Abstract
Organic nitrate vasodilators (ORN) exert their pharmacologic effects through the metabolic release of nitric oxide (NO). Mitochondrial aldehyde dehydrogenase (ALDH2) is the principal enzyme responsible for NO liberation from nitroglycerin (NTG), but lacks activity towards other ORN. Cytosolic aldehyde dehydrogenase (ALDH1a1) can produce NO from NTG, but its activity towards other ORN is unknown. Using purified enzymes, we showed that both isoforms could liberate NO from NTG, isosorbide dinitrate (ISDN), and nicrorandil, while only ALDH1a1 metabolized isosorbide-2-mononitrate and isosorbide-5-mononitrate (IS-5-MN). Following a 10-min incubation with purified enzyme, 0.1 mM NTG and 1 mM ISDN potently inactivated ALDH1a1 (to 21.9% ± 11.1% and 0.44% ± 1.04% of control activity, respectively) and ALDH2 (no activity remaining and 4.57% ± 7.92% of control activity, respectively), while 1 mM IS-5-MN exerted only modest inactivation of ALDH1a1 (reduced to 89% ± 4.3% of control). Cytosolic ALDH in hepatic homogenates incubated at the vascular EC50 concentrations of ORN was inactivated by NTG (to 45.1% ± 8.1% of control activity) while mitochondrial ALDH was inactivated by NTG and nicorandil (to 68.2% ± 10.0% and 78.7% ± 19.8% of control, respectively). Via site-directed mutagenesis, the active sites of ORN metabolism of ALDH2 (Cys-319) and ALDH1a1 (Cys-303) were found to be identical to those responsible for their dehydrogenase activity. Cysteine-302 of ALDH1a1 and glutamate-504 of ALDH2 were found to modulate the rate of ORN metabolism. These studies provide further characterization of the substrate selectivity, inactivation, and active sites of ALDH2 and ALDH1a1 toward ORN.
Key words: aldehyde dehydrogenase, nitric oxide, organic nitrate, site-directed mutagenesis
INTRODUCTION
The predominant role of mitochondrial aldehyde dehydrogenase (ALDH2) to bioactivate nitroglycerin (NTG) is now widely accepted (1). In addition to its dehydrogenase and esterase activity, ALDH2 possesses a nitrate reductase activity involving redox-sensitive thiol groups in the enzyme, producing nitrite ions and 1,2-glyceryl dinitrate (GDN) as the predominant metabolites. However, ALDH2 is not responsible for the metabolism of two other clinically used organic nitrates, viz., isosorbide dinitrate (ISDN) and isosorbide-5-mononitrate (IS-5-MN), which contain a lower number of nitrate moieties (2 and 1, respectively) vs. the three nitrate group resident in NTG (2). Recently, it has been reported that a cytosolic form of ALDH, ALDH1a1, has the ability to activate NTG to nitric oxide (NO) in vitro (3). It is presently unknown whether this isoform of ALDH can bioactivate these lower organic nitrates.
Inactivation of ALDH2 has been proposed as a principal mechanism for the development of nitrate tolerance (1,4). Since ISDN and IS-5-MN also produce therapeutic tolerance upon repeated administration (5–7) and cross-tolerance toward NTG (5,6), it is of interest to determine whether these organic nitrates can also inactivate ALDH2. Furthermore, if ALDH1a1 can activate these lower organic nitrates, it would be pertinent to examine whether it would also be inactivated by these compounds. The presence of such an inactivation would then provide a potential mechanism for the tolerance development of these nitrates.
ALDH1a1 and ALDH2 both exist as tetramers comprised of homologous subunits, which contain multiple cysteine residues known to be critical to their dehydrogenase and esterase activity. ALDH2 contains three cysteine residues spanning its active site at positions 318–320, with the cysteine at position 319 identified as its active site residue (8). ALDH1a1 possesses 2 cysteines in at sites 302 and 303 with cysteine 303 being identified as its active site (8). In addition, the site of co-factor binding in ALDH2 is subject to mutation which results in the replacement of glutamate with lysine at position 504 (E504K) on exon 12. This polymorphism commonly observed in Asians results in the loss of 94% of aldehyde dehydrogenase activity in Glu504/Lys504 heterozygotes and negligible activity in Lys504/Lys504 homozygotes (9) and has been linked to impaired NTG bioactivation leading to reduced potency (4) and poor angina control (10). It is not known at present whether alteration of these critical active sites would alter the nitrate reductase activity of ALDH2 and ALDH1a1.
In this study, therefore, we compared the selectivity of purified human ALDH1a1 and ALDH2 in their ability to activate various organic nitrates to NO. We then compared the inactivation of the dehydrogenase activity of purified human ALDH1a1 and ALDH2 by the same panel of organic nitrates. Because reactions with pure enzymes may be modified physiologically by co-factors that exist in tissues/organs, we performed the inactivation experiments also in rat liver homogenates which contain high physiological amounts of both ALDH1a1 and ALDH2. Finally, we constructed and purified several mutants of ALDH1a1 and ALDH2, and compared the relative roles of the critical cysteine residues in mediating their nitrate reductase and NO-producing activities.
MATERIAL AND METHODS
Materials
NTG in lactose (10%) was kindly provided by Copperhead Chemical (Tamaqua, PA, USA). IS-5-MN was purchased from LKT (St. Paul, MN, USA) and nicorandil was purchased from Tocris (Ellisville, MO, USA). IS-2-MN was a gift from Ayerst Labs (New York, NY, USA). Sephacryl S-300 HR was obtained from GE Healthcare (Piscataway, NJ, USA). All other chemicals were purchased from Sigma (St. Louis, MO, USA).
Protein Expression, Purification, and Enzyme Activity Assay
pT7.7 vectors encoding the hALDH2 or human ALDH1a1 (hALDH1a1) cDNA were transfected into Escherichia coli BL21(DE3) pLysS (Stratagene, La Jolla, CA, USA) for expression of the recombinant protein (11). The enzymes were purified following a previously published p-hydroxyacetophenone affinity chromatography method (12). The protein was further purified using a sephacryl-S-300 size-exclusion column, eluting in 100-mM MES buffer (pH 6.5) containing 0.2 mM MgCl2. The proteins were characterized by SDS-PAGE/Coomassie blue staining and dehydrogenase activity. The dehydrogenase activity was measured according to Chen et al. (1) with slight modifications. The assay medium consisted of 50 mM sodium phosphate buffer (pH 9.0), 1 mM 4-methylpyrazole, 1 mM propionaldehyde, and 1 mM NAD+. The reaction was initiated by addition of enzyme solution and was monitored by the appearance of NADH at 340 nm at room temperature with a Shimadzu UV-1700 spectrophotometer (Columbia, MA, USA). Protein content was determined by the method of Lowry (13).
Measurement of NO Release from Nitrates
NO production was measured with an InNO-T measuring system with an AmiNO-600 electrode (Innovative Instruments, Tampa, FL, USA). The system was calibrated prior to use with acidified nitrite, as specified by the manufacturer, and the assay was conducted according to Beretta et al. (3). All reactions were performed at 37° ± 1°C in 50-mM triethanolamine/HCl buffer (pH 7.4) containing 2 mM NAD+, 2 mM DTT, 1,000 U/ml superoxide dismutase, and 5 mM MgCl2. The selected organic nitrate was added and the electrode was allowed to equilibrate. Once a steady baseline was obtained, 0.15 mg/ml of pre-warmed hALDH was added. The NO signal was recorded from the time the enzyme was added until it returned to baseline value. Although the organic nitrate concentrations used in the NO electrode studies are supra-pharmacologic (0.1 and 1 mM), they were identical to those used in the first literature report which demonstrated this phenomenon (3), and are necessary to detect the unstable and elusive NO gas as a metabolite.
Preparation of Rat Hepatic Homogenate
An adult male Sprague Dawley Rat was anesthetized with 90 mg/kg ketamine and 9 mg/kg xylazine administered intramuscularly. The liver was excised and washed free of excess blood with ice-cold HEPES buffer (50 mM HEPES, 70 mM sucrose, 220 mM mannitol, and 1 mM EGTA, pH 7.4) containing a protease inhibitor cocktail. The liver was homogenized on ice in HEPES buffer using a motor-driven Teflon homogenizer. The crude homogenate was centrifuged at 3,000×g for 10 min to pellet large cellular debris. Following incubation of the homogenate with organic nitrates, as described below, the cytosolic and mitochondrial fractions were separated (14). Briefly, the homogenate was centrifuged at 15,000×g for 10 min to pellet the mitochondrial fraction, and the supernatant was retained as the cytosolic fraction. The mitochondrial pellet was washed and recentrifuged three times using HEPES buffer to remove the remaining cytosol. Prior to the determination of dehydrogenase activity in the cytosolic (ALDH1a1) and the mitochondrial (ALDH2) fraction, 2% triton X-100 in HEPES was used to solubilize each fraction. Cytochrome C oxidase activity was monitored spectrophotometrically to determine the enrichment of the mitochondrial fraction. All animal work was approved by the Institutional Animal Care and Use Committee of the University at Buffalo.
Inactivation of ALDH by Organic Nitrates
hALDH2 and hALDh1a1 were incubated with 0.1-mM NTG, 1-mM ISDN, and 1-mM IS-5-MN for 10 min at 37°C prior to the determination of activity. Rat homogenate was incubated with 1-mM NTG, ISDN, IS-5-MN, IS-2-MN, and nicorandil for 30 min at 37°C prior to cellular fractionation and determination of activity. Due to the extreme difference in potency between organic nitrates, a follow-up study was conducted in which rat homogenate was exposed to equipotent concentrations using EC50 values for rat aortic ring relaxation from the literature (2,15). Due to the lack of a value for IS-2-MN, its EC50 was assumed to be equivalent to that of its isomer IS-5-MN. Homogenate was incubated with 22 nM NTG, 5 μM ISDN, 170 μM IS-5-MN, 170 μM IS-2-MN, or 10 μM nicrorandil for 4 h at 37°C prior to determination of activity.
Site-Directed Mutagenesis Protein Expression and Purification
The vectors encoding the wild-type ALDH were used for mutagenesis. Oligonucleotide-directed mutagenesis was performed following instructions provided by the QuickChange® site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). The primer sequences for each mutant are summarized in Table I. Mutations were verified by DNA sequencing performed by Roswell Park DNA Sequencing Laboratory (Buffalo, NY, USA). The selected plasmids were then transformed into Escherichia coli strain BL21(DE3) pLysS (Stratagene, La Jolla, CA, USA) for expression. The mutant proteins were expressed, purified, and characterized in the same fashion as the wild-type isoforms. The ability of the mutants to liberate NO from organic nitrates and subsequent inactivation was investigated using the methods described earlier.
Table I.
Forward Primer Sequences for Site-Directed Mutagenesis of Human ALDH1a1 and ALDH2
| Isoform | Mutant | Forward primer sequences |
|---|---|---|
| ALDH1a1 | C302A | 5′-GTATTCTACCACCAGGGCCAGGCTTGTATAGCCGCATCCAGG-3′ |
| C303A | 5′-GTATTCTACCACCAGGGCCAGTGTGCTATAGCCGCATCCAGGATTTTTG-3′ | |
| ALDH2 | C319A | 5′-CTTCAACCAGGGCCAGTGCGCCTGTGCCGGCTCCCGGACC-3′ |
| E504K | 5′- GTACGGGCTGCAGGCATACACTAAAGTGAAAACTGTCACAGTCAAAG-3′ |
Dinitrate Formation from NTG
Glyceryl dinitrate formation from NTG from various ALDH enzymes was examined in a medium containing 0.1 μM NTG, 5.4 μg/ml hALDH wild-type (WT) or mutant, and cofactors (0.2 mM NAD+, 0.4 mM glutathione, and 0.4 mM dithiothreitol (DTT)) in PBS (pH 7.4). The reaction was started by the addition of enzyme solutions, and the mixture was incubated at 37°C. The samples were collected at 0, 2, 5, 10, 30, and 120 min and stored at −20°C. Control solution was obtained from boiling the enzyme solutions for 30 min, after which the activity was found to be negligible when compared to unboiled enzyme. Prior to analysis, the samples were diluted two times with 50% methanol and 50% water containing the internal standard 1,2,4-butanetriol-1,4-dinitrate. After centrifugation at 16,000×g at 4°C for 20 min, the samples were subjected to liquid chromatography–tandem mass spectrometry (LCMS) analysis as described previously (16). In a separate experiment, hALDH2 incubations were performed at 4°C, and samples were directly spiked with internal standard in methanol to quench the reaction.
Statistical Analysis
All data are presented as mean ± SD unless stated otherwise. Statistical differences between two sets of values were evaluated by the Student's t test. Differences with p < 0.05 were denoted as statistically significant. One-way and two-way ANOVA with Tukey's post hoc tests were carried out where appropriate with Minitab 16 (State College, PA, USA). Area-under-the-curves (AUCs) of the NO signal were calculated using the trapezoidal method in Excel (Microsoft, Redmond, WA, USA).
RESULTS
Bioactivation of Organic Nitrates by Purified WT hALDH1a1 and hALDH2
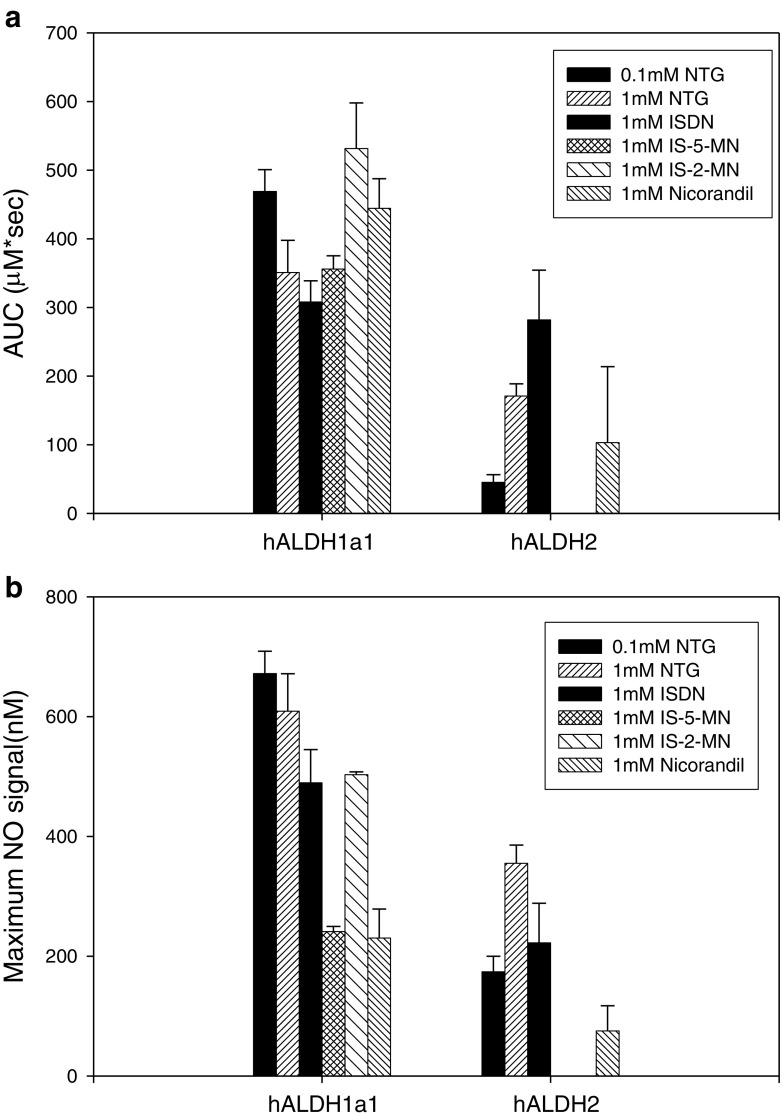
Using a NO-selective electrode, the ability of WT hALDH1a1 and hALDH2 to activate various organic nitrates to NO in vitro was compared, both in terms of cumulative NO production (Fig. 1a) and the peak signal observed (Fig. 1b). At the substrate concentrations chosen, both ALDH isoforms were able to liberate NO from NTG, ISDN, and nicorandil. However, only WT hALDH1a1 was able to generate measurable NO from IS-2-MN and IS-5-MN. Under the described experimental conditions, hALDH1a1 appeared to be more efficient in generating NO than ALDH2 for all organic nitrates.
Fig. 1.
a Cumulative and b maximal NO generation from the indicated organic nitrate from WT hALDH1a1 and hALDH2. Data are expressed as mean ± SD, n = 3–4
Inactivation of ALDH Isoforms by Organic Nitrates
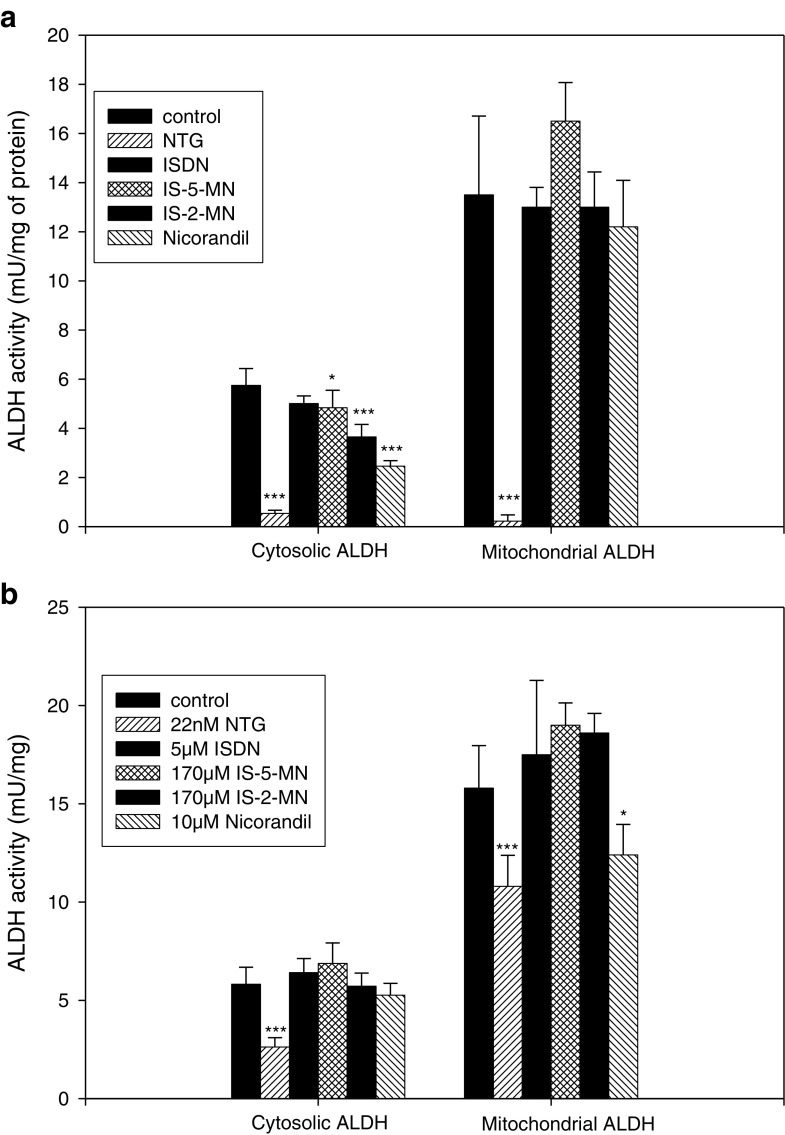
The dehydrogenase activity of purified hALDH1a1 and hALDH2 was significantly inactivated following a 10-min incubation of 0.1 mM NTG and 1 mM ISDN. In contrast, 1 mM IS-5-MN caused negligible inactivation of hALDH2, and only a slight inactivating effect on hALDH1a1 (Table II). The inactivating ability of various organic nitrates was further compared in the cytosolic and mitochondrial fractions of rat liver homogenate, reflecting tissue ALDH1a1 and ALDH2 activities, respectively. The differential centrifugation procedure resulted in 7.7-fold enrichment in the activity of cytochrome C oxidase in the mitochondrial fraction when compared to the cytosolic fraction (p < 0.001). Cytosolic ALDH activity was significantly inactivated by several organic nitrates, with a potency order of NTG > nicorandil > IS-2-MN > IS-5-MN, Interestingly, inactivation was not observed with 1 mM ISDN (Fig. 2a). Mitochondrial ALDH activity was only inactivated by 1 mM NTG, reducing it to <5% of the control. All other organic nitrates studied did not inactivate the enzyme under this condition.
Table II.
Inactivation of Dehydrogenase Activity by the Indicated Organic Nitrate Following a 10-min Incubation
| WT hALDH2 | WT hALDH1a1 | |
|---|---|---|
| Control | 100 ± 9 | 100 ± 13 |
| 0.1 mM NTG | NAD* | 22 ± 11* |
| 1 mM ISDN | 4.6 ± 7.9* | 0.44 ± 1.0* |
| 1 mM IS-5-MN | 102 ± 14 | 89 ± 4** |
Data are expressed as% of vehicle control ± SD, n = 3–4
NAD no activity detected
*p < 0.001 vs. WT, **p < 0.01
Fig. 2.
a Effects of 1 mM organic nitrate on ALDH activity in rat hepatic homogenate and b effects on ALDH at equipotent concentrations. Data are expressed as mean ± SD, n = 6 (*p < 0.05, ***p < 0.001 control vs. nitrate)
Because the organic nitrates examined exhibit highly diverse pharmacological potencies, their relative inactivation potency was also compared when they were incubated at concentrations reported as their EC50 values for dilating rat aortas. Because the nitrate concentrations used are much lower (e.g., for NTG, it was 45 times lower), the exposure period in this experiment was extended to 4 h, a period over which no loss in ALDH activities in either fraction was detected. Our results (Fig. 2b) showed that NTG maintained its ability to inactivate both cytosolic and mitochondrial ALDH, while nicorandil exhibited significant inactivation of mitochondrial ALDH under this condition. ISDN and both isosorbide mononitrates did not inactivate hepatic ALDH under these conditions.
Expression of hALDH2 and hALDH1a1 Mutants in E.coli
Two hALDH2 mutants (C319A and E504K) and two hALDH1a1 mutants (C302A and C303A) were expressed and purified successfully as determined by SDS-PAGE followed by protein staining, with a subunit molecular weight around 55 kDa, consistent with literature report for the WT (11). The dehydrogenase activity of the C319A and E504K mutants of ALDH2 was below detection limit and 3.12% ± 2.16%, respectively, when compared to WT (p < 0.01). The C302A and C303A mutants of ALDH1a1 exhibited 186% ± 62% and no detectable activity, respectively, when compared to the WT enzyme (p < 0.001). The inactivation of the C302A hALDH1a1 mutant by organic nitrates was examined. Following a 10-min incubation with either 0.1 mM NTG or 1 mM IS-5-MN, dehydrogenase activity was reduced to 10.5% ± 0.03% of WT (p < 0.001 vs. WT) and 92.0% ± 3.0% of control, respectively.
Comparison of Dinitrate Formation from NTG by WT vs Mutant ALDH Enzymes
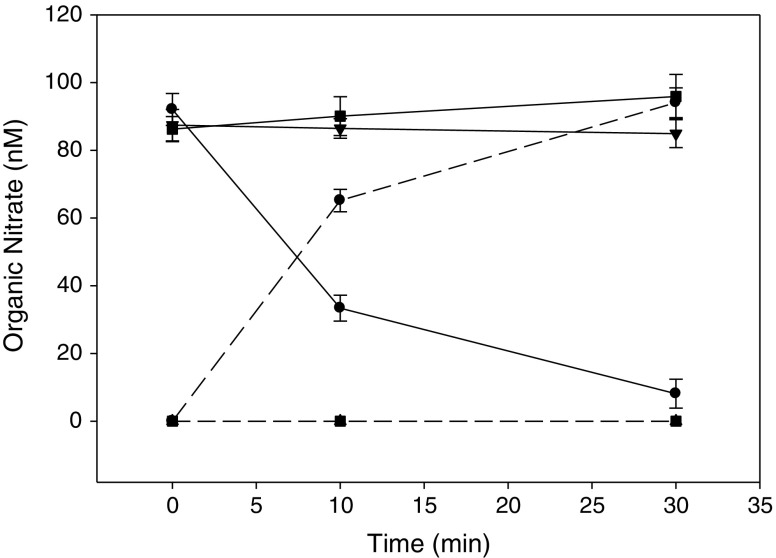
The conversion of NTG to its dinitrate metabolites by the hALDH2 WT and the two mutants was examined by LCMS. At 37°C, NTG was completely metabolized within 2 min into 1,2-GDN by WT, while no metabolite was produced by C319A. Metabolism of NTG by E504K was slower with negligible production of 1,2-GDN over 2 min. Over a longer period of incubation (2 h), about 25% of initial NTG was degraded, mostly to 1,2-GDN. The extent of 1,3-GDN production by E504K were similar to those observed with the boiled denatured enzyme (at 2 min, 1.4 ± 2.4 vs. 3.2 ± 2.8 nM; at 2 h, 6.0 ± 0.3 vs. 6.2 ± 0.5 nM for E504K and the boiled enzyme, respectively). The boiled control for the WT and the other mutants were inactive as negligible 1,2-GDN or 1,3 GDN formation was observed (data not shown). The reaction was repeated at 4°C to enable additional comparison of the rates of NTG metabolism at a slower rate. At this temperature (Fig. 3), WT hALDH2 completely converted NTG to 1,2-GDN in 30 min, while E504K and C319A did not produce any significant amounts of 1,2-GDN.
Fig. 3.
NTG degradation and 1,2-GDN production by hALDH2 WT or mutants at 4°C. Black circle represents WT, black square represents C319A, black down-pointing triangle represents E504K, solid lines represent NTG and dashed lines represent 1,2-GDN. Data are expressed as mean ± SD, n = 3. (***p < 0.001 WT vs. mutants for NTG and 1,2-GDN)
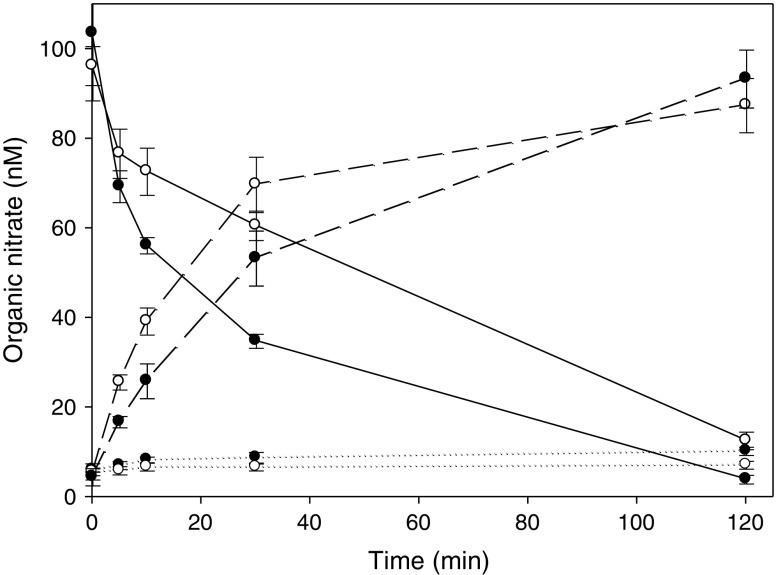
NTG was completely metabolized by hALDH1a1 following the 120-min incubation at 37°C producing 1,2-GDN as the predominant metabolite (90%) with a small quantity of 1,3-GDN produced (Fig. 4). The C302A mutant completely metabolized the NTG over the incubation period, albeit at a slower rate; as evident by the longer NTG disappearance half-life (42.5 ± 4.4 min for the C302A mutant vs. 27.5 ± 2.6 min for the WT, p < 0.01). Similar to the WT isoform, 1,2-GDN was the major metabolite formed accounting for 93% of metabolite formation and the 1,3-GDN metabolize accounting for the remaining 7% (Fig. 4). The C303A mutant failed to significantly metabolize NTG over the 120-min incubation period (data not shown) which is consistent with its lack of dehydrogenase activity.
Fig. 4.
NTG degradation and GDN production by hALDH1a1 WT or mutants at 37°C. Black circle represents WT, white circle represents C302A. Solid lines represent NTG, dashed lines represent 1,2-GDN, and dotted lines represent 1,3-GDN. Data are expressed as mean ± SD, n = 3. (***p < 0.001 WT vs. C302A for NTG, 1,2-GDN, and 1,3-GDN)
Comparison of NO Formation from NTG by WT vs. Mutant ALDH Enzymes
For the hALDH2 mutants, the time to maximum NO liberation by WT hALDH2 was quite rapid (within 1 min). In comparison, NO production from E504K exhibited a lower peak (77.2 ± 3.5 vs. 174 ± 26 nM for WT, p < 0.001), but was more sustained. C319A did not produce any detectable NO (p < 0.001 vs. WT). When the cumulative amounts of NO production were compared by AUC, the WT and E504K mutants were shown to produce a similar amount of NO (45.3 ± 11.0 and 55.0 ± 3.0 μM · s, respectively, p > 0.05) and no detectable NO was liberated by C319A (p < 0.001 vs. WT).
Like hALDH2, the time to peak NO liberation was quite rapid for the WT hALDH1a1 enzyme and its C302A isoform. The C302A mutant generated a lower maximum concentration of NO and cumulative NO from 10 μM NTG and 1 mM IS-5-MN compared to the WT. At a substrate concentration of 0.1 mM NTG, the amount and peak NO generated were similar between the two isoforms. The C303A mutant was unable to liberate NO from NTG or IS-5-MN (Table III).
Table III.
NO Generation from NTG and IS-5-MN and hALDH1a1 Mutants
| Cumulative NO generation (μM · s) | Peak NO generated (nM) | |||||
|---|---|---|---|---|---|---|
| hALDH1a1 isoform | WT | C302A | C303A | WT | C302A | C303A |
| 10 μM NTG | 447 ± 85 | 202 ± 23* | NA** | 629 ± 94 | 272 ± 29* | NA** |
| 0.1 mM NTG | 469 ± 32 | 502 ± 116 | NA** | 672 ± 31 | 646 ± 87 | NA** |
| 1 mM IS-5-MN | 356 ± 19 | 131 ± 24* | NA** | 241 ± 9 | 36.7 ± 6.3* | NA** |
Data are expressed as mean ± SD, n = 3–4
NA no activity as evident by negligible NO detection
*p < 0.01; **p < 0.001 WT vs. mutant
DISCUSSION
Investigative interests in the biochemical mechanism of organic nitrate bioactivation and tolerance have recently been renewed after the report that ALDH2 acts as the primary enzyme responsible the liberation of NO from NTG, and that inactivation of this enzyme is responsible for the development of NTG tolerance (1,2,17). However, ALDH2 is not involved in the activation of the other clinically used organic nitrates (ISDN and IS-5-MN) (2,18). The enzyme(s) responsible for NO release from these compounds, and for nicorandil and IS-2-MN, is(are) at present unknown. The demonstration that the major cytosolic isoform of hALDH (hALDH1a1) possesses the ability to metabolize NTG (3) raises the possibility that this isoform may be able to metabolize these lower organic nitrates. When compared to ALDH2, ALDH1a1 has a more flexible active site pocket which contains more hydrophilic residues and a larger solvent accessible volume (19,20), thus allowing the enzyme to metabolize a wider range of substrates.
Our results regarding NO production from hALDH2 and NTG were in agreement with literature findings (3). Interestingly, we demonstrated that this enzyme was able to liberate NO from ISDN despite previous ex vivo results suggesting that ALDH2 activity is not responsible for the activity of this agent (2,18). This is likely due to the presence of other proteins in the vascular tissue which may have greater affinity and activity towards ISDN. Our NO-electrode results also confirmed that both isomers of isosorbide mononitrate are poor substrates for ALDH2, consistent with literature reports (2).
Regarding hALDH1a1, we confirmed that it has the ability to metabolize NTG to liberate NO. In addition, this isoform preferentially produces 1,2-GDN vs. 1,3-GDN (at about a 9:1 ratio) as the major metabolite of NTG denitration (Fig. 3). Here, we showed that this enzyme also exhibits an ability to metabolize the lower organic nitrates (ISDN, IS-5-MN, IS-2-MN, and nicorandil) to liberate NO (Fig. 1), suggesting that it may contribute to the in vivo bioactivation of these agents. Our findings therefore represent the first demonstration of an enzyme with the ability to produce NO from ISDN, IS-5-MN, and IS-2-MN.
Mukerjee and Pietruszko demonstrated that ISDN inactivated ALDH1 and ALDH2 (21) in a time-dependent fashion via oxidation of critical cysteine sulfhydryl group. The mechanism-based inactivation of purified glutathione-S-transferase by NTG was also shown to be sulfhydryl-dependent (22). Here, we found that purified WT hALDH2 and hALDH1a1 was subjected to mechanism-based inactivation within 10 min of exposure to NTG, ISDN, and IS-5-MN (Table II) consistent with previous reports (21,23–25). We obtained similar results in rat hepatic homogenates which also revealed the ability of nicorandil to inactivate both isoforms and IS-2-MN to inactivate cytosolic ALDH (Fig. 2). Interestingly, ISDN had no significant effect on ALDH activity in the hepatic homogenate despite being a potent inactivator of the pure enzymes. This finding is consistent with that reported by Murphy et al. who examined ISDN-mediated inactivation of ALDH2 in isolated rat hepatic mitochondria (25). The reason for this discrepancy is presently unknown, and may be related to the specific time and concentrations chosen for the study. Further studies involving a range of concentrations and times of incubation should be carried out to further understand this phenomenon.
The high reactivity of hALDH towards NTG and other organic nitrates may be due to the high abundance of cysteine groups (7 in hALDH2 and 11 in hALDH1a1) in each subunit. In addition, the active site of each enzyme contains multiple cysteines (three for hALDH2 and two for hALDH1a1) (26). Intramolecular disulfide bond formation, S-nitrosylation (27,28), as well as S-glutathionylation (29,30) have been shown to affect ALDH activity. We recently showed that NTG oxidized cysteine residues in multiple cellular proteins, including extensive S-glutathionylation (31). In addition, it has been demonstrated that organic nitrate-mediated inactivation of ALDH is partially reversed by thiol-reducing agents (23,25) suggesting that oxidation of one or more cysteine residues at the active site is responsible for loss of nitrate reductase activity.
For ALDH2, Cys-319 has been shown to be the active site for its esterase and dehydrogenase activities. Here, we showed that this cysteine residue is also critical for its NO generating activity, because the C319A mutant was unable to bioactivate NTG to 1,2-GDN and NO (Fig. 3). Our results are consistent with a previous report (32) that replacement of Cys-319 with serine significantly reduced NTG reductase activity, by approximately 500-fold when compared to WT. For hALDH1a1, mutation of Cys-303 to alanine also abolished the enzyme's NO generating activity, as well as its dehydrogenase and nitrate reductase activity, suggesting that this residue is the sole active site for the enzyme.
To examine the role of the flanking site cysteine in the metabolism and bioactivation of organic nitrates, the cysteine at the 302 position of hALDH1a1 was mutated to alanine (C302A). Interestingly, the dehydrogenase activity of the C302A mutant was modestly, but significantly, higher than the WT isoform. The reason for this is unknown and warrants further investigations. Despite this increased dehydrogenase activity, the mutant exhibited reduced nitrate reductase activity toward NTG (Fig. 4) and reduced ability to generate NO from NTG and IS-5-MN (Table III). Nevertheless, 1,2-GDN remained as the major metabolite (∼90%) produced from the denitration of NTG by this mutant.
The substitution of a glutamate with lysine at position 504 (E504K) of hALDH2 is a commonly observed polymorphism in Asians and results in a 94% reduction of ALDH activity in heterozygous subjects and near compete abolishment of activity in the homozygotes (9). Presence of this mutation led to reduced forearm blood flow response when given NTG (4). The E504K mutant constructed in this study showed significantly lower dehydrogenase activity compared to WT, consistent with previous reports (32,33). It has been shown that in the absence of cofactor NAD, this enzyme had negligible activity, whereas in the presence of cofactors at saturated concentrations, the activity improved tremendously but not to the level of WT. This mutation affected cofactor binding and in turn resulted in a less stable structure at the active site as well as a lower reactivity of Cys-319. The present study employed the physiological concentration of NAD (2 mM), and under this condition, we were able to detect a reduced peak release of NO by NTG and similar cumulative AUC vs. WT, but low to almost negligible dehydrogenase activities. This result suggests that the mutation to 504 position affects the different enzyme activities to various extents. It is possible that the diminished peak release of NO is due to a lower nucleophilicity of Cys-319 in the mutated enzyme. In contrast to other mutants, measureable amounts of 1,3-GDN was also produced in addition to 1,2-GDN. Since both the boiled control and the mutated enzyme produced similar amounts of 1,3-GDN, this result suggests that this metabolite may arise from chemical degradation rather than enzymatic catalysis.
CONCLUSIONS
These studies demonstrate that ALDH1a1 and ALDH2 differed in their substrate selectivity to produce NO from several organic nitrates. While ALDH2 mediated bioactivation has been previously demonstrated with NTG, this is the first report showing that ALDH1a1 has the ability to liberate NO from ISDN, IS-2-MN, IS-5-MN, and nicorandil. Mechanism-based inactivation of the enzymes by these organic nitrates was demonstrated in purified enzyme preparations, as well as in liver homogenates. Site-directed mutagenesis studies showed that the active sites for NO generation are the same (i.e., Cys-319 for ALDH2 and Cys-303 for ALDH1a1) as those for their dehydrogenase activity. The E504K mutant of ALDH2 and C302A mutant for ALDH1a1 exhibited reduced NO-generating abilities. These findings suggest that ALDH1a1 should also be considered as a potential bioactivating enzyme for organic nitrates, particularly for ISDN, ISMN, and nicorandil. In addition, studies examining the relative expression of ALDH1a1 vs. ALDH2 in cardiovascular tissues should be carried out to determine the therapeutic relevance of these enzymes in the bioactivation of various ORN.
Acknowledgments
We thank Dr. George A. Garcia and Dr. Yi-Chen Chen (at the University of Michigan) for their technical support in the mutagenesis studies, Dr. Matteo Beretta (Karl-Franzens-Universitat Graz) for his assistance in the hALDH purification procedure and NO electrode experiment, and Dr. Bernd Mayer (Karl-Franzens-Universitat Graz) for providing the hALDH1a1 plasmid containing E. coli. This study was supported in part by the National Institutes of Health [Grant HL081580] awarded to H.-L.F., a Schering Plough Pre-doctoral Fellowship to P. T. and a Schering Plough-Rho-Chi-AFPE First Year Graduate School Scholarship to N.A.P.
Footnotes
Pei-Suen Tsou and Nathaniel A. Page contributed equally to this work.
References
- 1.Chen Z, Zhang J, Stamler JS. Identification of the enzymatic mechanism of nitroglycerin bioactivation. Proc Natl Acad Sci U S A. 2002;99:8306–11. doi: 10.1073/pnas.122225199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daiber A, Oelze M, Coldewey M, Bachschmid M, Wenzel P, Sydow K, et al. Oxidative stress and mitochondrial aldehyde dehydrogenase activity: a comparison of pentaerythritol tetranitrate with other organic nitrates. Mol Pharmacol. 2004;66:1372–82. doi: 10.1124/mol.104.002600. [DOI] [PubMed] [Google Scholar]
- 3.Beretta M, Gruber K, Kollau A, Russwurm M, Koesling D, Goessler W, et al. Bioactivation of nitroglycerin by purified mitochondrial and cytosolic aldehyde dehydrogenases. J Biol Chem. 2008;283:17873–80. doi: 10.1074/jbc.M801182200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackenzie IS, Maki-Petaja KM, McEniery CM, Bao YP, Wallace SM, Cheriyan J, et al. Aldehyde dehydrogenase 2 plays a role in the bioactivation of nitroglycerin in humans. Arterioscler Thromb Vasc Biol. 2005;25:1891–5. doi: 10.1161/01.ATV.0000179599.71086.89. [DOI] [PubMed] [Google Scholar]
- 5.Dalal JJ, Parker JO. Nitrate cross-tolerance: effect of sublingual isosorbide dinitrate and nitroglycerin during sustained nitrate therapy. Am J Cardiol. 1984;54:286–8. doi: 10.1016/0002-9149(84)90184-X. [DOI] [PubMed] [Google Scholar]
- 6.Thadani U, Manyari D, Parker JO, Fung HL. Tolerance to the circulatory effects of oral isosorbide dinitrate. Rate of development and cross-tolerance to glyceryl trinitrate. Circulation. 1980;61:526–35. doi: 10.1161/01.cir.61.3.526. [DOI] [PubMed] [Google Scholar]
- 7.Mertens HM, Ohlmeier H, Mannebach H, Kabelitz K, Gleichmann U. 4 different nitrate preparations with regard to the possible development of tolerance in long-term treatment. Herz. 1985;10:172–81. [PubMed] [Google Scholar]
- 8.Moon KH, Abdelmegeed MA, Song BJ. Inactivation of cytosolic aldehyde dehydrogenase via S-nitrosylation in ethanol-exposed rat liver. FEBS Lett. 2007;581:3967–72. doi: 10.1016/j.febslet.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh S, Fritze G, Fang BL, Harada S, Paik YK, Eckey R, et al. Inheritance of mitochondrial aldehyde dehydrogenase: genotyping in Chinese, Japanese and South Korean families reveals dominance of the mutant allele. Hum Genet. 1989;83:119–21. doi: 10.1007/BF00286702. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Zhang D, Jin W, Shao C, Yan P, Xu C, et al. Mitochondrial aldehyde dehydrogenase-2 (ALDH2) Glu504Lys polymorphism contributes to the variation in efficacy of sublingual nitroglycerin. J Clin Invest. 2006;116:506–11. doi: 10.1172/JCI26564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng CF, Wang TT, Weiner H. Cloning and expression of the full-length cDNAS encoding human liver class 1 and class 2 aldehyde dehydrogenase. Alcohol Clin Exp Res. 1993;17:828–31. doi: 10.1111/j.1530-0277.1993.tb00849.x. [DOI] [PubMed] [Google Scholar]
- 12.Ghenbot G, Weiner H. Purification of liver aldehyde dehydrogenase by p-hydroxyacetophenone-sepharose affinity matrix and the coelution of chloramphenicol acetyl transferase from the same matrix with recombinantly expressed aldehyde dehydrogenase. Protein Expr Purif. 1992;3:470–8. doi: 10.1016/1046-5928(92)90064-4. [DOI] [PubMed] [Google Scholar]
- 13.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–75. [PubMed] [Google Scholar]
- 14.Raha S, McEachern GE, Myint AT, Robinson BH. Superoxides from mitochondrial complex III: the role of manganese superoxide dismutase. Free Radic Biol Med. 2000;29:170–80. doi: 10.1016/S0891-5849(00)00338-5. [DOI] [PubMed] [Google Scholar]
- 15.Greenberg SS, Cantor E, Ho E, Walega M. Comparison of nicorandil-induced relaxation, elevations of cyclic guanosine monophosphate and stimulation of guanylate cyclase with organic nitrate esters. J Pharmacol Exp Ther. 1991;258:1061–71. [PubMed] [Google Scholar]
- 16.Miyayama T, Tsou PS, Fung SM, Fung HL. Simultaneous determination of nitroglycerin and dinitrate metabolites in metabolism studies using liquid chromatography-mass spectrometry with electrospray ionization. J Chromatogr. 2006;835:21–6. doi: 10.1016/j.jchromb.2006.02.059. [DOI] [PubMed] [Google Scholar]
- 17.Tsou PS, Addanki V, Fung HL. Dissociation between superoxide accumulation and nitroglycerin-induced tolerance. J Pharmacol Exp Ther. 2008;327:97–104. doi: 10.1124/jpet.108.138784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Z, Foster MW, Zhang J, Mao L, Rockman HA, Kawamoto T, et al. An essential role for mitochondrial aldehyde dehydrogenase in nitroglycerin bioactivation. Proc Natl Acad Sci U S A. 2005;102:12159–64. doi: 10.1073/pnas.0503723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore SA, Baker HM, Blythe TJ, Kitson KE, Kitson TM, Baker EN. Sheep liver cytosolic aldehyde dehydrogenase: the structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases. Structure. 1998;6:1541–51. doi: 10.1016/S0969-2126(98)00152-X. [DOI] [PubMed] [Google Scholar]
- 20.Wang MF, Han CL, Yin SJ. Substrate specificity of human and yeast aldehyde dehydrogenases. Chem Biol Interact. 2009;178:36–9. doi: 10.1016/j.cbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Mukerjee N, Pietruszko R. Inactivation of human aldehyde dehydrogenase by isosorbide dinitrate. J Biol Chem. 1994;269:21664–9. [PubMed] [Google Scholar]
- 22.Lee WI, Fung HL. Mechanism-based partial inactivation of glutathione S-transferases by nitroglycerin: tyrosine nitration vs sulfhydryl oxidation. Nitric Oxide. 2003;8:103–10. doi: 10.1016/S1089-8603(02)00183-0. [DOI] [PubMed] [Google Scholar]
- 23.Beretta M, Sottler A, Schmidt K, Mayer B, Gorren AC. Partially irreversible inactivation of mitochondrial aldehyde dehydrogenase by nitroglycerin. J Biol Chem. 2008;283:30735–44. doi: 10.1074/jbc.M804001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pietruszko R, Mukerjee N, Blatter EE, Lehmann T. Nitrate esters as inhibitors and substrates of aldehyde dehydrogenase. Adv Exp Med Biol. 1995;372:25–34. doi: 10.1007/978-1-4615-1965-2_4. [DOI] [PubMed] [Google Scholar]
- 25.Murphy TC, Arntzen R, Picklo MJ., Sr Nitrate-based vasodilators inhibit multiple vascular aldehyde dehydrogenases. Cardiovasc Toxicol. 2005;5:321–32. doi: 10.1385/CT:5:3:321. [DOI] [PubMed] [Google Scholar]
- 26.Hsu LC, Chang WC. Cloning and characterization of a new functional human aldehyde dehydrogenase gene. J Biol Chem. 1991;266:12257–65. [PubMed] [Google Scholar]
- 27.DeMaster EG, Redfern B, Quast BJ, Dahlseid T, Nagasawa HT. Mechanism for the inhibition of aldehyde dehydrogenase by nitric oxide. Alcohol (Fayetteville, NY) 1997;14:181–9. doi: 10.1016/S0741-8329(96)00142-5. [DOI] [PubMed] [Google Scholar]
- 28.Moon KH, Kim BJ, Song BJ. Inhibition of mitochondrial aldehyde dehydrogenase by nitric oxide-mediated S-nitrosylation. FEBS Lett. 2005;579:6115–20. doi: 10.1016/j.febslet.2005.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venkatraman A, Landar A, Davis AJ, Ulasova E, Page G, Murphy MP, et al. Oxidative modification of hepatic mitochondria protein thiols: effect of chronic alcohol consumption. Am J Physiol. 2004;286:G521–7. doi: 10.1152/ajpgi.00399.2003. [DOI] [PubMed] [Google Scholar]
- 30.Wenzel P, Hink U, Oelze M, Schuppan S, Schaeuble K, Schildknecht S, et al. Role of reduced lipoic acid in the redox regulation of mitochondrial aldehyde dehydrogenase (ALDH-2) activity. Implications for mitochondrial oxidative stress and nitrate tolerance. J Biol Chem. 2007;282:792–9. doi: 10.1074/jbc.M606477200. [DOI] [PubMed] [Google Scholar]
- 31.Tsou PS, Addanki V, Haas JA, Page NA, Fung HL. Role of glutaredoxin-mediated protein S-glutathionylation in cellular nitroglycerin tolerance. J Pharmacol Exp Ther. 2009;329:649–56. doi: 10.1124/jpet.108.149997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larson HN, Zhou J, Chen Z, Stamler JS, Weiner H, Hurley TD. Structural and functional consequences of coenzyme binding to the inactive asian variant of mitochondrial aldehyde dehydrogenase: roles of residues 475 and 487. J Biol Chem. 2007;282:12940–50. doi: 10.1074/jbc.M607959200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farres J, Wang X, Takahashi K, Cunningham SJ, Wang TT, Weiner H. Effects of changing glutamate 487 to lysine in rat and human liver mitochondrial aldehyde dehydrogenase. A model to study human (Oriental type) class 2 aldehyde dehydrogenase. J Biol Chem. 1994;269:13854–60. [PubMed] [Google Scholar]