Abstract
Curcumin (CUR), a major bioactive polyphenolic component from turmeric curry, Curcuma longa, has been shown to be a potent anti-cancer phytochemical with well-established anti-inflammatory and anti-oxidative stress effects. Chromatin remodeling-related epigenetic regulation has emerged as an important mechanism of carcinogenesis, chemoprevention, and chemotherapy. CUR has been found to inhibit histone acetyltransferase activity, and it was also postulated to be a potential DNA methyltransferase (DNMT) and histone deacetylase (HDAC) inhibitor. In this study, we show that when human prostate LNCaP cells were treated with CUR, it led to demethylation of the first 14 CpG sites of the CpG island of the Neurog1 gene and restored the expression of this cancer-related CpG-methylation epigenome marker gene. At the protein level, CUR treatment had limited effects on the expression of epigenetic modifying proteins MBD2, MeCP2, DNMT1, and DNMT3a. Using ChIP assay, CUR decreased MeCP2 binding to the promoter of Neurog1 dramatically. CUR treatment showed different effects on the protein expression of HDACs, increasing the expression of HDAC1, 4, 5, and 8 but decreasing HDAC3. However, the total HDAC activity was decreased upon CUR treatment. Further analysis of the tri-methylation of histone 3 at lysine 27 (H3K27me3) showed that CUR decreased the enrichment of H3K27me3 at the Neurog1 promoter region as well as at the global level. Taken together, our present study provides evidence on the CpG demethylation ability of CUR on Neurog1 while activating its expression, suggesting a potential epigenetic modifying role for this phytochemical compound in human prostate cancer cells.
KEY WORDS: curcumin, demethylation, hypermethylation, LNCaP, Neurog1
INTRODUCTION
Epigenetic regulation involves the alterations of the chromatin structure with certain genes being selectively activated while others inactivated transcriptionally. Epigenetic silencing of genes through CpG island hypermethylation has been accepted as an important potential carcinogenic mechanism in the development of human prostate cancers, and these modulations can be caused by the cross talk of DNA methylation, histone modification, loss of imprinting, and microRNA regulation (1). Inappropriate gene hypermethylation may represent an early event in cancer development, and it can serve as a marker of carcinogenesis. For instance, when methylation of glutathione-S-transferase (GST) pi is considered together with methylation of APC gene, the sensitivity to detecting cancer approached 98% and the specificity was 100% in prostate cancer (2–4).
Neurog1 belongs to the basic helix-loop-helix protein, and it is a single-exon transcription factor located in chromosome 5 in human (5). Neurog1 plays a critical role in neuronal differentiation, it controls neurogenesis in the mouse pallium, and it is expressed in the dorsal telencephalon of zebrafish (5). In addition, there are reports showing that Neurog1 is highly methylated and that its expression is perturbed in colorectal cancer (6–8) and prostate cancer (1,9) and has been selected as one of the cancer methylation markers.
Curcumin (CUR) is a major active polyphenolic component in the perennial herb Curcuma longa. As a potent cancer chemoprevention compound, CUR may target all steps of cancer development, including tumor initiation, promotion, and progression. In prostate and other cancer cell lines, a series of epigenetic and genetic alterations affecting oncogenes and tumor suppressor genes may be involved in the process of the anti-cancer effects of CUR: affecting cell proliferation and apoptosis along with the alteration of the activity of AKT/mTOR and downstream signaling (10,11); suppressing inflammation by inhibiting the activation of NF-kappa B and cyclooxygenase-2; counteracting the free radicals reactive oxygen and nitrogen species generated by certain carcinogens (12); and exerting antioxidant effect corresponding with the induction of some phase II drug metabolizing/antioxidant enzymes, such as GST (13), UDP-glucuronosyltransferase (14), and heme oxygenase-1, and the transcription activation of the transcription factor NF-E2-related factor 2 (15,16).
CUR has been reported to reduce histone acetylation through inhibition of histone acetyltransferase (HAT) activity (17–19). Interestingly, it has also been predicted to possess histone deacetylase (HDAC) inhibition ability at 50 and 500 μM concentrations (20). In addition, CUR could inhibit the activity of M.SssI, an analog of DNA methyltransferase 1 (DNMT1), by potentially blocking the catalytic thiol of C1226 in DNMT1 and induce global DNA hypomethylation in MV4-11, a leukemia cell line (21), suggesting that potential epigenetic regulation may also contribute to the overall anti-cancer effects of CUR. However, to date there are few studies demonstrating the effect of CUR on the CpG methylation of potential target biomarker genes in prostate cancer cells. Our present study provides evidence that CUR reverses the CpG methylation of the promoter region of Neurog1 in the human prostate cancer LNCaP cell and restores its mRNA and protein expression.
EXPERIMENTAL SECTION
Cell Culture and Treatment
LNCaP cell was maintained in RPMI-1640 with 10% fetal bovine serum (FBS, Gibco) and grown at 37°C in a humidified 5% CO2 atmosphere. All the chemicals, curcumin (CUR), 5-azadeoxycytidine (5-Aza), and trichostatin A (TSA), were purchased from Sigma (St. Louis, MO, USA). Cells were plated in 10 cm plates for 24 h and then treated with 0.1% DMSO, 5 μM CUR, or 2.5 μM 5-Aza with 1% FBS-containing medium. The medium was changed every 2 days. On the sixth day, for the 5-Aza and TSA combination treatment, 500 nM TSA was then added to the 5-Aza-containing medium, cultured for another 20 h, and then the cells were harvested for DNA, protein, and RNA analyses.
Protein Lyses Preparation and Western Blotting
For the samples used for protein assays, all cells were harvested using radioimmunoprecipitation assay (RIPA) buffer with protein inhibitor cocktail (Sigma). The protein concentrations of the cleared lysates were determined using the bicinchoninic acid (BCA) method (Pierce, Rockford, IL), and 20 μg of the total proteins from each sample were resolved by 4–15% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (Bio-Rad, Hercules, CA). After electrophoresis, the proteins were electro-transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA). After blocking with 5% fat-free milk in Tris-buffered saline–0.1% Tween 20, the PVDF membrane was sequentially incubated with specified primary antibodies and HRP-conjugated secondary antibodies. The blots were then visualized by SuperSignal enhanced chemiluminescence detection system and documented using a Gel Documentation 2000 system (Bio-Rad, Hercules, CA). The antibodies were purchased from different resources: anti-MeCP2 and anti-beta-actin from Santa Cruz Biotechnology(Santa Cruz, CA); anti-DNMT1 and anti-DNMT3a from IMGENEX (San Diego, CA); anti-MBD2 from Abcam (Cambridge, MA); anti-Neurog1 from OriGene (Rockville, MD); anti-HDAC1, 3, 4, and 5 from Cell Signaling (Boston, MA); and anti-HDAC8 from Proteintech (Chicago, IL).
DNA Extraction and Bisulfite Genomic Sequencing
Genomic DNA was isolated from the DMSO-, CUR-, or 5-Aza/TSA-treated LNCaP cells using the DNeasy tissue kit (QIAGEN, Valencia, CA). The extracted genomic DNAs were subjected to bisulfite conversion. The bisulfite conversion was carried out using 750 ng of genomic DNA and EZ DNA Methylation Gold Kits (Zymo Research Corp., Orange, CA) following the manufacturer’s instructions. The converted DNA was amplified by PCR using Platinum PCR SuperMix (Invitrogen, Carlsbad, CA) with two specific sets of primers. Two fragments of Neurog1 gene were amplified: fragment 1 included the CpG island from −671 to −307 with the translation start site referenced as +1, the forward and reverse primers were 5′-ATTATTAAGTAGTTGGAGAGGGGATG-3′ and 5′-ACTAACCTCAAAACCCCTTAAATAC-3′; fragment 2 included the CpG islands from +199 to +409, the forward and reverse primers used were 5′-GTTTGAGGTTTTAGGGGTATAGGA-3′ and 5′-CCAAAACCCAAATATAATTATAAAC-3′. Both PCR amplification conditions were: 94°C 3 min; 94°C 30 s, 60°C 45 s, 72°C 45 s, 35 cycles. The PCR products were cloned into pCR4 TOPO vector using a TOPO™ TA Cloning Kit (Invitrogen, Carlsbad, CA). Plasmids DNA from at least ten colonies of each treatment were prepared using QIAprep Spin Miniprep Kit (QIAGEN, Valencia, CA) and sequenced (Genwiz, Piscataway, NJ).
PCR Array
Human prostate cancer DNA methylation PCR array (MeAH-051) was purchased from SABioscience (now QIAGEN, Frederick, MD). The extracted RNA-free genomic DNA was aliquoted into four equal portions, each portion was mock-digested or digested with methylation-sensitive and methylation-dependent enzymes, separately or in combination, and then these samples were loaded onto 96- or 384-well plate for real-time PCR (qPCR) amplification. The relative amount of each DNA (methylated, intermediate methylated, and unmethylated) fraction was calculated using a standard delta Ct method; normalizing the amount of DNA of each digestion against the no-digestion input and the methylation status of a panel of 24 or 96 genes were examined simultaneously (the specific information regarding the enzymes and the primer sequences used were not revealed by SABioscience).
RNA Isolation and Reverse-Transcription PCRs
Total RNA was extracted from the treated cells using the RNeasy Mini Kit (QIAGEN, Valencia, CA). First-strand cDNA was synthesized from 1 μg of total RNA using SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. The cDNA was used as the template for real-time PCR (ABI7900HT system). The sequences of the primers used for cDNA amplification of Neurog1 were: 5′-GCCTTTCTATCTGTCCGTCG-3′ and 5′-GTCTGGCACAGTCTTCCTC-3′.
Methylation DNA Immunoprecipitation Analysis
For the methylation DNA immunoprecipitation (MeDIP) analysis modified from previous reports (22,23), 8-μg DNAs each extracted from control- and CUR-treated cells were used. Briefly, the DNAs were adjusted to 150 μl using TE buffer and subjected to sonication, and the size of DNA fragments (around 300 to 500 bp) was checked. About 1/10 of the fragmented DNAs were taken as inputs; the remaining DNA was followed by denaturation for 10 min, and immunoprecipitation (IP) was performed in 1× IP buffer (10 mM sodium phosphate pH 7.0, 140 mM NaCl, 0.25% Triton X-100) using anti-methylcytosine (mecyt, purchased from AnaSpec, Fremont, CA) antibody and negative control antibody (anti-cMyc, purchased from Santa Cruz, Santa Cruz, CA) for 2 h at 4°C. After the incubation, 30-μl magnetic beads (Cell Signaling, Boston, MA) were added and rotated at 4°C for another 2 h. The pulled-down DNA–beads complex was then washed four times using ice cold IP buffer, digested with proteinase K, and followed by DNA purification using Miniprep kit from QIAGEN (Valencia, CA). In the final step, the volume of inputs and the volume of precipitated DNA were adjusted to the same volume, and the relative amount of DNA precipitated was quantified by regular and real-time PCR (ABI7900HT). Regular PCR was performed using Platinum PCR SuperMix (Invitrogen, Carlsbad, CA) with the following conditions: 94°C 3 min; 94°C 30 s, 60°C 45 s, 72°C 45 s, 35 cycles. The PCR products were isolated by agarose gel electrophoresis and visualized by EB staining using a Gel Documentation 2000 system (Bio-Rad, Hercules, CA). For the real-time PCR (qPCR), the enrichment of the pulled-down DNA was calculated according to the calibration of the serial dilution of the inputs, and the percentage of the pulled-down DNA to the whole amount of IP DNA was used for further comparison. To amplify the Neurog1 fragment, a forward primer 5′-CAAAGTCACGCGACCACTAA-3′ and a reverse primer 5′-CGGAAACCTGGAGGGTACTT-3′ were used to amplify a 72-bp fragment which is 12 bp upstream of the first CpG site of Neurog1. As a negative control, a non-methylated sequence of ribosomal protein, large protein 0 (RPLP0) was amplified simultaneously, with the forward and reverse primer sequences: 5′-TTAGTTTGCTGAGCTCGCCAG-3′ and 5′-CTCTGAGCTGCTGCCACCTG-3′, respectively.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) assay was performed using Invitrogen kit (Invitrogen, CA, 49-024) following the manufacturer’s instructions. Briefly, LNCaP cells treated with DMSO, CUR, or 5-Aza/TSA for 7 days were washed with PBS and trypsinized. Followed by a PBS wash, chromatins in these cells (100,000 cells were used per IP) were then cross-linked with 1% formaldehyde for 10 min at room temperature, sonicated to generate ~200- to 500-bp DNA fragments in lysis buffer, and immunoprecipitated with 6 to 10 μl anti-MeCP2, anti-trimethyl-histone H3-Lys27 (H3K27me3) antibodies (Millipore, MA), or mouse IgG to capture antibody-DNA complex. The enrichment of the eluted DNA was quantified based on comparison with the inputs by qPCR; primers used for the amplification of Neurog1 were the same as the MeDIP analysis described above.
Histone H3 Methylated Lys27 ELISA
Acid extracts/crude histone proteins obtained from the treated LNCaP cells were prepared using the protocol provided by Active Motif (Carlsbad, CA). Cells were washed with PBS, resuspended in five volumes of lysis buffer (0.4 M HCL), and incubated on ice for 30 min. After centrifugation, the supernatant was neutralized by adding two fifth of the total volume of neutralization buffer (1 M sodium phosphate, dibasic, pH 12.5; 2.5 mM DTT; 10 mM PMSF). 5 μg of crude histone protein from each treatment was used for histone trimethyl-lys27 (H3K27me3) ELISA (Active Motif, Carlsbad, CA).
HDAC Activity Assay
Nuclear extracts and acid extracts of proteins prepared from LNCaP cells treated with DMSO, CUR, or 5-Aza/TSA combination were used for HDAC activity assay using EpiQuik Nuclear Extract Kit (Epigentek, NY), following the manufacturer’s instructions. For this HDAC activity assay, 4 μg of nuclear extract and 1 μg of crude acid extract of proteins were used, respectively.
RESULTS
Neurog1 Is One of the Genes Demethylated in the PCR Array Analysis
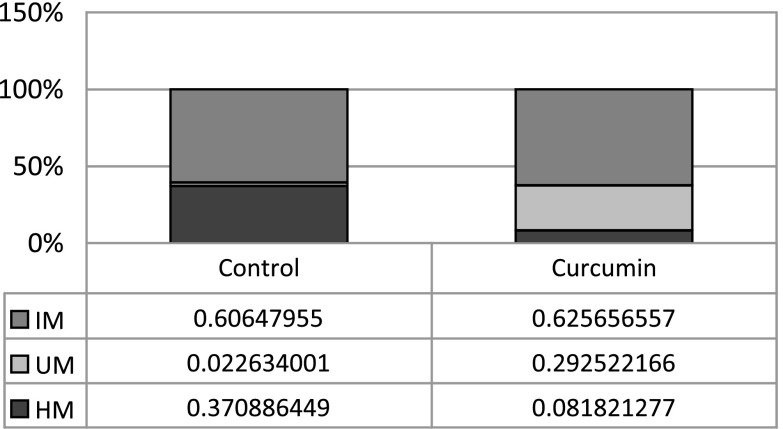
To examine whether CUR has any demethylation effect in prostate cancer cells, a methyl-profiler qPCR assay (SABioscience, QIAGEN, MD) with a collected panel of 24 genes or 96 genes selected from their reported hypermethylation was performed in two different experiment sets. For the LNCaP cell treated with 5 μM CUR for 5 days, some of the genes showed a decrease in methylation level (data not shown). In the next panel experiment, we treated the LNCaP cell with 5 μM CUR for 7 days. Using the PCR array panel of the 96 genes, six genes including Neurog1 displayed methylation level decreased by more than 20% (for Neurog1, see Fig. 1). According to the instruction of the PCR array (www.sabioscience.com), the total amount of hypermethylated (Hm, > 60% methylated) and unmethylated (Um, 0% methylated) DNA in the sample was each directly and reliably detected. The total amount of DNA minus the hypermethylated and unmethylated DNA amounts yields the amount of intermediate-methylated DNA (IM, between 0% and 60% methylated). Compared with the DMSO-treated cells, the hypermethylation level of Neurog1 for CUR treatment decreased from 37% to 8%, corresponding with a significant increase of unmethylation level from 2% to 29%, while the intermediate-methylation level remained unchanged.
Fig. 1.
Curcumin treatment demethylated CpG sites of Neurog1 CpG island. LNCaP cells were treated with 5 μM CUR for 7 days. A PCR array consisting of a panel of 96 genes of human prostate cancer DNA methylation PCR assay (MeAH-3050, 384 wells) was performed following the manufacturer’s instructions. The data were uploaded and analyzed online (www.sabioscience.com), and the results of Neurog1 were shown. According to instructions, the total hypermethylated (Hm, > 60% methylated) and unmethylated (Um, 0% methylated) amounts of DNA in the sample were directly and reliably detected. The total amount of DNA minus the hypermethylated and unmethylated DNA yields the amount of intermediate methylated (IM) DNA (between 0% and 60% methylated)
Among the six demethylated genes by CUR, Neurog1 has been reported and selected as a cancer methylation marker in retinoblastoma and prostate cancers (1,9), and therefore we next performed further investigation on the methylation status of the Neurog1 gene.
CUR Partially Demethylated CpG Sites of Neurog1 in LNCaP Cell
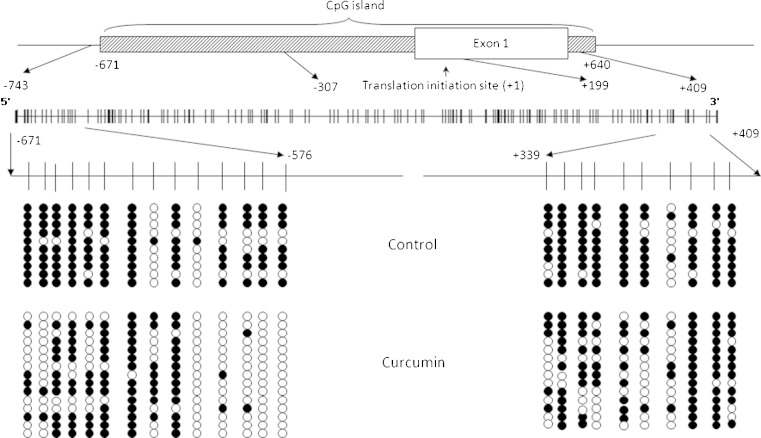
Neurog1 is a single-exon gene with the CpG island of around 1.3 kb (www.sabioscience.com), starting from −671 bp (with the translation initiation site designated as +1). To investigate the methylation status of the CpG islands of Neurog1, we designed primers to amplify the bisulfite converted CpG island containing fragments. The first fragment (fragment 1) covers the region from −671 to −307 containing 47 CpG islands, while the second fragment (fragment 2) covers the region from +199 to +409 containing 35 CpG islands. Interestingly, for the fragment 1, only the first 14 CpGs (−671 to −575) were highly methylated in the DMSO-treated control cell, with the average proportion of methylation around 73.6% (Fig. 2), while the remaining 33 CpG sites were rarely methylated (data not shown). For instance, after the first 14th CpGs, the remaining 12 continuous CpGs (−572 to −498) have no methylation detected at all in the 10 clones sequenced. The proportion of the average methylation level for the whole 47 CpGs sequenced is 26%, while it is only 3.9% for the remaining 33 CpGs.
Fig. 2.
Bisulfite genomic sequencing of Neurog1 CpG islands. DNAs extracted from 5 μM CUR- and DMSO-treated control LNCaP cells for 7 days were bisulfite converted, and two fragments of the Neurog1 gene were amplified using two methylation specific primers and cloned into the pCR4 TOPO vector for sequencing: fragment 1 covered −671 to −307, with the translation start site referenced as +1. The first 14 CpG sites (−671 to −575) were highly methylated and dramatically decreased by CUR are shown in the left panel, while the CpG sites (−572 to −307) were not methylated and not shown; fragment 2 covered +199 to +409, and CUR has limited effect on demethylation, a representative of 10 CpG sites (from +339 to +409) are shown. The white dots represent unmethylated and the black dots represent methylated CpG sites. Detailed procedure and sequence information are described in “Experimental Section”
Upon 5 μM CUR treatment for 7 days, the first 14 CpGs of fragment 1 were dramatically demethylated. The proportion of methylated CpGs decreased from 73.6% to 40.0% (Fisher exact test, following the same, p value < 0.0001), and the changes were the most significant for the first two and the last four CpGs (Fig. 2, left panel). The next downstream 33 CpGs of fragment 1 were least affected by CUR treatment; the proportion of methylation levels were 3.9% and 1.6% respectively, both at very low levels. A representative circle plot of the last 10 CpG sites of the fragment 2 is shown in Fig. 2, right panel.
With regards to fragment 2 (+199 to +409), the average methylation level was 74% for all 35 CpG sites sequenced. CUR treatment showed similar pattern with the control group, and it had limited effects on the overall average proportion of methylation (p value = 0.879). However, when examining the partial region of the CpG sites (+339 to +409), as shown in Fig. 2, right panel, CUR demethylated these CpG sites significantly (p value = 0.0004 for the CpG sites +339 to +409).
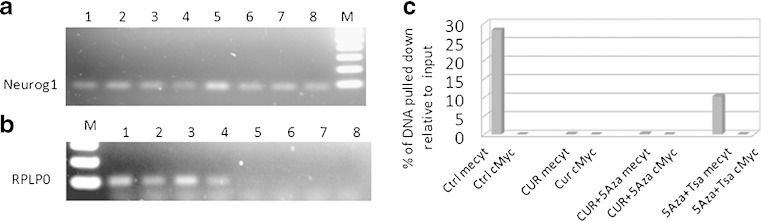
Methylated DNA can be detected unbiasedly by immunoprecipitation (IP) using anti-methyl cytosine (mecyt) antibody, and this methylated DNA immunoprecipitation (MeDIP) method has been established to show the level of DNA enrichment increased in a linear manner with the number of methylated cytosines (24,25). LNCaP cells were treated with DMSO, CUR, CUR/5-Aza, or 5-Aza/TSA for 7 days, followed by DNA extraction and MeDIP analysis. A 72-bp fragment located 12 bp upstream from the first CpG site (−755 to −683) was amplified to analyze the mecyt antibody binding, and 5-Aza/TSA-treated sample was used as a positive demethylation control. Corresponding with the TA cloning results above, the CUR-treated groups had much less amplification compared to the control group (Fig. 3a, lanes 5–7). A real-time PCR was performed to quantify the immunoprecipitated DNA products with their inputs. Both CUR alone and the CUR/5-Aza combination dramatically decreased the anti-mecyt antibody binding (Fig. 3c), whereas the cMyc antibody used as a non-specific binding control gave very low or non-detectable amplification in this MeDIP system. In contrast, a house keeping gene, RPLP0, which is ummethylated (26), was not detected in the immunoprecipitated DNA (Fig. 3b, lanes 5–8).
Fig. 3.
MeDIP analysis of Neurog1 methylation. Eight-microgram genomic DNAs extracted from control (Ctrl), curcumin (CUR), curcumin/5-Aza-2-deoxycytidine (CUR/5-Aza), 5-Aza/Trichostatin A (5-Aza/TSA, TSA was added 20 h before harvested) were sonicated, denatured, and subjected to DNA IP with anti-methyl cytosine (mecyt) antibody. a, b Semi-quantitative PCR comparing the immunoprecipitated DNA with its inputs; lanes 1–4 input DNA of (1 = Ctrl, 2 = CUR, 3 = CUR/5-Aza, 4 = 5-Aza/TSA); lanes 5–8 Mecyt-IP DNA of (5 = Ctrl; 6 = CUR; 7 = CUR/5-Aza; 8 = 5-Aza/TSA). M marker. Primers used are a Neurog1 and b RPLP0. c qPCR was performed to quantify the IP DNA with its inputs. The inputs were diluted from 2×, 4×, 8×, 16×, and 32×, based on the standard curve of ΔCT value, and the relative amount of IP DNA was calculated. The cMyc antibody was considered as non-specific binding control
CUR Treatment Reactivates Neurog1 in LNCaP Cells
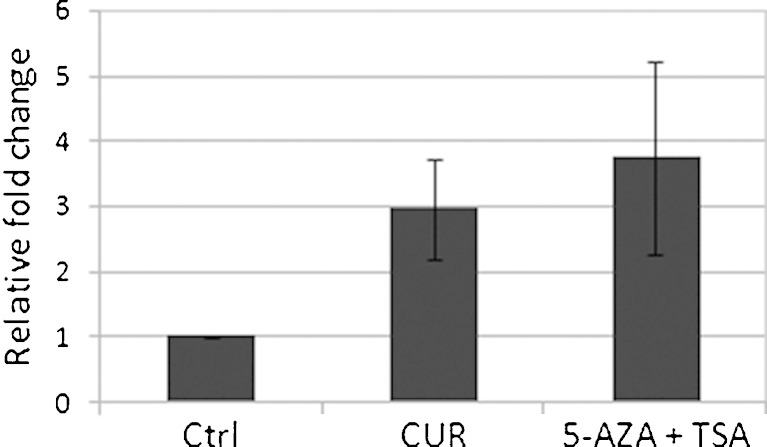
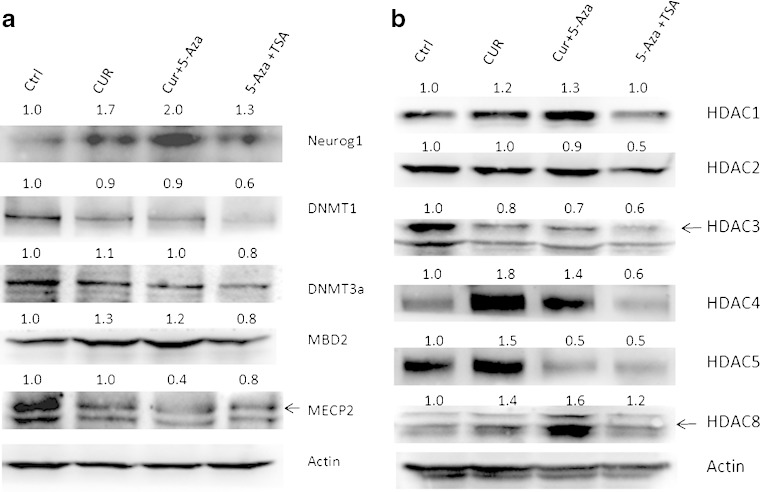
One of the possible consequences of promoter demethylation of the gene is the transcription activation of that gene. From the samples described above, mRNAs were extracted and reverse-transcribed, and the cDNAs were used to perform qPCR to determine the mRNA level of Neurog1. Consistent with the demethylation of CpG sites, CUR treatment increased the mRNA level of Neurog1 (Fig. 4). Similarly, when Western blotting was performed to measure the protein expression of Neurog1, CUR alone or CUR/5-Aza combinations increased the protein level of Neurog1 to 1.7- and 2.0-fold of control, respectively (Fig. 5a, top panel).
Fig. 4.
CUR-activated Neurog1 mRNA expression. Total RNA extracted from the cells treated for 7 days was reverse-transcribed and quantified by real-time PCR (qPCR). Two parallel RNAs were prepared and each was duplicated for the qPCR: control (Ctrl), curcumin (CUR), 5-Aza/Trichostatin A (5-Aza/TSA, TSA was added 20 h before harvest). Primer sequences are shown in the “Experimental Section”
Fig. 5.
CUR-regulated Neurog1 and chromatin remodeling proteins expression. Control and CUR-treated cells were harvested using a RIPA buffer with protein inhibitor cocktail (Sigma); the protein concentrations of the cleared lysates were determined using the BCA method and 20 μg of total proteins each was resolved by 4–15% SDS-polyacrylamide gel electrophoresis, followed by immunoblotting with different antibodies. Neurog1 and methyl-binding proteins and DNMTs were shown in a followed by the image quantification with ImageJ (NIH) and normalized using anti-actin b; HDACs (1–5,8) are shown and quantified in c. The different sources of the antibodies are described in “Experimental Section”
CUR Treatment Has Various Effects on Epigenetic Modifying Proteins
Since CUR has been reported to be a DNMT inhibitor, we next examined whether CUR can alter the protein level of DNMTs. When normalized with actin, we did not see any significant decrease in the expression of DNMT1 and DNMT3a. In contrast, 5-Aza/TSA combination treatment decreased DNMT1 and DNMT3a expression by almost 40% and 20%, respectively (Fig. 5a). Two major methyl DNA binding proteins, MBD2 and MeCP2, were also checked for their expression. CUR alone had little effect on their expressions except for CUR/5-Aza (Fig. 5a).
CUR has been reported to inhibit histone acetyltransferase (HAT) activity, and it was also postulated to inhibit some HDACs. We therefore performed a series of Western blots to determine whether CUR had any effect on the expression level of HDAC1–5 and HDAC8. Interestingly, when normalized with actin, expressions of HDAC1, HDAC4, and HDAC8 increased with CUR alone and CUR/5-Aza combination treatments. HDAC5 expression was increased by CUR treatment alone but decreased in CUR/5-Aza combination treatment, while HDAC3 expression level was decreased around 20% whereas HDAC2 was not affected (Fig. 5b). In contrast, treatment with 5-Aza/TSA decreased most of the HDACs expression (HDAC2-5) (Fig. 5b).
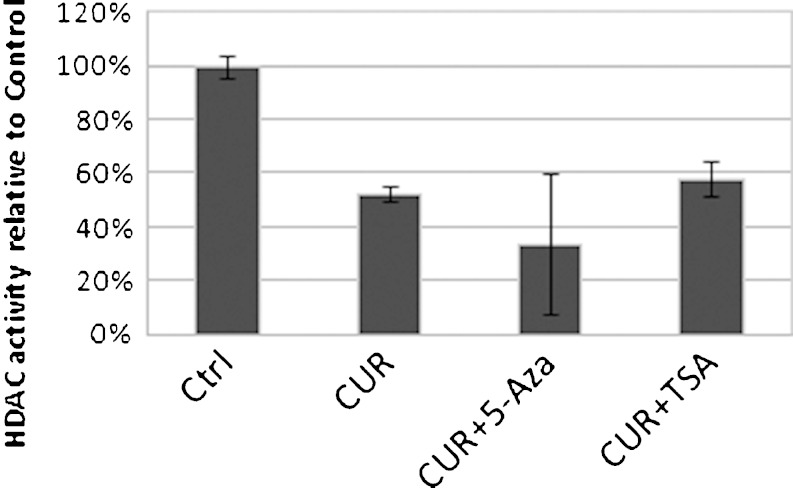
CUR Treatment Decreases the Total HDAC Activity
To investigate whether CUR has any effect on HDAC activity, nuclear extracts of cells with different treatments were measured for the total HDAC activities. Compared to the control group, in CUR treatment alone and combination treatments of CUR with 5Aza or CUR with TSA, the total HDAC activities were decreased significantly (Fig. 6). When acid extracts were used for the same test, similar results were obtained in that CUR treatment decreased HDAC activity (data not shown).
Fig. 6.
CUR treatment decreased total HDAC activity. Four-microgram nuclear extracts were prepared from LNCaP cells treated with DMSO, 5 μM CUR, 5 μM CUR with 2.5 μM 5-Aza, and 5 μM CUR with 500 nM TSA for 7 days (TSA was added 20 h before harvest) using Epiquik Nuclear Extraction Kit (Epigentek, NY, OP-0002-1), and the HDAC activity assay was performed following the manufacturer’s (Epigentek, P-4002) instructions
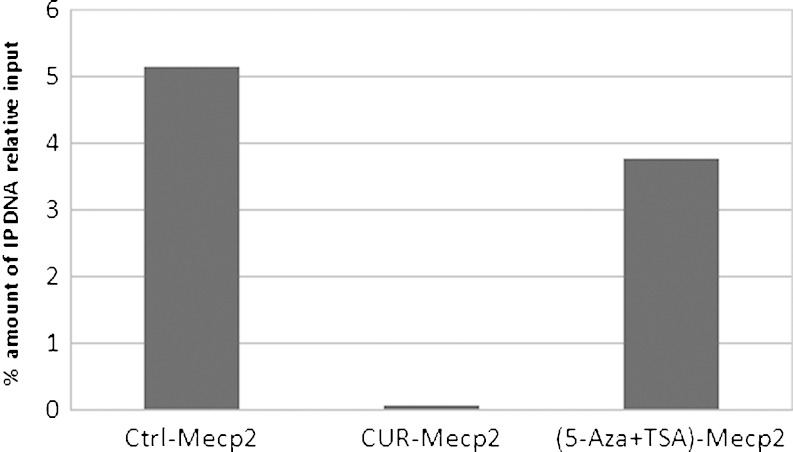
Methylated CpG binding protein 2 (MeCP2) is one of the methyl DNA binding proteins. This nuclear protein is involved in histone modification-related transcriptional repression of specific genes (27). MeCP2 binds to both methylated and unmethylated DNA, but it associates preferentially with methylated regions and its genome-wide binding tracks with methyl-CpG density (28). To determine whether LNCaP cells treated with CUR would cause the methylation status to change, a ChIP assay was performed. CUR treatment decreased the MeCP2-Neurog1 binding dramatically, and the pulled-down Neurog1 fragment was even less than the 5-Aza/TSA combination treatment (Fig. 7). This result further confirms that CUR was able to demethylate the Neurog1 promoter region and also suggests that the decreased binding of MeCP2 to the methylated CpG promoter of Neurog1 could lead to an inhibited transcription repression.
Fig. 7.
CUR treatment decreased Neurog1 enrichment pulled down by anti-MeCP2 antibody. The ChIP assay was performed following the manufacturer’s instruction (Invitrogen, CA, 49-024). LNCaP cells treated with DMSO, CUR, and 5-Aza/TSA (TSA was added 20 h before harvest) for 7 days were washed with PBS and trypsinized. Chromatins in these cells (100,000 cells were used per IP) were then cross-linked, sonicated, and immunoprecipated with MeCP2 antibody. The eluted DNA was quantified and compared to the inputs based on Q-PCR. Primers used for the amplification of Neurog1 were the same as MeDIP analysis
CUR Treatment Decreases the Enrichment of H3K27me3 at Neurog1 Promoter and at Global Level
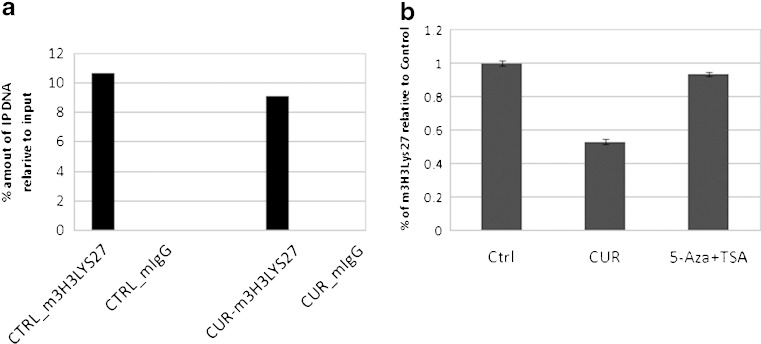
Many epigenetic regulations of gene expression are controlled by histone modifications, particularly histone methylation (29). Tri-methylation of histone 3 at lysine 27 (H3K27me3) is associated with the repression of transcription. This methylation was catalyzed by Polycomb Regressive Complex 2, and therefore the methylation state of lysine 27 is an interesting marker of transcriptional activity (30). To analyze whether CUR treatment had any effect on the H3K27me3, ChIP assay was performed using H3K27me3 antibody. It revealed that CUR treatment decreased the binding of H3K27me3 to the promoter of the Neurog1 gene (Fig. 8a).
Fig. 8.
CUR decreased the enrichment of tri-methylation of histone 3 lys 27 (H3K27me3) at Neurog1 promoter region and global level. a For the LNCaP cells treated with DMSO and CUR for 7 days, ChIP assay was performed to pull down the antibody–chromatin complex using anti-H3K27me3 antibody. The details were described in “Experimental Section” and quantified by Q-PCR. b For LNCaP cells treated with DMSO, CUR, and 5-Aza/TSA (TSA was added 20 h before harvest), global enrichment of H3Lys27me3 was measured by histone H3 methylated lys27 ELISA (Active Motif, Carlsbad, CA) following the manufacturer’s instructions
To determine whether CUR treatment had any global effect on tri-methylation of histone 3 lys 27, an H3K27me3 ELISA was performed to detect the total amount of H3K27me3 enrichment in LNCaP cells. Acid extract/crude histone proteins were extracted from the control-, CUR- and 5-Aza/TSA-treated LNCaP cells, and the tri-methylation of histone H3-specific antibody was used for ELISA. Interestingly, CUR treatment decreased the total amount of H3K27me3 enrichment, even more dramatically than the 5-Aza/TSA combination treatment (Fig. 8b), suggesting the potential activation of other genes by CUR in addition to Neurog1.
DISCUSSION
CUR is a potent anti-cancer polyphenolic phytochemical, and its anti-inflammatory and anti-oxidative effects are well established (11,31). Regarding the molecular mechanism of CUR on epigenetic regulation, although there are some reports suggesting that it may possess potential DNMT inhibition (20), there was a lack of direct experimental evidence to show the epigenetic effect of CUR in prostate cancer cells. In our current study, we present evidence that CUR was able to demethylate a specific gene, Neurog1, in human LNCaP cells. Neurog1 has been extensively studied in neuronal cells; it interacts with CREB-binding protein (CBP)/p300 and smad1 by sequestering this transcription complex and thereby inhibiting astrocyte differentiation (32). Furthermore, Neurog1 is hypermethylated in prostate and colon cancers and it has been accepted as a cancer methylation marker (8,9). Recently, it was reported that methylation of Neurog1 in serum could be used as a sensitive marker to detect early colorectal cancer (33). Although the relationship between Neurog1 and prostate cancer has not been established, an interference transcription activity of CBP–smad 1 transcription complex could be potentially involved. Demethylation of Neurog1 by CUR would provide an alternative anti-cancer mechanism beyond the conventional anti-inflammatory and anti-oxidative effects of this compound.
CUR treatment did not reduce the protein level of the methyl DNA binding proteins, MBD2 and MeCP2. To date, there are still no clear correlations between MBD expression and tumor risk or aggressiveness (34). Similar results were also observed for the protein expression level of DNA methyltransferases DNMT1 and DNMT3a, CUR did not alter their protein expression levels significantly. These results are consistent with previous reports showing that CUR could be involved in the demethylation process through inhibition of DNMT activity (21) instead of acting through protein expression.
CUR has been shown to regulate the HAT activity (17–19). However, this is somewhat puzzling, since inhibition of HAT activity would normally lead to inhibition of gene transcription activity that is typically associated with hypermethylation of the promoter of the gene (35). Currently, several histone deacetylation (HDAC) inhibitors such as vorinostat (SAHA) are being discovered in cancer therapy (www.clinicaltrial.gov/ct2/results?term=saha). Hypoacetylation caused by inhibition of the HAT could potentially exert other side effects including cell viability, differentiation, and apoptosis (18). Both HAT and HDAC proteins are involved in the chromatin remodeling process by targeting histone and non-histone proteins and serving as co-activators and co-repressors, respectively (36). The balance between histone acetylation and deacetylation may determine the fate of the cells. The specific contribution of HAT and HDAC to the overall chromatin remodeling currently remains unclear. An aberrant activation or inhibition of HATs or HDACs activities could impact on the carcinogenesis process. In the context of DNA methylation, hypermethylation on some specific genes may not represent a “closed” state of the whole chromatin. For instance, in prostate adenocarcinoma, it has been reported that hypermethylation of ASC, COX2, RARB, TNFRSF10c, and many other methylation marker genes, including Neurog1, is associated with lower methylation levels of the repetitive elements Alu and LINE-1(9). In this regard, CUR could demethylate and activate some specific methylation marker or tumor suppressor genes, and the HAT inhibition activity of CUR may contribute to maintaining the stability of the whole genome. However, further studies would be needed to address the capability of CUR on the methylation status of the whole genome.
Beyond HAT inhibition activity, CUR has also been reported to possess HDAC inhibition ability using molecular docking (20). We performed Western blots to test the protein expression level of HDACs of class I and II. Interestingly, we found that some HDACs, such as HDAC4, 5, and 8 can be upregulated upon CUR treatment. In some way, this could be due to a direct and/or indirect inhibition on HAT activity. On the other hand, the expression level of HDAC3 was decreased, suggesting that CUR could regulate different HDACs expression possibly through different mechanisms. Further HDAC activity test showed that CUR treatment caused a decrease of the overall HDAC activity. Furthermore, H3Lys27me3 ChIP assay and ELISA analysis showed that CUR treatment decreased H3Lys27me3 binding with the promoter of Neurog1 and significantly decreased the total amount of H3Lys27me3. These results appear to corroborate with previous reports showing that tri-methylation of Histone 3 at lysine 27 (H3K27me3) is associated with the repression of genes transcription (30). Although the exact mechanism as to how Neurog1 is demethylated and transcriptionally activated, requires further study, our results above show that CUR is indeed a strong modulator of DNA demethylation and transcriptionally activate Neurog1 gene.
As positive control, 5-Aza/TSA combination treatment dramatically decreased DNMT1, DNMT3a, and multiple HDAC proteins (HDAC2-5). Interestingly, the reactivation of Neurog1 protein expression for 5-Aza/TSA combination treatment was not as dramatic as CUR alone or CUR/5-Aza combination treatments (Fig. 5a). This appears to be consistent with our observations above that CUR treatment has more dramatic effect on the MeCP2-Neurog1 binding and global H3Lys27m3 enrichment, which contribute in part to the higher Neurog1 expression. It also potentially suggests that the specificity of CUR involved in epigenetic regulation of genes such as Neurog1.
In conclusion, in this study, we showed that CUR was able to demethylate the Neurog1 promoter and reactivate its mRNA and protein expression in human LNCaP prostate cancer cells. Hypermethylation of the promoter regions of this methylation marker has been associated with epigenetic changes in human tumors (37). In general, the therapeutic control of cancers through epigenetic regulations is considered to be more amenable than irreversible genetic changes (38). In this context, our current study with CUR suggests that some phytochemicals may be selected for epigenetic therapy of prostate cancers. Given that CUR can demethylate some specific promoters of genes as well as maintaining/promoting the stability of the whole chromatin structure, future detailed study of the methylation status of the whole genome will be interesting.
ACKNOWLEDGMENTS
We thank all the members in Dr. Tony Kong’s lab for their helpful discussion and preparation of this manuscript. This work was supported in part by Institutional Funds.
REFERENCES
- 1.Cohen Y, Merhavi-Shoham E, Avraham RB, Frenkel S, Pe’er J, Goldenberg-Cohen N. Hypermethylation of CpG island loci of multiple tumor suppressor genes in retinoblastoma. Exp Eye Res. 2008;86(2):201–206. doi: 10.1016/j.exer.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 2.Cooper CS, Foster CS. Concepts of epigenetics in prostate cancer development. Br J Cancer. 2009;100(2):240–245. doi: 10.1038/sj.bjc.6604771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeronimo C, Henrique R, Hoque MO, Mambo E, Ribeiro FR, Varzim G, et al. A quantitative promoter methylation profile of prostate cancer. Clin Cancer Res. 2004;10(24):8472–8478. doi: 10.1158/1078-0432.CCR-04-0894. [DOI] [PubMed] [Google Scholar]
- 4.Yu S, Khor TO, Cheung KL, Li W, Wu TY, Huang Y, et al. Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice. PLoS One. 2010;5(1):e8579. doi: 10.1371/journal.pone.0008579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blader P, Lam CS, Rastegar S, Scardigli R, Nicod JC, Simplicio N, et al. Conserved and acquired features of neurogenin1 regulation. Development. 2004;131(22):5627–5637. doi: 10.1242/dev.01455. [DOI] [PubMed] [Google Scholar]
- 6.Ogino S, Kawasaki T, Kirkner GJ, Suemoto Y, Meyerhardt JA, Fuchs CS. Molecular correlates with MGMT promoter methylation and silencing support CpG island methylator phenotype-low (CIMP-low) in colorectal cancer. Gut. 2007;56(11):1564–1571. doi: 10.1136/gut.2007.119750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shima K, Morikawa T, Baba Y, Nosho K, Suzuki M, Yamauchi M, et al. MGMT promoter methylation, loss of expression and prognosis in 855 colorectal cancers. Cancer Causes Control. 2011;22(2):301–309. doi: 10.1007/s10552-010-9698-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogino S, Cantor M, Kawasaki T, Brahmandam M, Kirkner GJ, Weisenberger DJ, et al. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006;55(7):1000–1006. doi: 10.1136/gut.2005.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho NY, Kim BH, Choi M, Yoo EJ, Moon KC, Cho YM, et al. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J Pathol. 2007;211(3):269–277. doi: 10.1002/path.2106. [DOI] [PubMed] [Google Scholar]
- 10.Srivastava RK, Chen Q, Siddiqui I, Sarva K, Shankar S. Linkage of curcumin-induced cell cycle arrest and apoptosis by cyclin-dependent kinase inhibitor p21(/WAF1/CIP1) Cell Cycle. 2007;6(23):2953–2961. doi: 10.4161/cc.6.23.4951. [DOI] [PubMed] [Google Scholar]
- 11.Yu S, Shen G, Khor TO, Kim JH, Kong AN. Curcumin inhibits Akt/mammalian target of rapamycin signaling through protein phosphatase-dependent mechanism. Mol Cancer Ther. 2008;7(9):2609–2620. doi: 10.1158/1535-7163.MCT-07-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thangapazham RL, Sharma A, Maheshwari RK. Multiple molecular targets in cancer chemoprevention by curcumin. AAPS J. 2006;8(3):E443–E449. doi: 10.1208/aapsj080352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma RA, Ireson CR, Verschoyle RD, Hill KA, Williams ML, Leuratti C, et al. Effects of dietary curcumin on glutathione S-transferase and malondialdehyde-DNA adducts in rat liver and colon mucosa: relationship with drug levels. Clin Cancer Res. 2001;7(5):1452–1458. [PubMed] [Google Scholar]
- 14.Volak LP, Ghirmai S, Cashman JR, Court MH. Curcuminoids inhibit multiple human cytochromes P450, UDP-glucuronosyltransferase, and sulfotransferase enzymes, whereas piperine is a relatively selective CYP3A4 inhibitor. Drug Metab Dispos. 2008;36(8):1594–1605. doi: 10.1124/dmd.108.020552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goel A, Aggarwal BB. Curcumin, the golden spice from Indian saffron, is a chemosensitizer and radiosensitizer for tumors and chemoprotector and radioprotector for normal organs. Nutr Cancer. 2010;62(7):919–930. doi: 10.1080/01635581.2010.509835. [DOI] [PubMed] [Google Scholar]
- 16.Shen G, Xu C, Hu R, Jain MR, Gopalkrishnan A, Nair S, et al. Modulation of nuclear factor E2-related factor 2-mediated gene expression in mice liver and small intestine by cancer chemopreventive agent curcumin. Mol Cancer Ther. 2006;5(1):39–51. doi: 10.1158/1535-7163.MCT-05-0293. [DOI] [PubMed] [Google Scholar]
- 17.Balasubramanyam K, Varier RA, Altaf M, Swaminathan V, Siddappa NB, Ranga U, et al. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J Biol Chem. 2004;279(49):51163–51171. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- 18.Kang J, Chen J, Shi Y, Jia J, Zhang Y. Curcumin-induced histone hypoacetylation: the role of reactive oxygen species. Biochem Pharmacol. 2005;69(8):1205–1213. doi: 10.1016/j.bcp.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 19.Kang SK, Cha SH, Jeon HG. Curcumin-induced histone hypoacetylation enhances caspase-3-dependent glioma cell death and neurogenesis of neural progenitor cells. Stem Cells Dev. 2006;15(2):165–174. doi: 10.1089/scd.2006.15.165. [DOI] [PubMed] [Google Scholar]
- 20.Bora-Tatar G, Dayangac-Erden D, Demir AS, Dalkara S, Yelekci K, Erdem-Yurter H. Molecular modifications on carboxylic acid derivatives as potent histone deacetylase inhibitors: activity and docking studies. Bioorg Med Chem. 2009;17(14):5219–5228. doi: 10.1016/j.bmc.2009.05.042. [DOI] [PubMed] [Google Scholar]
- 21.Liu Z, Xie Z, Jones W, Pavlovicz RE, Liu S, Yu J, et al. Curcumin is a potent DNA hypomethylation agent. Bioorg Med Chem Lett. 2009;19(3):706–709. doi: 10.1016/j.bmcl.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 22.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37(8):853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 23.Cheung HH, Lee TL, Davis AJ, Taft DH, Rennert OM, Chan WY. Genome-wide DNA methylation profiling reveals novel epigenetically regulated genes and non-coding RNAs in human testicular cancer. Br J Cancer. 2010;102(2):419–427. doi: 10.1038/sj.bjc.6605505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37(8):853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 25.Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38(2):149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 26.Muller I, Wischnewski F, Pantel K, Schwarzenbach H. Promoter- and cell-specific epigenetic regulation of CD44, Cyclin D2, GLIPR1 and PTEN by methyl-CpG binding proteins and histone modifications. BMC Cancer. 2010;10:297. doi: 10.1186/1471-2407-10-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen JC, Ghosh RP, Woodcock CL. Binding of the Rett syndrome protein, MeCP2, to methylated and unmethylated DNA and chromatin. IUBMB Life. 2010;62(10):732–738. doi: 10.1002/iub.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, et al. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37(4):457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14(2):155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 30.Young MD, Willson TA, Wakefield MJ, Trounson E, Hilton DJ, Blewitt ME, et al. ChIPseq analysis reveals distinct H3K27me3 profiles that correlate with transcriptional activity. Nucleic Acids Res. 2011. doi:10.1093/nar/gkr416 [DOI] [PMC free article] [PubMed]
- 31.Kamat AM, Tharakan ST, Sung B, Aggarwal BB. Curcumin potentiates the antitumor effects of Bacillus Calmette-Guerin against bladder cancer through the downregulation of NF-kappaB and upregulation of TRAIL receptors. Cancer Res. 2009;69(23):8958–8966. doi: 10.1158/0008-5472.CAN-09-2045. [DOI] [PubMed] [Google Scholar]
- 32.Sun Y, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, et al. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104(3):365–376. doi: 10.1016/S0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- 33.Herbst A, Rahmig K, Stieber P, Philipp A, Jung A, Ofner A, et al. Methylation of NEUROG1 in serum is a sensitive marker for the detection of early colorectal cancer. Am J Gastroenterol. 2011;106(6):1110–1118. doi: 10.1038/ajg.2011.6. [DOI] [PubMed] [Google Scholar]
- 34.Berger J, Bird A. Role of MBD2 in gene regulation and tumorigenesis. Biochem Soc Trans. 2005;33(Pt 6):1537–1540. doi: 10.1042/BST0331537. [DOI] [PubMed] [Google Scholar]
- 35.Verdone L, Agricola E, Caserta M, Di Mauro E. Histone acetylation in gene regulation. Brief Funct Genomic Proteomic. 2006;5(3):209–221. doi: 10.1093/bfgp/ell028. [DOI] [PubMed] [Google Scholar]
- 36.Lafon-Hughes L, Di Tomaso MV, Mendez-Acuna L, Martinez-Lopez W. Chromatin-remodelling mechanisms in cancer. Mutat Res. 2008;658(3):191–214. doi: 10.1016/j.mrrev.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 37.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3(6):415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 38.Rennie PS, Nelson CC. Epigenetic mechanisms for progression of prostate cancer. Cancer Metastasis Rev. 1998;17(4):401–409. doi: 10.1023/A:1006121219097. [DOI] [PubMed] [Google Scholar]