7 genes the sequence variation was generally distributed. In contrast, the variation in the UL9, UL43, UL46, and UL48 genes was concentrated at certain points in the coding sequence. For example, with the UL43 gene, most of the variation was concentrated in the middle of the coding sequence, whereas in UL46, the variation was near the carboxyl terminus. Several of the intergenic regions—for example, the area between US1 and US2—also exhibited sequence dissimilarity.
Figure 7.
Similarity plots of selected areas of the HSV-1 genome compared to strain 17. Areas of the genome featuring genes with variance greater than two standard deviations from Figure 4 (UL1, UL11, UL43, UL49A, US4, and US7) were analyzed with Simplot. The analysis includes plots of HSV-1 strains TFT401, 134, CJ311, CJ360, CJ394, CJ970, OD4, F, and H129. The UL1 (A), UL11 (B), UL43 (C), UL49A (C), US4 (D), and US7 (D) genes are highlighted in red.
Evidence of Recombination
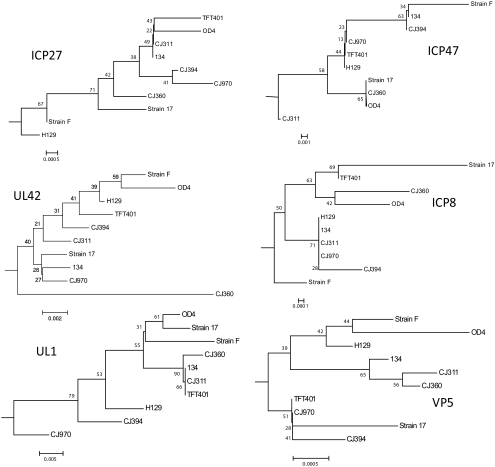
Phylogenetic analysis using single genes can detect whether recombination has occurred between strains. Therefore, we repeated the phylogenetic analysis for all the sequenced strains using two proteins each from the immediate-early (α), early (β), and late (γ) kinetic expression groups. These were the ICP27 (UL54), ICP47 (US12), UL42, ICP8 (UL29), glycoprotein L (UL1), and VP5 (UL19) proteins. HSV-2 HG52 was included in the analysis as an outgroup. Consensus trees generated from the neighbor-joining trees (Fig. 8) revealed that the branching patterns and the placement of each virus within the branches was different depending on the protein being analyzed. The branching and strain placement also differ significantly from the whole genome analysis presented above. The only tree that recovered a genomic clade from Figure 3B was VP5, which recovered genomic clade B. The fact that different trees were generated depending on the protein used for analysis suggests that recombination has occurred.40
Figure 8.
Consensus bootstrap neighbor-joining trees of selected α, β, and γ proteins. The nucleotide sequences of ICP27 (UL53), ICP47 (US12), UL42, ICP8 (UL29), glycoprotein L (UL1), and VP5 (UL19) from HSV-1 strains 17, F, H129, TFT401, 134, CJ311, CJ360, CJ394, CJ970, and OD4 were each translated into their inferred amino acid sequences and then aligned with ClustalW. Consensus bootstrap (1000 replicates) neighbor-joining trees were then generated. The relative phylogenetic distance marker is located near the bottom of each tree.
Correlation between SNPs and Disease Phenotype
To determine whether the SNPs are correlated with a disease phenotype, we partitioned the strains according to the presence or absence of a given SNP, and ask whether the binary partition clearly separates the phenotypic values. Due to the small number of strains examined, however, we were not able to detect statistically significant associations with this approach. As an alternative, we clustered SNPs by gene to test association with a disease phenotype. Specifically, we ordered strains by disease phenotype and then asked whether this ordering is consistent with the ordering induced by count of SNPs in each gene. Taking into account multiple comparisons, this approach failed to identify any significant associations (P > 0.05).
Discussion
High-Throughput Sequencing and Genome Assembly
Our data, showing that multiplex sequencing of seven HSV-1 genomes in a single lane is feasible, will increase throughput and reduce the cost of sequencing HSV genomes and facilitate studies on the population structure, virulence traits, and structure–function studies. We found that de novo assembly with this sequence data set yielded no clear advantage over reference alignment and as a result, the reference alignment was presented. It is not clear why de novo assembly did not provide an advantage over reference alignment. With both single-lane and multiplex sequencing, the VNTRs were still problematic. Szpara et al.,23 used VNTR sequences from strain 17 to assemble entire genomes, but this strategy has to be validated. The difficulty in sequencing the large repeat and VNTR regions due to GC bias represents a limitation of the current technology. Technological challenges notwithstanding, our results show that multiplex high-throughput sequencing is an effective method of rapidly and successfully sequencing multiple viral genomes.
Phylogenetic Grouping
The genomes of our seven isolates were aligned with the previously published genomes of HSV-1 strains 17 and H129 along with HSV-2 strain HG52. The subsequent phylogenetic analysis demonstrated that the HSV-1 strains formed a single species with no apparent genomic contribution from HSV-2 in any of the HSV-1 strains (Fig. 3A). The neighbor-joining tree in Figure 3B, showed that the HSV-1 strains group into three main clades: A, B, and C. The placement of strain TFT401 in the neighbor-joining tree was somewhat unstable, with this strain grouping in clade C or with strain CJ970, depending on the phylogenetic parameters that were used. The reason is not clear but may be the result of recombination events in strain TFT401 that affect the placement within the tree.
Previous work using genes in the unique short-coding region reported three clade groupings,39,41 and our whole genome phylogenetic analysis supports this conclusion. Strain CJ970 appears to be an outlier and may represent a fourth clade, although the bootstrap values are low. Sequencing of additional HSV-1 strains may clarify the status of strain CJ970. The single-gene neighbor-joining trees shown in Figure 8 exhibited a more complicated pattern. The VP5 tree recovered a three-clade pattern, whereas the branching configurations in the remaining trees were highly variable. Further, the placement of the viral strains within the branches was also inconsistent. The variable groupings in the phylogenetic trees, using single proteins, strongly suggests that recombination has occurred and that each of the viral genomes is a unique mosaic.40
SNPs and Their Distribution across the Genomes
The ability to rapidly and inexpensively sequence the entire HSV-1 genome will change how we study the role of sequence differences in both virulence and structure and function of viral proteins. For example, we previously characterized the virulence properties of several recombinant viruses between strains OD4 and CJ39425 and OD4 and 99426,42 and have quantitative data on the severity of neuro- and ocular virulence for these recombinants. Previously, the identification of virulence mutations required tedious cloning and construction of recombinant viruses, followed by animal testing, and it was difficult to look at the effect of multiple changes on the virulence phenotype. We found that each of the strains had approximately 700 SNPs and 200 coding SNP differences compared with strain 17 (Table 3), and more important, these SNPs were relatively evenly distributed across the entire genome (Fig. 5). These data are encouraging and indicate that it is now feasible to sequence all recombinants and identify SNPs that segregate with specific virulence traits.
Gene Variability
To determine whether specific HSV-1 genes were hypervariable, we calculated the total average number of amino acid changes per protein and then corrected this for the length of the protein. We then identified proteins whose mean substitution values were two standard deviations from the mean of all proteins in the genome.
Variable Proteins
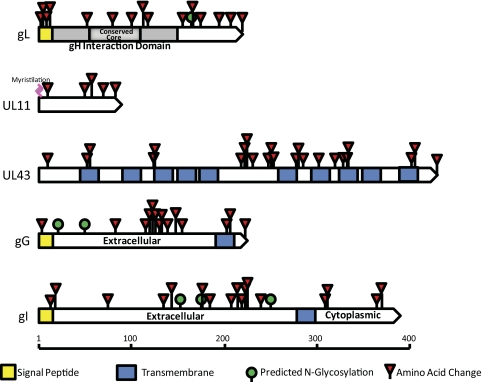
Six proteins were identified as being hypervariable: UL1, UL11, UL43, UL49A, UL4, and UL7. Figure 9 shows schematic diagrams of each protein with the locations of the substitutions. The identification of UL49.543,44 as hypervariable is somewhat misleading, as all the strains except 17 have the same sequence, thus this artifact arose because we chose strain 17 as the baseline. For some of these proteins, such as gL, the function or functions have been identified45 but for others, little is known. Their properties also differ, as some are membrane associated and others are not.46–50 They also differ in length, thus there does not appear to be a commonality between variability and properties of the proteins.
Figure 9.
High variability proteins showing the locations of sequence differences using strain 17 as the baseline. UL49.5 is not shown, although it was identified as having higher variability.
Conserved Proteins
Two proteins, UL35 and UL53, were highly conserved across all the strains. Szpara et al.,23 in comparing strains F, H129, and 17, found that 10 proteins were completely conserved including UL35 and UL53. Thus, the combined data set suggests that only two genes exhibit extreme sequence conservation. The UL53 gene is dispensable for growth in cell culture and so the conservation of sequence suggests the gene encodes a protein with important functions in vivo. As with the variable proteins, there does not seem to be any commonality in function or length of the conserved proteins.51–55 Antibodies to gK are responsible for antibody-dependent enhancement of infection,56 and sequence conservation suggests that gK may be more important than previously realized in the context of in vivo infection.
Premature Stop Mutations
Two proteins were found to have been affected by nucleotide deletions: UL13 and UL55. The CJ311, CJ970, and OD4 strains were all found to contain the same nucleotide deletion resulting in a frameshift at amino acid 117 in the UL13 kinase. It appears that this same mutation was reported in an isolate of HSV-1 strain F (Szpara et al.23). UL13 is a serine–threonine kinase that phosphorylates the virulence protein ICP22, is required for virus-induced modification of RNAPii, and is dispensable in cell culture.57–59 Szpara et al. suggest that the mutation is an artifact of tissue culture passaging. Although the hypothesis may be correct, it is interesting that the premature stop in which none of the kinase domain is encoded appears to have little to no affect on ocular or neurovirulence, as CJ970 and CJ311 are highly neurovirulent (Fig. 1). Another frameshift resulting in a significant premature stop is the UL55 protein in strain 134. The UL55 gene has been shown to be dispensable for growth in cell culture and has been reported to be dispensable for infection in mice.60 Since the virulence phenotype of strain 134 has not been determined, it is difficult to comment on the significance of this mutation.
The UL41 virion host shutoff protein in strain OD4 contains a base change resulting in a stop codon at amino acid 49. The UL41 gene has been shown to be dispensable in cell culture and is not surprising, given the avirulent phenotype of the OD4 strain.61,62
Virulence Determinants
We have previously characterized the neuro- and ocular virulence phenotypes of all the strains sequenced in this article except for strain 134,24 raising the possibility that virulence determinants could be identified from the genomic sequences. The fact that each strain has approximately 200 amino acid sequence differences compared with strain 17, and multiple differences from each other, complicates this type of analysis and makes it difficult to identify possible virulence determinants, given this small sample size. When we attempted to identify SNPs that were correlated with disease phenotypes (blepharitis, stromal disease, vascularization, and mortality) no significant associations were found due to the small number of strains that were analyzed. Sequencing our panel of recombinants between OD4 and CJ39425 and OD4 and 994,26 will potentially be more useful in identifying such determinants, and such studies are in progress.
Conclusions
In this study, we have sequenced the partial genomes of seven clinical isolates using multiplex high-throughput sequencing. Whole genome phylogenetic analysis of the available HSV-1 genomes demonstrated that the strains group into three main clades. In addition, each of the genomes contained significant numbers of SNPs that, for the most part, were evenly distributed across the genome. Finally, several genes with significantly different variability or a lack of variability were identified. These data will be very useful for structure–function analysis of HSV-1 proteins and the even distribution of SNPs across the genomes will be valuable for studying the genetic basis of virulence.
Supplementary Material
Acknowledgments
The authors thank John Chandler for providing the ocular isolates sequenced in this study.
Footnotes
Supported by a grant from the NIH EY07336 (CRB), Core Grant for Vision Research P30EY016665, a Research to Prevent Blindness (RPB) Senior Scientist Award (CRB), and an unrestricted grant to the Department of Ophthalmology and Visual Sciences from RPB, Inc.
Disclosure: A.W. Kolb, None; M. Adams, None; E.L. Cabot, None; M. Craven, None; C.R. Brandt, None
References
- 1. Liesegang TJ. Herpes simplex virus epidemiology and ocular importance. Cornea. 2001;20:1–13 [DOI] [PubMed] [Google Scholar]
- 2. Whitley RJ. Herpes simplex viruses. In: Fields BN, Knipe DM, Howley PM, eds. Fields Virology. 3rd ed Vol. 2 Philadelphia: Lippincott-Raven; 1996:2297–2342 [Google Scholar]
- 3. Bhattacharjee PS, Neumann DM, Foster TP, et al. Effect of human apolipoprotein E genotype on the pathogenesis of experimental ocular HSV-1. Exp Eye Res. 2008;87:122–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burgos JS, Ramirez C, Sastre I, Valdivieso F. Effect of apolipoprotein E on the cerebral load of latent herpes simplex virus type 1 DNA. J Virol. 2006;80:5383–5387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Han X, Lundberg P, Tanamachi B, et al. Gender influences herpes simplex virus type 1 infection in normal and gamma interferon-mutant mice. J Virol. 2001;75:3048–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kastrukoff LF, Lau AS, Puterman ML. Genetics of natural resistance to herpes simplex virus type 1 latent infection of the peripheral nervous system in mice. J Gen Virol. 1986;67:613–621 [DOI] [PubMed] [Google Scholar]
- 7. Lopez C. Genetics of natural resistance to herpesvirus infections in mice. Nature. 1975;258:152–153 [DOI] [PubMed] [Google Scholar]
- 8. Lundberg P, Welander P, Openshaw H, et al. A locus on mouse chromosome 6 that determines resistance to herpes simplex virus also influences reactivation, while an unlinked locus augments resistance of female mice. J Virol. 2003;77:11661–11673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sørensen LN, Reinert LS, Malmgaard L, et al. TLR2 and TLR9 synergistically control herpes simplex virus infection in the brain. J Immunol. 2008;181:8604–8612 [DOI] [PubMed] [Google Scholar]
- 10. Stulting RD, Kindle JC, Nahmias AJ. Patterns of herpes simplex keratitis in inbred mice. Invest Ophthalmol Vis Sci. 1985;26:1360–1367 [PubMed] [Google Scholar]
- 11. Zhang SY, Jouanguy E, Ugolini S, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317:1522–1527 [DOI] [PubMed] [Google Scholar]
- 12. Koelle DM, Corey L. Recent progress in herpes simplex virus immunobiology and vaccine research. Clin Microbiol Rev. 2003;16:96–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pollara G, Katz DR, Chain BM. The host response to herpes simplex virus infection. Curr Opin Infect Dis. 2004;17:199–203 [DOI] [PubMed] [Google Scholar]
- 14. Doymaz MZ, Rouse BT. Immunopathology of herpes simplex virus infections. Curr Top Microbiol Immunol. 1992;179:121–136 [DOI] [PubMed] [Google Scholar]
- 15. Streilein JW, Dana MR, Ksander BR. Immunity causing blindness: five different paths to herpes stromal keratitis. Immunol Today. 1997;18:443–449 [DOI] [PubMed] [Google Scholar]
- 16. Thomas J, Rouse BT. Immunopathogenesis of herpetic ocular disease. Immunol Res. 1997;16:375–386 [DOI] [PubMed] [Google Scholar]
- 17. Brandt CR. Virulence genes in herpes simplex virus type 1 corneal infection. Curr Eye Res. 2004;29:103–117 [DOI] [PubMed] [Google Scholar]
- 18. Brandt CR. The role of viral and host genes in corneal infection with herpes simplex virus type 1. Exp Eye Res. 2005;80:607–621 [DOI] [PubMed] [Google Scholar]
- 19. McGeoch DJ, Dalrymple MA, Davison AJ, et al. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574 [DOI] [PubMed] [Google Scholar]
- 20. McGeoch DJ, Dolan A, Donald S, Rixon FJ. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J Mol Biol. 1985;181:1–13 [DOI] [PubMed] [Google Scholar]
- 21. McGeoch DJ, Dolan A, Donald S, Brauer DH. Complete DNA sequence of the short repeat region in the genome of herpes simplex virus type 1. Nucleic Acids Res. 1986;14:1727–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Perry LJ, McGeoch DJ. The DNA sequences of the long repeat region and adjoining parts of the long unique region in the genome of herpes-simplex virus type-1. J Gen Virol. 1988;69:2831–2846 [DOI] [PubMed] [Google Scholar]
- 23. Szpara ML, Parsons L, Enquist LW. Sequence variability in clinical and laboratory isolates of herpes simplex virus 1 reveals new mutations. J Virol. 2010;84:5303–5313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grau DR, Visalli RJ, Brandt CR. Herpes simplex virus stromal keratitis is not titer-dependent and does not correlate with neurovirulence. Invest Ophthalmol Vis Sci. 1989;30:2474–2480 [PubMed] [Google Scholar]
- 25. Brandt CR, Grau DR. Mixed infection with herpes simplex virus type 1 generates recombinants with increased ocular and neurovirulence. Invest Ophthalmol Vis Sci. 1990;31:2214–2223 [PubMed] [Google Scholar]
- 26. Kintner RL, Allan RW, Brandt CR. Recombinants are isolated at high frequency following in vivo mixed ocular infection with two avirulent herpes simplex virus type 1 strains. Arch Virol. 1995;140:231–244 [DOI] [PubMed] [Google Scholar]
- 27. Brandt CR, Kolb AW, Shah DD, et al. Multiple determinants contribute to the virulence of HSV ocular and CNS infection and identification of serine 34 of the US1 gene as an ocular disease determinant. Invest Ophthalmol Vis Sci. 2003;44:2657–2668 [DOI] [PubMed] [Google Scholar]
- 28. Kintner RL, Brandt CR. Rapid small-scale isolation of Herpes simplex virus DNA. J Virol Methods. 1994;48:189–196 [DOI] [PubMed] [Google Scholar]
- 29. Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 30. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 31. Altshuler D, Pollara VJ, Cowles CR, et al. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature. 2000;407:513–516 [DOI] [PubMed] [Google Scholar]
- 32. Brockman W, Alvarez P, Young S, et al. Quality scores and SNP detection in sequencing-by-synthesis systems. Genome Res. 2008;18:763–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791 [DOI] [PubMed] [Google Scholar]
- 34. Saitou N, Nei M. On the maximum likelihood method for molecular phylogeny Jpn J Genet. 1987;62:547–548 [Google Scholar]
- 35. Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526 [DOI] [PubMed] [Google Scholar]
- 36. Brudno M, Do CB, Cooper GM, et al. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13:721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martin DP, Lemey P, Lott M, et al. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120 [DOI] [PubMed] [Google Scholar]
- 39. Norberg P, Bergstrom T, Rekabdar E, Lindh M, Lijeqvist J. Phylogenetic analysis of clinical herpes simplex virus type 1 isolates identified three genetic groups and recombinant viruses. J Virol. 2004;78:10755–10764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Walsh MP, Seto J, Tirado D, et al. Computational analysis of human adenovirus serotype 18. Virology. 2010;404:284–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kolb AW, Schmidt TR, Dyer DW, Brandt CR. Sequence variation and phosphorylation sites in the herpes simplex virus US1 ocular virulence determinant. Invest Ophthalmol Vis Sci. 2011;52:4630–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brandt CR. Mixed ocular infections identify strains of herpes simplex virus for use in genetic studies. J Virol Methods. 1991;35:127–135 [DOI] [PubMed] [Google Scholar]
- 43. Barker DE, Roizman B. The unique sequence of the herpes-simplex virus-1 L component contains an additional translated open reading frame designated UL49.5. J Virol. 1992;66:562–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Barnett BC, Dolan A, Telford EAR, Davison AJ, McGeoch DJ. A novel herpes-simplex virus gene (UL49A) encodes a putative membrane-protein with counterparts in other herpesviruses. J Gen Virol. 1992;73:2167–2171 [DOI] [PubMed] [Google Scholar]
- 45. Westra DF, Glazenburg KL, Harmsen MC, et al. Glycoprotein H of herpes simplex virus type 1 requires glycoprotein L for transport to the surfaces of insect cells. J Virol. 1997;71:2285–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Baines JD, Jacob RJ, Simmerman L, Roizman B. The herpes-simplex virus-1 U(L) 11 proteins are associated with cytoplasmic and nuclear-membranes and with nuclear-bodies of infected-cells. J Virol. 1995;69:825–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Baines JD, Roizman B. The UL11 gene of herpes-simplex virus-1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J Virol. 1992;66:5168–5174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Maclean CA, Clark B, McGeoch DJ. Geene UL11 of herpes-simplex virus type-1 encodes a virion protein which is myristoylated. J Gen Virol. 1989;70:3147–3157 [DOI] [PubMed] [Google Scholar]
- 49. Dingwell KS, Brunetti CR, Hendricks RL, et al. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J Virol. 1994;68:834–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ward PL, Barker DE, Roizman B. A novel herpes simplex virus 1 gene, U(L)43.5, maps antisense to the U(L)43 gene and encodes a protein which colocalizes in nuclear structures with capsid proteins. J Virol. 1996;70:2684–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Booy FP, Trus BL, Newcomb WW, et al. Finding a needle in a haystack-detection of a small protein (the 12-KDA VP26) in a large complex (the 200-MDA capsid of herpes-simplex virus). Proc Natl Acad Sci U S A. 1994;91:5652–5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Trus BL, Homa FL, Booy FP, et al. Herpes-simplex virus capsids assembled in insect cells infected with recombinant baculoviruses-structural authenticity and location of VP26. J Virol. 1995;69:7362–7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou ZH, He J, Jakana J, et al. Assembly of VP26 in herpes-simplex virus inferred from structures of wild-type and recombinant capsids. Nat Struct Biol. 1995;2:1026–1030 [DOI] [PubMed] [Google Scholar]
- 54. Hutchinson L, Johnson DC. Herpes-simplex virus glycoprotein-K promotes egress of virus particles. J Virol. 1995;69:5401–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jayachandra S, Baghian A, Kousoulas KG. Herpes simplex virus type 1 glycoprotein K is not essential for infectious virus production in actively replicating cells but is required for efficient envelopment and translocation of infectious virions from the cytoplasm to the extracellular space. J Virol. 1997;71:5012–5024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ghiasi H, Perng GC, Nesburn AB, Wechsler SL. Antibody-dependent enhancement of HSV-1 infection by anti-gK sera. Virus Res. 2000;68:137–144 [DOI] [PubMed] [Google Scholar]
- 57. Asai R, Ohno T, Kato A, Kawaguchi Y. Identification of proteins directly phosphorylated by UL13 protein kinase from herpes simples virus 1. Microb Infect. 2007;9:1434–1438 [DOI] [PubMed] [Google Scholar]
- 58. Long MC, Leong V, Schaffer PA, Spencer CA, Rice SA. ICP22 and the UL13 protein kinase are both required for herpes simplex virus-induced modification of the large subunit of RNA polymerase II. J Virol. 1999;73:5593–5604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Purves FC, Roizman B. The UL13 gene of herpes simplex virus 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein a22. Proc Natl Acad Sci U S A. 1992;89:7310–7314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nash TC, Spivack JG. The UL55 and UL56 genes of herpes simplex virus type 1 are not required for viral replication, intraperitoneal virulence, or establishment of latency in mice. Virology. 1994;204:794–798 [DOI] [PubMed] [Google Scholar]
- 61. Read GS, Frenkel H. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of alpha (immediate-early) polypeptides. J Virol. 1983;67:489–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Strelow LI, Lieb DA. Role of the virion host shutoff (vhs) of herpes simplex virus type 1 in latency and pathogenesis. J Virol. 1995;69:6779–6786 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.