Abstract
The detection of acute kidney injury (AKI) and the monitoring of chronic kidney disease (CKD) is becoming more important in industrialized countries. Because of the direct relation of kidney damage to the increasing age of the population, as well as the connection to other diseases like diabetes mellitus and congestive heart failure, renal diseases/failure has increased in the last decades. In addition, drug-induced kidney injury, especially of patients in intensive care units, is very often a cause of AKI. The need for diagnostic tools to identify drug-induced nephrotoxicity has been emphasized by the ICH-regulated agencies. This has lead to multiple national and international projects focusing on the identification of novel biomarkers to enhance drug development. Several parameters related to AKI or CKD are known and have been used for several decades. Most of these markers deliver information only when renal damage is well established, as is the case for serum creatinine. The field of molecular toxicology has spawned new options of the detection of nephrotoxicity. These new developments lead to the identification of urinary protein biomarkers, including Kim-1, clusterin, osteopontin or RPA-1, and other transcriptional biomarkers which enable the earlier detection of AKI and deliver further information about the area of nephron damage or the underlying mechanism. These biomarkers were mainly identified and qualified in rat but also for humans, several biomarkers have been described and now have to be validated. This review will give an overview of traditional and novel tools for the detection of renal damage.
Electronic supplementary material
The online version of this article (doi:10.1208/s12248-011-9301-x) contains supplementary material, which is available to authorized users.
KEY WORDS: Kim-1, nephrotoxicity, Predictive Safety Testing Consortium (PSTC), toxicogenomics, urinary protein biomarkers
INTRODUCTION
Minimizing toxicity remains one of the major barriers to bringing a drug to market. Approximately 92% of all developed compounds fail because of adverse effects of the candidate during clinical development (1).
In the pharmaceutical industry, the kidney is one of the routinely assessed organs during preclinical safety evaluations. The crucial role of the kidney in drug excretion and detoxification makes it one of the major organs evoking drug-related toxic responses and an important target of toxicological studies. Renal excretion within the nephron starts with the filtration of the blood by the glomerulus. After passing this filtration barrier, the primary urine enters the tubular system. The filtrate then moves through the proximal tubule, where important compounds are re-absorbed, the loop of Henle, for urine concentration, the distal tubule, and finally the collecting duct where the final urinary concentration is tuned and the urine finally removed. In drug development, the major focus in renal damage research is on proximal tubule toxicity, where the majority of drug metabolite reabsorption occurs.
The extraordinary exposition of the kidney to high levels of drugs and/or metabolites can be followed by cell damage primarily due to high blood flow, clearance and xenobiotic metabolism (2). However, only 7% of new drug candidates fail in preclinical trials because of nephrotoxicity (1) while the incidence of patients in intensive care units developing acute kidney injury (AKI) is about 30–50% (3). This discrepancy may help to explain the underestimation of nephrotoxicity in preclinical trials. To minimize the risk of (sub) clinical AKI or the initiation of chronic kidney disease, the data generated during preclinical toxicity testing and provided to clinicians for the different phases of the clinical development, has to be as detailed and informative as possible.
In the past, the monitoring of patients in clinical trials was primarily based on serum creatinine, blood urea nitrogen (BUN), and the detection of urinary components like electrolytes, enzymes, and other waste products. However, the diagnostic value of all of these parameters is poor and often only delivered false-positive or false-negative results which finally lead to such a late detection of renal damage that only dialysis for the patient is possible.
In the last few decades, many consortia have focused on this specific problem and ran many large Omics studies to identify specific biomarkers for nephrotoxicity and to discover the underlying mechanism of different kinds of kidney insult. Initially, rats were used as a model system, which lead to a large variety of biomarkers for the use in preclinical studies. By working together with regulatory authorities, like the United States Food and Drug Administration (US-FDA) and the European Medicines Agency (EMA), some of the identified markers have now been accepted for use in preclinical rodent toxicity studies with relevance for drug submission (4). This was unique in the history of toxicology and lead to an important enhancement in safety assessment and also to the use of new technologies, such as Omics profiling, in the identification of new biomarkers.
TRADITIONAL DETECTION OF NEPHROTOXICITY IN PRECLINICAL AND CLINICAL TRIALS
In toxicology, the “gold standard” for assessing a compound’s effect on an animal system (and identifying target organs) is histopathalogical observation. In the kidney, the area and intensity of renal insult can be directly observed and characterized. However, detailed information when damage occurs in the kidney is not feasible. To generate this information, it would be necessary to use many more animals for histopathalogical observation at different time points, which would lead to an increase in animals, costs, and time of the drug development process. It is also ethically unrealistic and only possible under restricted conditions to use renal tissue in clinical trials for diagnostic purposes.
Kidney damage usually affects both kidneys. If the ability of the kidneys to filter the blood is strongly reduced, waste products and excess fluid may build up in the body. A reduction in the reabsorption capability of the kidney for endogenous components, e.g., small proteins, sugars, or metabolites after a renal insult can lead to the increase of these components in urine. Although many forms of Drug Induced Kidney Injury (DIKI) damage do not produce symptoms until late in the course of the insult, there are six general/traditional warning signs:
BUN and creatinine levels in blood outside the normal range (reference values for both parameters can vary between different species and gender)
A lowered glomerular filtration rate (GFR) (<60%)
Blood and/or protein in the urine
High blood pressure
More frequent or painful urination
Swelling of hands and feet, puffiness around the eyes
It is clear that all of these symptoms are not specific for kidney damage but are also reported for other organ damage or diseases. For many reasons, these “markers” cannot identify specifically renal damage.
For the classification of renal injury in clinical trials, the risk, injury, failure, loss and end-stage renal disease (RIFLE) scheme is used. It is based on specific cutoff values for serum creatinine, GFR, and the urinary output. Another often used decision tree is the acute kidney injury network (AKIN) scheme (see supplementary material Fig. 1). The AKIN has, based on the RIFLE classification, developed a definition of acute kidney injury. The AKI stages define the whole spectrum of AKI from moderate to severe deterioration of kidney function.
Serum Creatinine and BUN
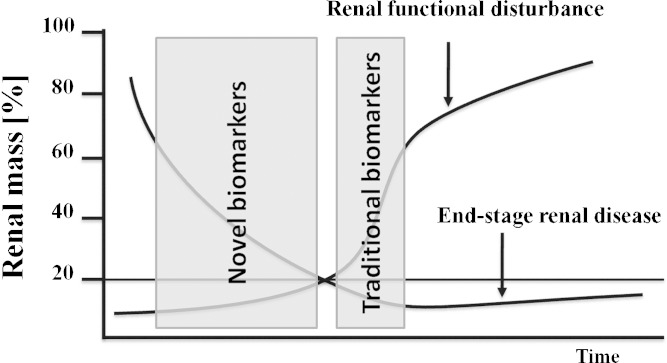
The clinical–chemical standard parameters in preclinical and clinical trials for the detection and monitoring of renal function are serum creatinine and BUN. These markers have been described since 1904 and 1952, respectively (5,6), and are still the “gold standard” in the minimal invasive clinical chemical analysis. Because of the great capability of the kidney to compensate renal mass loss and to recover after acute insult, the sensitivity of serum creatinine and BUN is very poor. It has been observed that a reduction of renal functionality occurs only after approximately two thirds of renal biomass has been lost (7). Figure 1 gives an overview of the correlation of renal mass loss and renal functional disturbance with the known and expected field of verifiability of traditional and novel biomarkers.
Fig. 1.
Only substantial loss of renal biomass leads to a lack of renal functionality. Traditional clinical–chemical parameters like serum creatinine or BUN only detected this decrease in kidney function. The novel urinary biomarkers are accepted to detect renal damage earlier because not the function but the damage of tissue was predicted
These two parameters can easily be influenced by many physiological functions/mechanisms. The serum level of creatinine, a breakdown product of muscle tissue, depends on age, gender, muscle mass, and weight. It also has been reported that gastrointestinal bleeding can lead to an increase in serum creatinine without any negative impact on the kidney. BUN also shows an increase in serum in other pathological processes, like during enhanced protein catabolism. A detailed list of factors influencing serum creatinine and BUN is shown below.
Nonrenal-related causes of alteration in BUN levels:
Congestive heart failure (CHF) (8)
Heart attack (8)
Excessive protein levels in the gastrointestinal tract (9)
Gastrointestinal bleeding (10)
Hypovolemia (11)
Shock (12)
Dehydration (13)
Nonrenal-related causes of alteration in serum creatinine levels:
Traditional Urinary Parameters for the Detection of Drug-Induced Renal Injury
Standard observations in urinalysis are shown in Table I. All of these markers are not present in the urine of healthy subjects. However, urinary markers have the great disadvantage that they are often influenced by other organ toxicities, as is the case with blood markers. Major changes in blood parameters can influence what markers are seen in the urine. Glucose for example, is normally 100% reabsorbed in the proximal convolute. If the plasma glucose level rises, like in diabetic mellitus, the reabsorptive transport capacity of the proximal tubular cells is saturated and the urinary glucose level increases. Also, other factors such as special diets or other physiologic/pathologic conditions can directly influence these parameters.
Table I.
Traditional Urine Parameters and their Intra- and Extrarenal Causes
| Parameters | Causes |
|---|---|
| Urine coloration | |
| Colorless | Diluted urine |
| Cloudy | Phosphaturia, hyperoxaluria, lipiduria |
| Brown/black | Myo-/methemoglobin, bile pigments, melanin |
| Red | Hematuria in general |
| Orange | Bile pigments |
| Yellow | Concentrated urine |
| Green or blue | Biliverdin, pseudomonal urinary tract infection |
| Parameters of urinalysis | |
| Blood | Urinary tract infection, kidney stones, Berger’s disease, nephritic syndrome, prostate inflammation |
| Glucose | Enhanced plasma glucose level (tubular saturation), proximal tubular damage |
| Ketones | T1DM, glycosuria, glucogen storage disease, starvation, fasting, prolonged vomiting, hyperthyroidism, pregnancy |
| Leukocyte esterase | Urinary tract infection, varginal contamination |
| Nitrites | Urinary tract infection, gross hematuria |
| Protein | Glomerular or proximal tubular damage |
| Specific gravity | Hypovolumia, dehydration |
| Phosphates | Renal dysfunction, hyperthyroidism |
| Urinary casts and crystals | |
| Hyaline | Chronic renal disease, pyelonephritis |
| Erythrocyte | Glomerulonephritis, urinary tract infection |
| Leukocyte | Renal inflammatory processes, interstitial nephritis, glomerulonephritis, pyelonephritis |
| Epithelial | Acute tubular necrosis, interstitial nephritis, nephritic syndrome, renal disease, eclampsi, heavy metal ingestion |
| Granular | Advanced renal disease |
| Waxy | Advanced renal disease |
| Fatty | Nephrotic syndrome, renal disease, hypothyroidism |
| Broad | End-stage renal disease |
This list is not intended to be exhaustive
GFR and Clearance
The GFR is a calculated value to describe the flow rate of filtered fluid through the kidney. To estimate the GFR, the clearance rate traditionally is used. The creatinine clearance rate (CCr) describes the volume of plasma that is cleared of creatinine per unit time. Both GFR and CCr may be accurately calculated by comparative measurements of substances in the blood and urine. This so-called clearance equation can be used to estimate the GFR and to assess the manner in which the kidney handles a multitude of substances.
The results can be used to assess the excretory function of the kidneys. For example, grading of chronic renal insufficiency and dosage of drugs that are excreted primarily via urine are based on GFR or creatinine clearance (see supplementary material Table I).
In most cases, GFR determination based on the measurement of cystatin C delivers comparable results to creatinine based calculations. Better results have been produced by using cystatin C clearance instead of creatinine clearance in select patient groups, such as patients with reduced muscle mass (17), children (18), and the elderly (19). This molecule is now also used as an approved urinary toxicity biomarker and will be described in more detail later.
Another calculated value often used to diagnose renal alteration is the fractional excretion from electrolytes, for example sodium. This parameter is focused on substances predominantly secreted by the tubular cells and which can lead to a clearance greater than Ccr. The benefit of the fractional excretion is that because VS is canceled out, a timed urine collection, as is needed for the calculation of GFR, is not necessary.
Overall, these calculated values can help to monitor the progression of established renal damage. However, the early detection of a developing insult is not possible.
INTERNATIONAL PROJECTS WITH FOCUS ON BIOMARKER DISCOVERY
The ongoing technological/scientific progress on the field of synthesis of chemical compounds and biologicals and the related enhancement in the early phases of drug development has resulted in significant new challenges in toxicology. The particular requirements for approval of new drug candidates, as well as the increase in compounds for routine testing, has lead to a substantial increase in costs of clinical and preclinical studies, longer development times, and an increase in failed drugs in late phases of drug development. This, in part, leads the US-FDA to publish a White paper in 2004 to address this problem and to encourage the integration of new technologies into the discovery/developmental process. In 2007, the report “Toxicity Testing in the 21st Century: A Vision and a Strategy” was published by the National Research Council, which brought further acceptance to the use of these new technologies. Also on the European side, an Innovation Task Force was founded by the EMA to support the use of novel technologies in Drug Discovery and Development and to advise and inform pharmaceutical companies on regulatory issues (www.ema.europa.eu).
Omics technologies are now accepted to be an essential methodological part of systems or integrative biology, by which it is possible to observe global changes of transcripts (genomics), proteins (proteomics), or metabolites (metabolomics) in cellular systems or tissues. By parallel detection of changes in gene expression patterns of thousands of genes (transcriptomics), it is feasible to observe and identify gene interactions and early changes in specific gene expression caused by compounds, pathogens, or environmental factors in one sample. This enables the analysis of complex mechanistic interactions without any variations caused by repeated measurements of single samples or repeated experiments. Toxicogenomics, a relatively new (but now well established) area of toxicology, relates to substance-induced changes of the transcriptome to identify cellular and subcellular mechanisms. It is possible, by measuring the gene expression patterns before and after compound treatment, to study interactions between structure and activity of the whole genome and adverse biological effects with exogenous stressors (20). Genes that consistently exhibit increased or decreased expression during these toxic responses in model systems serve as markers to predict potential adverse (pre-)clinical outcomes. The overall aim of toxicogenomics analysis is the identification of the mode of action by which a compound induces a toxic/adverse effect (21). Additional to the identification of characteristic molecular pathways, it has been reported that specific transcripts can be used as mechanistic or predictive biomarker for organ-specific toxic alterations (22).
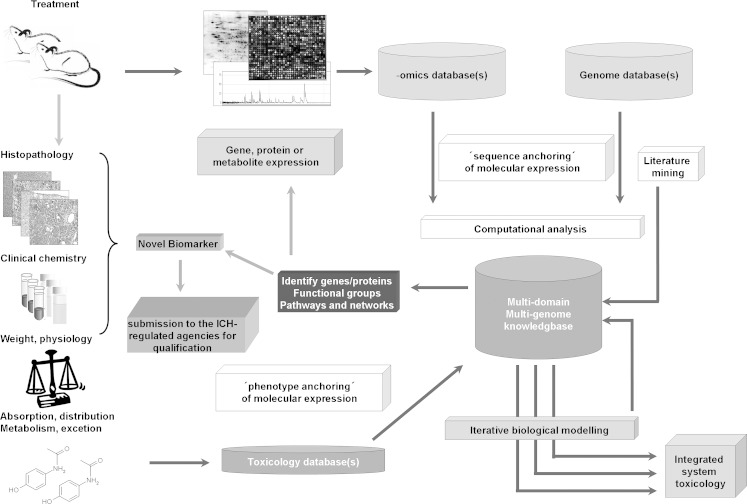
In 2005, the FDA published the FDA’s Pharmacogenomics Data Submissions Guidance which should lead to an enhanced use of this technology in pharmaceutical industry in practice (www.FDA.gov). A suggestion of the integration of toxicogenomics analysis in toxicological routine studies is shown in Fig. 2.
Fig. 2.
Shown is an integrated approach for the inclusion of novel Omics data in the toxicological routine safety assessment and the identification and implementation of novel biomarkers into the standard battery or else for the submission to the FDA/EMA qualification process. Figure adapted by (45)
All of these forces from regulatory agencies for the use of novel Omics technologies/biomarkers to enhance safety assessment in preclinical and clinical trials lead to many international projects being established.
International Consortia Projects
In the last decade, many international projects were established to examine the capability of Omics technologies to enhance safety assessment and to discover new biomarkers for hepato-, neuro-, cardio-, and nephrotoxicity.
The first consortium which intensively studied nephrotoxicity by using new technologies was the “Nephrotoxicity working group” of the International Life Sciences Institute Health/Environmental Services Institute (ILSI/HESI) Genomics Committee. The data generated since 1999 were reported in 2004 (23–25). These findings included details of the mechanism of cisplatin, gentamycin, and puromycin toxicity and the identification of several transcriptional biomarkers determined on multiple platforms. A panel of 44 markers identified in theses study included: Clusterin, Vimentin, Lipocalin 2, Cyclin G1, and Hmox-1 (25). The successful application of toxicogenomics and the founding of a collaboration with the European Bioinformatics Institute of the European Molecular Biology Laboratory to develop a database (26) was also part of this cooperation.
Another joint project of leading pharmaceutical companies, with focus on the regulatory decision-making process in respect to the use of biomarkers in preclinical studies, was initialized by the nonprofit Critical Path Institute, with a focus on compound induced renal damage (Predictive Safety Testing Consortium; PSTC). This project leads to the discovery of 23 urinary protein biomarkers and a large number of transcriptional biomarkers by a combination of proteomics and genomics technologies (4,27,28). Subsequently, seven of these markers were validated by testing different model compounds for up to 14 days in rats. These seven biomarkers are now qualified by all ICH regulatory agencies, FDA (USA), EMA (Europe), and Pharmaceuticals and Medical Devices Agency (Japan), which significantly promotes the use of these biomarkers in a global setting.
Further projects on national and international levels, which focused on nephrotoxicity, are given in supplementary material Table II.
Following the discovery, validation, and finally regulatory acceptance, several companies now offer test systems, based on multiplexing technologies, to determine these biomarkers in urine. In addition, exploratory biomarkers on the transcriptional level are also still being used. These mostly real-time quantitative polymerase chain reaction (qPCR)-based methods give specific and/or predictive values for the early detection of nephrotoxicity (29).
RAT KIDNEY TOXICITY BIOMARKERS
Protein-Based Biomarkers
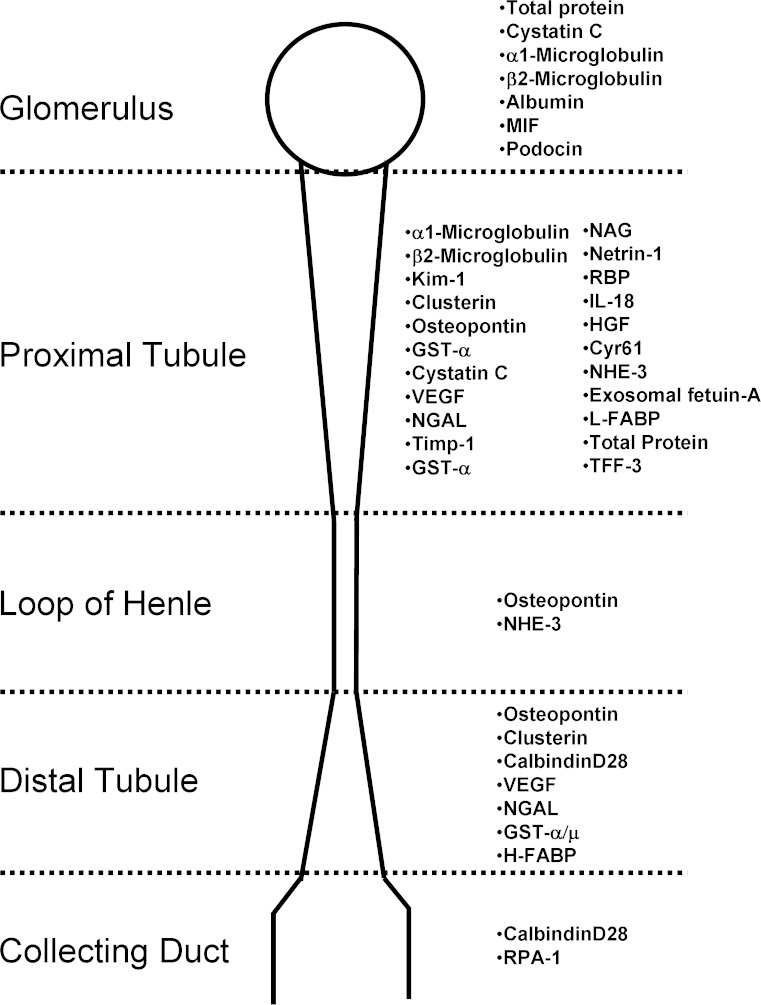
Urinary protein biomarkers have the great benefit of the easy availability of urine and the lack of sample preparation. Beside the previously mentioned 23 urinary protein biomarkers identified by the PSTC together with their partners from pharmaceutical industry, many other biomarkers were detected in recent years, many showing good predictive power, even if not qualified by regulatory agencies. It is not possible to describe all these proteins in detail. To give an overview, Fig. 3 gives some of the markers with their diagnostic area within the nephron. However, it should be noted that the majority of urinary protein biomarkers are predictive for both glomerular and tubular damage and rarely for one specific part of the nephron. Biomarkers for the loop of henle, like osteopontin or NHE-3, are neither specific for this part of the nephron nor are they yet qualified by the US-FDA or EMA. This is also true for the distal tubule, were only Clusterin and TFF-3, both specific for tubular damage in general, have been qualified to date. For the collecting duct only RPA-1, which already is qualified based on the study of the ILSI/HESI nephrotoxicity working group, and CalbindinD28 are described as a potential biomarkers. The areas where most damage is usually observed, i.e., within the nephron, the glomerulus, and the proximal tubule, are covered by well-known proteins, such as Kim-1, CystatinC, and albumin. There are further qualification efforts ongoing to enhance the sensitivity and specificity of these biomarkers for toxicological testing in preclinical trials.
Fig. 3.
Shown are several urinary biomarkers with their predictive area within the nephron. These biomarkers include qualified as well as exploratory once
A more detailed overview of the seven originally accepted biomarkers, plus the new accepted biomarker, RPA-1, is given in Table II.
Table II.
Overview of the Accepted Biomarkers for the Detection of Nephrotoxicity Including their Physiological Function, the Area of Toxicity Prediction, as well as Preclinical and Clinical Data Available
| Protein | Comments | Presumed marker of | Experimental support | Clinical support | References |
|---|---|---|---|---|---|
| Cystatin C | Inhibitor of extracellular cysteine proteinase widely expressed by nucleated cells | GFR (serum) PT damage | Increased in diabetic rats and in response to subtotal or unilateral nephrectomy Elevated during chromium nephropathy | Increased in patients with acute kidney injury Increased in patients with tubular dysfunction | (17,46–50) |
| Lysosomal degradation and reabsorption by proximal Glomerular filtration, reabsorption and lysosomal degradation by proximal tubule cells | |||||
| KIM-1 | Transmembrane protein expressed by tubule epithelial cells in response to injury | PT damage | Elevated in response to gentamicin, mercury, chromium, cadmium Increased mRNA expression in kidneys of rats treated with gentamicin, ochratoxin A, sevoflurane, cisplatin, cisplatin, bacitracin | Increased in renal transplant recipients and patients with acute kidney injury of various cause | (29,50–57) |
| Acts as a phosphatidylserine receptor and confers phagocytic capacity to clear cell debris | |||||
| β2-microglobulin | Protein found on the surfaces of all nucleated cells, shed into blood | PT damage | Increased in response to ochratoxin A, depleted uranium, cisplati, chlorotrifluoroethylene, 1,1-dichloro- 2,2-difluoroethylene | Elevated in patients with idiopathic membranous nephropathy, in patients suffering from with tenofovir disoproxil fumarate induced kidney damage, in transplant patients with acute tubulointerstitial rejection | (29,48,58–63) |
| Glomerular filtration and reabsorption by proximal tubule cells | |||||
| Albumin | It is the most abundant protein in blood plasma (comprises about half of the blood serum protein) | Glomerulus | Increased exposure of proximal tubular cells to Albumin can induce apoptosis by fatty acid boundary Elevated in response to cisplatin (Platinol), gentamicin, carbapenem A, thioacetamide, hexachlorobutadiene (HCB) and D-serinein rat | Pathological urinary excretion of albumin > 20 mg/l after 100– 150 mg/m2 Cisplatin treatment was detected in 39 out of 54 patients. Regression of microalbuminuria in patients with T1DM is associated with lower levels of urinary tubular injury biomarkers, Kim-1 and NAG | (64–70) |
| Its size and negative electric charge exclude it from excretion in the glomerulus | |||||
| Total protein | The total amount of protein in urine from healthy subjects is reduced to a minimum | Glomerulus PT damage | Elevated in response to cisplatin, gentamicin, vancomycin, tacrolimus, puromycin and doxorubicin | Total proteinuria perform as predictor of renal outcomes and mortality in patients with CDK. A correlation of increase was shown for types I + II diabetic, Hypertension, geratic and myoblobinuria | (48,71,72) |
| Large and strongly charged proteins were not filtered by the glomerulus | |||||
| Small proteins go free though the glomerular barrier and become re-absorbed by the proximal tubular | |||||
| Tff 3 | Small peptide hormones secreted by mucus-producing cells, and by epithelial cells of multiple tissues, in mammals | T damage | The sensitive value of TFF3 was shown by the elevated decrease after cisplatin, gentamicin, carbapenem A, thioacetamide, hexachlorobutadiene and D-serinein treatment | N/A | (69,73–75) |
| Tff3 plays essential functions in both mucosal surface maintenance and restitution | Tff3 mRNA shows strong abundance of the outer medulla of healthy kidneys | ||||
| Clusterin | In a wide range of tissue expressed secreted glycoprotein | PT + DT damage | Increased following renal ischemia, unilateral urethral obstruction or in response to various nephrotoxins, e.g., Increased mRNA expression in kidneys of rats treated with gentamicin, ochratoxin A, sevoflurane, cisplatin, vancomycin, bacitracin | Over expressed in human renal diseases | (29,48,54,56,76–79) |
| Extracellular chaperone | |||||
| Involved in cell adhesion, membrane recycling, tissue remodeling, cell cycle regulation apoptosis and DNA repair | |||||
| RPA-1 | Discovered by immunohistocalical screening (histomics) | CD damage | RPA-1 show increased amount in urine and tissue after treatment of rats with BEA, propyleneimine, and indomethacin | N/Aa | (80–82) |
| The protein is not doubtless identified and characterized |
aArgutus Medical Ltd. is involved in the SAFE-T IMI Project in an effort to qualify clinical renal injury markers with the health authorities.
These eight biomarkers are all accepted for preclinical studies in rodents, but they are also accepted for clinical observations, under certain circumstances. The individual qualification status is shown in Table III. However, in clinical trials, the use of novel urinary biomarkers is still in the discovery phase with little information available in the literature.
Table III.
The Current Status of the Originally Qualified Seven Urinary Protein Biomarkers and There Voluntary Usability in Clinical Trials Recommended by the ICH-Regulated Agencies
| Urinary biomarker | Qualified preclinical | Adds inform-SCr and BUN | Outperforms SCr and/or BUN | Qualified clinical |
|---|---|---|---|---|
| Kim-1 | + | +a | +a | + |
| Albumin | + | +a | +a | + |
| Clusterin | + | +a | +a | Pending |
| TFF3 | + | +a | − | Pending |
| Total Protein | + | +b | +b (SCr) | + |
| Cystatin C | + | +b | +b (SCr) | + |
| ß2 μ glob | + | +b | +b (SCr) | + |
aAcute renal tubular damage
bAcute glomerular damage with acute tubular re-absorption impairment
SCr Biomarker out performed serum creatinine
Modified by http://www.c-path.org/pdf/PMDAReportPSTCtranslation.pdf
Single-Plex Immunoassays
ELISA
For the detection of single analytes, enzyme-linked immunosorbet assays (ELISA) are commonly used. Many assays by different providers currently are available. However, not all are optimized and validated for the use with urine samples. For example, ELISAs for the most promising markers such as Kim-1 (e.g., Argutus Medical Ltd.) or Clusterin (BioVendor R&D) are available. BioPorto® specializes in ELISAs and test kits for clinical chemistry platforms with the detection of NGAL in pig, dog, monkey, mouse, rat, and human being available. However, all these ELISAs have the disadvantage of detecting only one biomarker at a time within the sample. The measurement of only one biomarker for the diagnosis of renal damage is, because of the heterogenicity of the nephron and the complexity of the kidney, not recommendable. So the use of more than one ELISA is necessary which makes it more time-consuming and expensive. Therefore, the use of multiplexing platforms, such as the Luminex® xMAP® or the MesoScale Discovery technology, is now well-established in most testing labs.
Dipstick Assay
BioAssay Works®, a founding partner of Kirkegaard and Perry Laboratories have developed a rapid and easy to use Dipstick assay for Kim-1 (rat) and KIM-1 (human) (30). Kim-1, one of the most commonly used and predictive urinary protein biomarkers, is of importance due to its translatability between preclinical and clinical trials. BioAssay Works® and its partner Argutus Medical Ltd. now provides the Kim-1 and KIM-1 Dipstick assay under the name RENA®-Strips and is based on gold–sol conjugates and lateral flow assays.
Multiplex Assays
Luminex® xMAP®
The Luminex® xMAP® technology is based on two proven, existing technologies: flow cytometry of color-coded, tiny microspheres in combination with a detection system which is characterized by its flexibility and open-architecture design (immunoassay in the case of nephrotoxicity biomarkers; www.luminexcorp.com).
Specific antibodies to an analyte are coated on one microsphere bead type. The beads float freely around enhancing the sensitivity of this technology. After incubation with a streptavidin–pyroerythrin-labeled secondary antibody against the analyte, the bead–analyte–flourophore complex is detected within the Luminex compact analyzer. In this way, the xMAP® Technology in theory allows the multiplexing of up to 100 unique assays within a single sample.
The detection of nephrotoxicity specific biomarkers can be performed either by rules based medicine (RBM), a service provider that was part of the PSTC-driven initiative which lead to the discovery and acceptance of the already described markers. In cooperation with RBM, Merck-Millipore (formally Merck-Chemicals) has commercialized a large number of kits with different combinations of nephrotoxicity markers. An overview of kits available for the detection of protein biomarkers for the detection of renal damage based on the Luminex® xMAP® and the MesoScale Discovery® platform is shown in Table IV.
Table IV.
Summary of Commercial Available Multiplex Assays Based on the Luminex® xMAP® Technology and the MULTI-Array® Technology from MesoScale Discovery®
| Assay name | Species | Matrix | Analytes | Provider/company |
|---|---|---|---|---|
| Luminex® xMAP® | ||||
| Widescreen™ Rat Kidney Toxicity Panel 1 | Rat | Urine (serum) | α-GST | Merck-Millipore formerly Merck-Chemicals |
| Timp-1 | ||||
| Kim-1 | ||||
| β-2-Microglobulin | ||||
| VEGF | ||||
| Widescreen™ Rat Kidney Toxicity Panel 1 | Rat | Urine (serum) | Calbindin | Merck-Millipore formerly Merck-Chemicals |
| Clusterin | ||||
| NGAL | ||||
| Cystatin C | ||||
| Osteopontin | ||||
| Rat Kidney MAP v. 1.0 | Rat | Urine (serum) | β-2-Microglobulin | Rules-based Medicine |
| Calbindin | ||||
| Clusterin | ||||
| Cystatin-C | ||||
| EGF | ||||
| α-GST | ||||
| m-GST | ||||
| Kim-1 | ||||
| NGAL | ||||
| Osteopontin | ||||
| Timp-1 | ||||
| VEGF-A | ||||
| MILLIPLEX® MAP Human Kidney Toxicity Panel 1 | Human | Serum | KIM-1 | Merck-Millipore formerly Millipore |
| Osteopontin | ||||
| Renin | ||||
| TFF-3 | ||||
| MILLIPLEX® MAP Human Kidney Toxicity Panel 2 | Human | Serum | β-2-Microglobulin | Merck-Millipore formerly Millipore |
| Clusterin | ||||
| Cystatin C | ||||
| MILLIPLEX® MAP Human Kidney Toxicity Panel 3 | Human | Urine | KIM-1 | Merck-Millipore formerly Millipore |
| Renin | ||||
| TFF-3 | ||||
| MILLIPLEX® MAP Human Kidney Toxicity Panel 4 | Human | Urine | Albumin | Merck-Millipore Formerly Millipore |
| β-2-Microglobulin | ||||
| Clusterin | ||||
| Cystein C | ||||
| Osteopontin | ||||
| Human Kidney MAP v. 1.0 | Human | Urine (serum) | α-2-Microglobulin | Rules-based medicine |
| β-2-Microglobulin | ||||
| Calbindin | ||||
| Clusterin | ||||
| CTGF | ||||
| Creatinine | ||||
| Cystatin-C | ||||
| α-GST | ||||
| Kim-1 | ||||
| Microalbumin | ||||
| NGAL | ||||
| Osteopontin | ||||
| Uromodulin | ||||
| Timp-1 | ||||
| TFF-3 | ||||
| VEGF-A | ||||
| OPG | ||||
| MesoScale Discovery® | ||||
| Kidney Injury Panel 1 | Rat | Urine | Albumin | MesoScale discovery |
| Kim-1 | ||||
| NGAL | ||||
| Osteopontin | ||||
| Kidney Injury Panel 2a | Rat | Urine | Albumin | MesoScale discovery |
| Kim-1 | ||||
| NGAL | ||||
| Osteopontin | ||||
| α-GST | ||||
| Clusterin | ||||
| Argutus AKI Test® | Rat | Urine | α-GST | MesoScale Discovery/Argutus Medical Ltd. |
| GSTYb1 | ||||
| RPA-1 | ||||
aCommercialization of the Kidney Injury Panel 2 is planned for end of 2011
Mesoscale Discovery®
Mesoscale Discovery® (MSD) provides an alternative multiplexing technology platform, able to detect novel protein biomarkers. The MULTI-ARRAY® technology is a combination of electrochemiluminescence detection and patterned arrays (31). In contrast to the liquid array of the Luminex®-based technology, the detection is performed on a solid phase. On a carbon surface in single spots, on the bottom of a multi-well plate (maximum 10 spots in a 96-well plate, 4 spots in a 384-well plate, and 100-spots in a 24-well plate) antibodies against specific analytes are coated. MSD’s instruments use sensitive photodetectors to collect and quantitatively measure light emitted from the microplates. Because of the short measurement time of only some minutes, this technology is more suited to high-throughput screening. MSD offers beside single-plex assays, one 4-plex Kit related to nephrotoxicity biomarkers (see Table IV). This can be extended by an additional kit based on the MSD technology provided by Argutus Medical Ltd.
Mosaique Diagnosis
Mosaiques Diagnostics is a provider of highly innovative “Clinical Proteomics” services for pharmaceutical companies. The detection is based on urinary proteins which are not directly associated with the previously described biomarkers, but on a specific proteome expression pattern. The capillary electrophoresis coupled to mass spectrometry (CE-MS) technology is used to diagnose severe diseases, including renal damage at early development stages (32). The generated pattern, based on an underlying online database, provides individual health status of an organism. The CE-MS technology enables the detection of renal damage as well the identification of surrogate markers and endpoints in clinical and potentially preclinical trials which allow evaluation of novel drug candidates on a small number of animal models (33). Because no antibodies are used, this technology is in principle capable of detecting renal damage by measurement of urine from different species (for those included in the database) without major restriction. Measurement of the proteome in combination with online database matching is the key to an all-in-one diagnosis for a wide spectrum of clinical and preclinical applications.
Mosaiques diagnostics has successfully implemented its technology in clinical trials in cooperation with pharmaceutical companies such as Roche Pharma and Bayer Healthcare (http://mosaiques-diagnostics.com)
Transcript-Based Biomarkers
In addition to protein biomarkers, tissue-based transcriptional biomarkers have been identified and described (23). Most of the proteins transcribed from these genes are not described as urinary biomarkers. However, some of the previously described biomarkers, like Kim-1, Timp-1, and Clusterin, can be used on both mRNA and protein levels. A systematic comparison of biomarkers detectable on both levels has not yet been reported.
The acceptance and the qualification status of these biomarkers are not yet as high as for the urinary protein biomarkers. On the one hand, the limitation in accessibility (tissue availability) and on the other hand the enhanced sample preparation effort compared to the use of immunoassays to detect urinary proteins, are the major points which limit the routine use of transcriptional biomarkers. However, several markers were identified, e.g., by the ILSI/HESI consortium (23–25), and are the bases for several assays commercially available from the detection of single transcript biomarkers.
Compugen Ltd
Compugen Ltd. is a company based in Tel Aviv, Israel which focuses on discovery and licensing of product candidates to the drug and diagnostic industry. Beside many other products (34), Compugen Ltd. have identified and validated biomarkers for the early detection of drug-induced nephrotoxicity by using model nephrotoxicants, microarray analysis, statistical, and machine learning tools. A set of four biomarkers were identified and validated on real-time PCR to predict drug-induced nephrotoxicity in the rat. The discovery process was based on the use of Compugen’s nucleic acid testing Discovery Platform.
The combination of biomarkers together with a random forest classification model, also provided by Compugen Ltd., is used for the detection of drug toxicity in the preclinical rat toxicity studies in the drug development process (http://www.cgen.com).
Althea DX4
AltheaDx is a company focused on supporting pharmaceutical and diagnostic companies with the translation of novel preclinical biomarkers into clinical use. The company provides a wide range of expertise on genetic diseases, biomarker discovery, diagnostic development, and testing as well as regulatory processes through a consultative mechanism.
Beside many other organ and disease-specific products, AltheaDx has developed a multiplexed qRT-PCR (XP™-PCR) product, whereby 24 genes for nephrotoxicity can be monitored within a single PCR reaction. The product is licensed to Beckman Coulter and used to create the Beckman GeXP platform. The selected genes are a combination of gene expression markers from the ILSI/HESI Health Perspective (23–25) and further published biomarkers (35). Thirty-three genes, including housekeeping genes are shown in Table V.
Table V.
33 Genes Provided by Althea DX4 for the qPCR-Based Detection of Renal Insult in Rat
| Symbol | Description | Accession no. |
|---|---|---|
| Actb | Actin-beta | NM_031144 |
| Agt2 | Beta-alanine-pyruvate aminotransferase | AA818440 |
| Bhmt | betain homocystein s-transferase | AA901407 |
| Ccng1 | CyclinG1 | X70871 |
| Clu | Clusterin | NM_053021 |
| Cyp2d18 | Cytochrom P450 2 d18 | AA997886 |
| Egf | preproepidermal growth factor | AA901327 |
| EST AA 925553 | N/A | AA925553 |
| EST AA 957270 | N/A | AA957270 |
| EST AA819101 | N/A | AA819101 |
| EST AA819209 | N/A | AA819209 |
| EST AA819244 | N/A | NM_053625 |
| EST AA858892 | N/A | AA858892 |
| EST AA899472 | N/A | AA899472 |
| EST AA899737 | N/A | AA899737 |
| EST AA901025 | N/A | AA901025 |
| EST AA925957 | N/A | AA925957 |
| Gc | Vitamin D binding protein | AA818706 |
| Hmox-1 | Heme Oxygenase-1 | J02722 |
| Idh1 | Isocitrate dehydrogenase 1 | NM_031501 |
| Igfbp3 | Insulin-like factor binding protein | AA819611 |
| Ipmk | Inositol polyphosphate multikinase | AA901117 |
| Kim-1 | Kidey injury molecule-1 | AF035963 |
| Ngfg | Nerve Growth Factor | AA925291 |
| Pctp | glyceraldehyde phosphate dehydrogenase | NM_0117008 |
| Ppib | CycliphilinB | AF071225 |
| Rbp4 | Retinol-binding protein | M10934 |
| Spp1 | Osteopontin | M99252 |
| Syngry2 | Synaptogyrin 2 | Al058493 |
| Tmsb4x | Thymosin beta-4 | AA819102 |
| Tubb5 | Class I beta-tubulin | AA858888 |
| Ugt2b5 | UPD-glucuronosyl transferase | Y00156 |
| Vim | Vimentin | BC061847 |
This list was kindly provided by Althea Technologies Inc
SABiosciences Corporation
The SYBR® green-based panel of up to 84 genes is available in 96-, 384-, and 100-well disk plates. The genes, shown in Table VI for the detection of early effects of renal damage, are freely selectable by the customer. The nephrotoxicity RT2Profiler™ PCR arrays are available for rat, human, and mouse, whereby the genes vary between the species.
Table VI.
84 Genes Specific for Rat, Provided by SABiosciences Corp. Provided qPCR Arrays Specific for Mouse or Human Can Include Different Genes (Not Shown)
| Symbol | Description | Accession no. |
|---|---|---|
| A2m | Alpha-2-macroglobulin | NM_000014 |
| Aass | Aminoadipate–semialdehyde synthase | NM_005763 |
| Abcb1 | ATP-binding cassette, subfamily B (MDR/TAP), member 1 | NM_000927 |
| Abcc2 | ATP-binding cassette, subfamily C (CFTR/MRP), member 2 | NM_000392 |
| Aldh1a1 | Aldehydede hydrogenase 1 family, member A1 | NM_000689 |
| Angptl4 | Angiopoietin-like4 | NM_001039667 |
| Anxa5 | AnnexinA5 | NM_001154 |
| Atf3 | Activating transcription factor3 | NM_001674 |
| Bhmt | Betaine-homocysteine methyltransferase | NM_001713 |
| Bmp1 | Bone morphogenetic protein 1 | NM_006129 |
| Bmp4 | Bone morphogenetic protein 4 | NM_130851 |
| Btg2 | BTG family, member 2 | NM_006763 |
| Calb1 | Calbindin 1,28 kDa | NM_004929 |
| Cat | Catalase | NM_001752 |
| Ccl3 | Chemokine (C-Cmotif) ligand3 | NM_002983 |
| Ccnd1 | Cyclin D1 | NM_053056 |
| Ccng1 | Cyclin G1 | NM_004060 |
| Ccs | Copper chaperone for superoxide dismutase | NM_005125 |
| CD24 | CD24 molecule | NM_013230 |
| CD44 | CD44 molecule (Indian blood group) | NM_000610 |
| Cdkn1A | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | NM_000389 |
| Clu | Clusterin | NM_001831 |
| Cp | Ceruloplasmin (ferroxidase) | NM_000096 |
| Cst3 | Cystatin C | NM_000099 |
| Ctss | Cathepsin S | NM_004079 |
| Cxcl1 | Chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) | NM_001511 |
| Cxcl10 | Chemokine (C-X-C motif) ligand 10 | NM_001565 |
| Cyp2C19 | Cytochrome P450, family 2, subfamily C, polypeptide 19 | NM_000769 |
| Cyp2D6 | Cytochrome P450, family 2, subfamily D, polypeptide 6 | NM_000106 |
| Cyr61 | Cysteine-rich, angiogenic inducer, 61 | NM_001554 |
| EGF | Epidermal growth factor (beta-urogastrone) | NM_001963 |
| FGB | Fibrinogen beta chain | NM_005141 |
| Fmo2 | Flavin containing monooxygenase 2 (non-functional) | NM_001460 |
| Fn1 | Fibronectin1 | NM_002026 |
| G6pc | Glucose-6-phosphatase, catalytic subunit | NM_000151 |
| G6pd | Glucose-6-phosphate dehydrogenase | NM_000402 |
| Gadd45A | Growth arrest and DNA damage-inducible, alpha | NM_001924 |
| Gamt | Guanidino acetate N-methyl transferase | NM_000156 |
| Gatm | Glycine amidino transferase (l-arginine: glycine amidino transferase) | NM_001482 |
| Gc | Group-specific component (vitamin D binding protein) | NM_000583 |
| Ghr | Growth hormone receptor | NM_000163 |
| Glul | Glutamate-ammonialigase (glutamine synthetase) | NM_002065 |
| Gpnmb | Glycoprotein (transmembrane) nmb | NM_002510 |
| Gpx8 | Glutathioneperoxidase 8 (putative) | NM_001008397 |
| Gstk1 | Glutathione S-transferase kappa1 | NM_015917 |
| Gstp1 | Glutathione S-transferase pi1 | NM_000852 |
| Havcr1 | Hepatitis A virus cellular receptor 1 | NM_012206 |
| Hmox1 | Hemeoxygenase (decycling) 1 | NM_002133 |
| Hmox2 | Hemeoxygenase (decycling) 2 | NM_002134 |
| Hsp90AA1 | Heat shock protein 90 kDa alpha (cytosolic), class A member 1 | NM_001017963 |
| Idh1 | Isocitrate dehydrogenase 1 (NADP+), soluble | NM_005896 |
| Igfbp1 | Insulin-like growth factor binding protein 1 | NM_000596 |
| Igfbp3 | Insulin-like growth factor binding protein 3 | NM_000598 |
| Ipmk | Inositol polyphosphate multikinase | NM_152230 |
| Klk1 | Kallikrein 1 | NM_002257 |
| Lcn2 | Lipocalin 2 | NM_005564 |
| Lgals3 | Lectin, galactoside-binding, soluble, 3 | NM_002306 |
| Mcm6 | Minichromosome maintenance complex component 6 | NM_005915 |
| Mgp | Matrix Glaprotein | NM_000900 |
| Mt1A | Metallothionein 1A | NM_005946 |
| Nox4 | NADPH oxidase 4 | NM_016931 |
| Nphs2 | Nephrosis 2, idiopathic, steroid-resistant (podocin) | NM_014625 |
| Nqo1 | NAD(P)H dehydrogenase, quinone1 | NM_000903 |
| Oat | Ornithine amino transferase (gyrateatrophy) | NM_000274 |
| Odc1 | Ornithinede carboxylase 1 | NM_002539 |
| Rgn | Regucalcin (senescence marker protein-30) | NM_004683 |
| Rtn4 | Reticulon 4 | NM_007008 |
| Scd | Stearoyl-Co adesaturase (delta-9-desaturase) | NM_005063 |
| Slc22A1 | Solute carrier family 22 (organic cation transporter), member 1 | NM_003057 |
| Slc22A5 | Solute carrier family 22 (organic cation/carnitine transporter), member 5 | NM_003060 |
| Slc22A6 | Solute carrier family 22 (organic anion transporter), member 6 | NM_004790 |
| Socs3 | Suppressor of cytokine signaling 3 | NM_003955 |
| Sod2 | Superoxide dismutase 2, mitochondrial | NM_000636 |
| Sod3 | Superoxide dismutase 3, extracellular | NM_003102 |
| Spp1 | Secreted phosphoprotein 1 | NM_000582 |
| Sprr1A | Small proline-rich protein 1A | NM_005987 |
| Timp1 | TIMP metallopeptidase inhibitor 1 | NM_003254 |
| Tmsb10 | Thymosin beta 10 | NM_021103 |
| Tnfrsf12A | Tumor necrosis factor receptor superfamily, member 12A | NM_016639 |
| Uchl1 | Ubiquitin carboxyl-terminal esterase L1 (ubiquitinthiol esterase) | NM_004181 |
| Ugt1A1 | UDP glucuronosyl transferase 1 family, polypeptide A1 | NM_000463 |
| Ugt1A6 | UDP glucuronosyl transferase 1 family, polypeptide A6 | NM_001072 |
| Vcam1 | Vascular cell adhesion molecule 1 | NM_001078 |
| Vim | Vimentin | NM_003380 |
| b2m | Beta-2-microglobulin | NM_004048 |
Metabolite-Based Biomarkers
Beside genomic and proteomic approaches, metabolomic technologies have the capability of providing translational diagnostic and prognostic biomarkers specific for early stages of nephrotoxicity. The benefit compared to genomics, and in part to proteomics, evaluations is the easy sample preparation, data acquisition, and, in the case of clinical trials, the use of body fluids collected through minimal invasive methods. The method is based on the detection of metabolites, including breakdown products, which are correlated to specific toxic insults.
The measurement for metabolomic-specific analytes normally is performed by analytical methods such as liquid chromatography (LC)–nuclear magnetic resonance, LC–mass spectrometry (MS), or gas chromatography–MS. One of the largest metabolomics studies performed to date was that by Imperial College (London, UK) in collaboration with six pharmaceutical companies. The Consortium of Metabonomic Toxicology built prediction models for nephrotoxicity, with a sensitivity of 67% and a specificity of 41% (36). Some metabolites were reported to show early and/or late responses in urine measured by metabolomic technologies, including glucosamine, monoethanolamine, phosphate, 3-hydroxyphenylacetate, hippurate, and riboflavin (37). In addition, other amino acids and peptides, as well as polyamines, were also able to identify and/or predict drug induced renal damage.
FURTHER PERSPECTIVES
Micro Rnas
The field of microRNA (miRNA) research is one of the most exciting and rapidly expanding fields in biosciences, including medical biology and (pre) clinical research. The role of miRNAs within the kidney has not yet been studied intensively. However, because of the high conservation of miRNAs between different species (supplementary material Fig. 2), their central role in gene expression regulation and in their response to toxins, miRNA are of growing interest for safety assessment.
Experiments with Dicer knockout mice have suggested a critical role of specific miRNAs in orchestrating kidney development and maintaining the functional and structural integrity of the tubular and glomerular barrier. miRNAs with a high abundance within the kidney or which are up regulated after an insult are shown in Table VII.
Table VII.
Overview of miRNAs Described in Relation to Renal Diseases and/or Damage (Also Shown are the Reexpected Function and Target mRNAs Identified so Far)
| MicroRNAs linked to urinary tract and kidney disease | |||
|---|---|---|---|
| miRNA | Function/pathway | Targets | Ref. |
| Bladder cancer | |||
| miR-129 | Signal transduction/protein expression | SOX4 GALNT1 | (44) |
| miR-221 | Apoptosis | TRAIL pathway | (83) |
| miR-1/133a/218 | Cytoskeleton | LASP1 | (84) |
| miR-19a | Apoptosis/mTOR pathway | PTEN | (85) |
| miRs-30a-3p/133a/199a | Differentiation | KRT7 | (86) |
| miR-34a | Cell cycle control | CDK6 | (87) |
| miR-99a/100 | Proliferation | FGFR3 | (88,89) |
| miR-101 | Gene expression | EZH2 | (90) |
| miR-125b | Apoptosis/proliferation | E2F3 | (91) |
| miRs-145/133a | Cytoskeleton | FSCN1 | (84) |
| miR-145 | Signal transduction | CBFB PPP3CA CLINT1 | (92) |
| miR-200/205 | EMT | ZEB 1 and 2 | (93,94) |
| miR-200 family | EMT | ERRFI-1/EMT process | (95) |
| Renal cancer | |||
| miR-34a | Apoptosis | Sirt1 | (87) |
| miR-23b | Modulation of TGF-ß1 signaling/ apoptosis and hypoxic signaling | POX | (96) |
| OncomiR-1a (Wilms’ tumor) | Proliferation response to E2F3 | PTEN | (97) |
| miR-200c | EMT | SIP1 | (98) |
| ZEB2 | |||
| miR-141 | EMT | SIP1 | |
| ZEB2 | |||
| Diabetic nephropathy | |||
| miR-21 | Inhibitor of apoptosis/cell proliferation | PTEN | (99) |
| miR-23b | Modulation of TGF-ß1 signaling/apoptosis and hypoxic signaling | POX | (96) |
| miR-30 family | Loss in podocyte specific dicer knockout | CTGF Xlim1/Lhx1 | (100) |
| miR-192 | Collagen synthesis | SIP1, ZEB 1 and 2 | (101) |
| miR-216 | Modulation of TGF-ß1 signaling | YB-1 PTEN | (102) |
| miR-377 | Enhanced expression by high glucose in mesangial cells/synthesis of fibronectin | PAK1 SOD1/2 | (103) |
| Polycystic kidney disease | |||
| miR-17 | Promotion of proliferation | PKD2 | (104) |
| miR-15a | Progression cell cycle | Cdc25A | (105) |
This list is not intended to be exhaustive.
a Synonyme miR 17–92 cluster, miR microRNA, PAK1 p21-activated kinase 1, SIP1 Smad-1 interacting protein, SOD superoxide dismutase, TGF-β1 transforming growth factor beta 1, YB-1 Y-box protein-1, PTEN phosphatase and tensin homologue, mTOR mammalian target of rapamycin, EMT epithelial-to-mesenchymal transition
The expression profile of miRNAs within the healthy kidney has provided a general idea of miRNA constitutive expression and which miRNA serves a specific functional role. Several miRNAs were identified to be potentially involved in renal dysfunction and disease. For example, miR-15a has been suggested to be involved in the outcome of cystic kidney disease while miR-17-92 seems to be linked to the growth of Wilms’ tumors. Some miRNA in the kidney were induced by transforming growth factor β-1 in models of diabetic nephropathy. Others, like miR-192 and miR-377, lead to matrix deposition or epithelial to mesenchymal transition (miR-200 and miR-2005) (38).
More interesting for the field of preclinical and clinical safety assessment is the role of miRNAs in urinary exosomes (39,40). Novel miRNAs identified could serve as biomarkers in humans and because of the highly conserved function across experimental animals, it could also be used to detect renal damage during toxicity testing.
The first study reported to use miRNA for the detection of cisplatin-induced nephrotoxicity has been published. miR-34a was induced by p53 and was reported to be involved by the outcome of cisplatin-induced renal damage (41).
In Vitro Systems
In vitro systems for nephrotoxicity testing are of major interest as supportive tools in two main areas. Firstly, the investigation and understanding of the mechanism of toxicity and secondly the development of an early high throughput screening tool for the prediction of nephrotoxicity earlier in the drug discovery process.
For the assessment of nephrotoxicity in vitro, different test systems are available. Isolated perfused kidneys, precision cut slices, and isolated fragments and cells (glomeruli and tubules) are relatively near to the in vivo situation but the isolation and cultivation is complex and the time of cultivation and treatment is strongly limiting. In the past, the use of cell culture, both primary and cell lines, has become the most popular method for screening. General cytotoxic properties of the test compound should be identified in order to distinguish general acute toxic responses from the more relevant kidney specific toxicological effects. For compounds identified in cytotoxicity assays as potentially harmful, additional investigations should be performed. Such investigations can include measurement of clinically relevant enzyme activities, barrier function, transport activities, energy metabolism, and xenobiotic metabolism. In the future, gene and/or protein biomarkers will be used, but these still need some level of prevalidation for the specific cells used. Subsequently, more detailed investigations relevant to mechanisms of nephrotoxic action will be necessary. Also the uses of epithelial barrier models are promising for the extrapolation to proximal tubule damage in vivo.
The recommendations by the European Centre for the Validation of Alternative Methods include the development of in vitro models employing cells and tissues from rats and mice, so that the in vitro and in vivo data can be compared and the possibility for extrapolating in vitro results to the in vivo situation in man can be determined.
Summary
For the first time, new Omics based (discovered by and based on) safety biomarkers have been accepted by the regulators. Now, the industry/users need to generate data to further validate these markers in real-life cases, as well as their use in other subchronic and chronic animal studies. A lot of questions remain unanswered. To date, only limited data are available showing the release of the urinary protein biomarkers after a recovery period. Differences between rat strains and genders in the basal level of urinary proteins and their response after drug induced renal insults are also not fully understood. The paradigm of using only a few biomarkers which give a complete diagnosis of the status quo of an organ has to be critically questioned (and rejected) as the new data for the new urinary biomarkers show. It would be wrong to focus only on Kim-1, the Holy Grail for the detection of nephrotoxicity, because of the limitation of even this common marker. The limitation of Kim-1, specific for damages in the pars recta (S3), was shown by comparing urinary Kim-1 excretion in different renal pathologies, including ischemic acute tubular necrosis, contrast nephropathy, and renal allograft rejection (42). This critical work underscores the limitations of Kim-1 (43) as a marker of renal damage in areas of the nephron other than the S3 segment.
In practice, a large panel of biomarkers are needed to discover the different sites and mechanisms of potential toxicity. Therefore, further studies focused on the qualified as well as the exploratory markers have to be performed to determine their applicability and to deliver further information which could be relevant for the safety profile of a new drug candidate or chemical. The translation of these biomarkers to humans (including their use in human cell systems) is of most importance in the near future. A revision of the RIFLE and AKIN schemes has to be contemplated for the future to optimize the decision points of therapy for patients with acute and chronic renal failure (and to follow potential DIKI during clinical trials). Additionally, the success of this biomarker discovery process, as well as the development of specific assays to measure them, will aid and speed up the discovery and acceptance of markers for other target organ systems (for example, liver and heart toxicities).
Beside the urinary protein biomarkers, many transcriptional biomarkers have been described and can promote the identification and maybe the prediction of renal injury. A systematic comparison of traditional toxicological approaches together with the novel urinary protein biomarkers and transcriptional biomarkers over different time points and doses is needed to classify the advantages and disadvantages of both types of biomarkers. The fact that toxicogenomics is becoming more and more important, also for the regulatory agencies, will influence their decision making in the future and consequently becomes of major interest in the pharmaceutical/chemical industry and in the clinical environment. There is a very clear benefit of a non-invasive sample collection for the detection of urinary markers, however specifically in the case of transcriptional biomarkers, the disadvantage of the need of biopsy material could be the major limiting factor for clinical purposes (unless surrogate transcriptional markers are identified in the blood). One methodology often used in clinical studies (40) is the use of formalin-fixed and paraffin-embedded tissue for genomics and proteomics analysis. This technology can reduce the invasive sample (i.e., tissue) collection to a minimum, or at least utilize samples from necropsies or biopsies which have already been taken and stored (often for up to several years). A younger, but not less exciting field of research, is the use of urinary miRNAs, which has the potential to combine the benefit of non-invasive sample collection with the sensitivity and predictivity of transcriptional changes.
CONCLUSION
Currently, nephrotoxicity is a much discussed topic that has to be studied in much more detail. Many different methods, qualified and exploratory, are available so far and now have to be proven under real-life conditions. The great successes so far achieved in the field of toxicology have to be used as a starting point for further discovery and more detailed analysis for an improved safety assessment. There is a major need for new biomarkers which are better, cheaper, and more efficient than existing ones. Likewise, it is clear that the concern of pharmaceutical industries in relation to the use of these novel biomarkers is an additional burden, which still needs to be addressed. Furthermore, to remove lingering doubts about the use of novel Omics-based biomarkers, there is a need for regulatory agencies to encourage their use within the pharmaceutical and chemical industries, even when these novel biomarkers are not novel or not yet qualified.
ELECTRONIC SUPPLEMENTARY MATERIALS
Below is the link to the electronic supplementary material.
Overview of RIFLE and AKIN processes for assessing DIKI/AKI. Patients who receive renal replacement therapy (RRT) are classified as stage 3 in the AKIN schema (Cr serum creatinine, GFR glomerular filtration rate, AKI acute kidney injury; JPEG 25 kb)
The overlap of human, mouse, and rat miRNAs identified in renal tissue shows the conservation of miRNA over different species. The total number of miRNAs determined per species is shown under the name of species. Out of these, a number of 73 miRNAs were detectable in all three species. Adapted from (44) (JPEG 9 kb)
The five kidney damage stages discerned by the GFR (DOC 29 kb)
An overview of international projects focused on the identification of biomarkers for the detection of nephrotoxicity using novel technologies like toxicogenomic and proteomics (DOC 32 kb)
Contributor Information
Tobias Christian Fuchs, Phone: +49-6151-728589, Email: tobias.fuchs@merck.de.
Philip Hewitt, Phone: +49-6151-722927, Email: philip.hewitt@merck.de.
REFERENCES
- 1.Frost & Sullivan (2007) Rang, HP (eds). Drug discovery and development. Churchill Livingstone: Elsevier
- 2.Werner M, Costa MJ, Mitchell LG, Nayar R. Nephrotoxicity of xenobiotics. Clin Chim Acta. 1995;237(1–2):107–154. doi: 10.1016/0009-8981(95)06068-O. [DOI] [PubMed] [Google Scholar]
- 3.Devarajan P. Novel biomarkers for the early prediction of acute kidney injury. Cancer Therapy. 2005;3:477–488. [Google Scholar]
- 4.Dieterle F, Sistare F, Goodsaid F, Papaluca M, Ozer JS, Webb CP, et al. Renal biomarker qualification submission: a dialog between the FDA-EMEA and Predictive Safety Testing Consortium. Nat Biotechnol. 2010;28(5):455–462. doi: 10.1038/nbt.1625. [DOI] [PubMed] [Google Scholar]
- 5.Delanghe JR, Speeckaert MM. Creatinine determination according to Jaffe—what does it stand for? NDT Plus. 2011;4(2):83–86. doi: 10.1093/ndtplus/sfq211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bateman JC, Barberio JR, Grice P, Klopp CT, Pierpont H. Fatal complications of intensive antibiotic therapy in patients with neoplastic disease. AMA Arch Intern Med. 1952;90(6):763–773. doi: 10.1001/archinte.1952.00240120038004. [DOI] [PubMed] [Google Scholar]
- 7.Pfaller W, Gstraunthaler G. Nephrotoxicity testing in vitro—what we know and what we need to know. Environ Health Perspect. 1998;106(Suppl 2):559–569. doi: 10.1289/ehp.98106559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schrier RW. Blood urea nitrogen and serum creatinine: not married in heart failure. Circ Heart Fail. 2008;1(1):2–5. doi: 10.1161/CIRCHEARTFAILURE.108.770834. [DOI] [PubMed] [Google Scholar]
- 9.Sanderson IR, Walker WA. Development of the gastrointestinal tract. 1. Ontario, Canada: B.C. Decker; 1999. [Google Scholar]
- 10.Urashima M, Toyoda S, Nakano T, Matsuda S, Kobayashi N, Kitajima H, et al. BUN/Cr ratio as an index of gastrointestinal bleeding mass in children. J Pediatr Gastroenterol Nutr. 1992;15(1):89–92. doi: 10.1097/00005176-199207000-00014. [DOI] [PubMed] [Google Scholar]
- 11.Berk L, Rana S. Hypovolemia and dehydration in the oncology patient. J Support Oncol. 2006;4(9):447–454. [PubMed] [Google Scholar]
- 12.Walid MS. Blood urea nitrogen/creatinine ratio in rhabdomyolysis. Indian J Nephrol. 2008;18(4):173–174. doi: 10.4103/0971-4065.45295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendelssohn DC, Barrett BJ, Brownscombe LM, Ethier J, Greenberg DE, Kanani SD, et al. Elevated levels of serum creatinine: recommendations for management and referral. CMAJ. 1999;161(4):413–417. [PMC free article] [PubMed] [Google Scholar]
- 14.Sibai BM, Stella CL. Diagnosis and management of atypical preeclampsia-eclampsia. Am J Obstet Gynecol. 2009;200(5):481. doi: 10.1016/j.ajog.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 15.Martin JN, Jr, May WL, Magann EF, Terrone DA, Rinehart BK, Blake PG. Early risk assessment of severe preeclampsia: admission battery of symptoms and laboratory tests to predict likelihood of subsequent significant maternal morbidity. Am J Obstet Gynecol. 1999;180(6 Pt 1):1407–1414. doi: 10.1016/S0002-9378(99)70026-8. [DOI] [PubMed] [Google Scholar]
- 16.Grossman RA, Hamilton RW, Morse BM, Penn AS, Goldberg M. Nontraumatic rhabdomyolysis and acute renal failure. N Engl J Med. 1974;291(16):807–811. doi: 10.1056/NEJM197410172911601. [DOI] [PubMed] [Google Scholar]
- 17.Uchida K, Gotoh A. Measurement of cystatin-C and creatinine in urine. Clin Chim Acta. 2002;323(1–2):121–128. doi: 10.1016/S0009-8981(02)00177-8. [DOI] [PubMed] [Google Scholar]
- 18.Bokenkamp A, Domanetzki M, Zinck R, Schumann G, Byrd D, Brodehl J. Cystatin C—a new marker of glomerular filtration rate in children independent of age and height. Pediatrics. 1998;101(5):875–881. doi: 10.1542/peds.101.5.875. [DOI] [PubMed] [Google Scholar]
- 19.Fliser D, Ritz E. Serum cystatin C concentration as a marker of renal dysfunction in the elderly. Am J Kidney Dis. 2001;37(1):79–83. doi: 10.1053/ajkd.2001.20628. [DOI] [PubMed] [Google Scholar]
- 20.Aardema MJ, MacGregor JT. Toxicology and genetic toxicology in the new era of "toxicogenomics": impact of "-omics" technologies. Mutat Res. 2002;499(1):13–25. doi: 10.1016/S0027-5107(01)00292-5. [DOI] [PubMed] [Google Scholar]
- 21.Edwards SW, Preston RJ. Systems biology and mode of action based risk assessment. Toxicol Sci. 2008;106(2):312–318. doi: 10.1093/toxsci/kfn190. [DOI] [PubMed] [Google Scholar]
- 22.Zidek N, Hellmann J, Kramer PJ, Hewitt PG. Acute hepatotoxicity: a predictive model based on focused illumina microarrays. Toxicol Sci. 2007;99(1):289–302. doi: 10.1093/toxsci/kfm131. [DOI] [PubMed] [Google Scholar]
- 23.Amin RP, Vickers AE, Sistare F, Thompson KL, Roman RJ, Lawton M, et al. Identification of putative gene based markers of renal toxicity. Environ Health Perspect. 2004;112(4):465–479. doi: 10.1289/ehp.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramer JA, Pettit SD, Amin RP, Bertram TA, Car B, Cunningham M, et al. Overview on the application of transcription profiling using selected nephrotoxicants for toxicology assessment. Environ Health Perspect. 2004;112(4):460–464. doi: 10.1289/ehp.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson KL, Afshari CA, Amin RP, Bertram TA, Car B, Cunningham M, et al. Identification of platform-independent gene expression markers of cisplatin nephrotoxicity. Environ Health Perspect. 2004;112(4):488–494. doi: 10.1289/ehp.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mattes WB, Pettit SD, Sansone SA, Bushel PR, Waters MD. Database development in toxicogenomics: issues and efforts. Environ Health Perspect. 2004;112(4):495–505. doi: 10.1289/ehp.6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozer JS, Dieterle F, Troth S, Perentes E, Cordier A, Verdes P, et al. A panel of urinary biomarkers to monitor reversibility of renal injury and a serum marker with improved potential to assess renal function. Nat Biotechnol. 2010;28(5):486–494. doi: 10.1038/nbt.1627. [DOI] [PubMed] [Google Scholar]
- 28.Vaidya VS, Ozer JS, Dieterle F, Collings FB, Ramirez V, Troth S, et al. Kidney injury molecule-1 outperforms traditional biomarkers of kidney injury in preclinical biomarker qualification studies. Nat Biotechnol. 2010;28(5):478–485. doi: 10.1038/nbt.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rached E, Hoffmann D, Blumbach K, Weber K, Dekant W, Mally A. Evaluation of putative biomarkers of nephrotoxicity after exposure to ochratoxin a in vivo and in vitro. Toxicol Sci. 2008;103(2):371–381. doi: 10.1093/toxsci/kfn040. [DOI] [PubMed] [Google Scholar]
- 30.Vaidya VS, Ford GM, et al. A rapid urine test for early detection of kidney injury. Kidney Int. 2009;76(1):108–114. doi: 10.1038/ki.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexopoulos LG, Saez-Rodriguez J, Espelin CW. High-throughput protein-based technologies and computational models for drug development, efficacy, and toxicity. In: Ekins S, Xu JJ, editors. Drug efficacy, safety, and biologics discovery: emerging technologies and tools. Hoboken, NJ, USA: Wiley; 2008. [Google Scholar]
- 32.Jantos-Siwy J, Schiffer E, Brand K, Schumann G, Rossing K, Delles C, et al. Quantitative urinary proteome analysis for biomarker evaluation in chronic kidney disease. J Proteome Res. 2009;8(1):268–281. doi: 10.1021/pr800401m. [DOI] [PubMed] [Google Scholar]
- 33.Mischak H, Delles C, Klein J, Schanstra JP. Urinary proteomics based on capillary electrophoresis-coupled mass spectrometry in kidney disease: discovery and validation of biomarkers, and clinical application. Adv Chronic Kidney Dis. 2010;17(6):493–506. doi: 10.1053/j.ackd.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Zangvil E. Compugen, Ltd. Pharmacogenomics. 2007;8(10):1461–1463. doi: 10.2217/14622416.8.10.1461. [DOI] [PubMed] [Google Scholar]
- 35.Thukral SK, Nordone PJ, Hu R, Sullivan L, Galambos E, Fitzpatrick VD, et al. Prediction of nephrotoxicant action and identification of candidate toxicity-related biomarkers. Toxicol Pathol. 2005;33(3):343–355. doi: 10.1080/01926230590927230. [DOI] [PubMed] [Google Scholar]
- 36.Ebbels TM, Keun HC, Beckonert OP, Bollard ME, Lindon JC, Holmes E, et al. Prediction and classification of drug toxicity using probabilistic modeling of temporal metabolic data: the consortium on metabonomic toxicology screening approach. J Proteome Res. 2007;6(11):4407–4422. doi: 10.1021/pr0703021. [DOI] [PubMed] [Google Scholar]
- 37.Boudonck KJ, Mitchell MW, Nemet L, Keresztes L, Nyska A, Shinar D, et al. Discovery of metabolomics biomarkers for early detection of nephrotoxicity. Toxicol Pathol. 2009;37(3):280–292. doi: 10.1177/0192623309332992. [DOI] [PubMed] [Google Scholar]
- 38.Saal S, Harvey SJ. MicroRNAs and the kidney: coming of age. Curr Opin Nephrol Hypertens. 2009;18(4):317–323. doi: 10.1097/MNH.0b013e32832c9da2. [DOI] [PubMed] [Google Scholar]
- 39.Melkonyan HS, Feaver WJ, Meyer E, Scheinker V, Shekhtman EM, Xin Z, et al. Transrenal nucleic acids: from proof of principle to clinical tests. Ann N Y Acad Sci. 2008;1137:73–81. doi: 10.1196/annals.1448.015. [DOI] [PubMed] [Google Scholar]
- 40.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6(3):267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 41.Bhatt K, Zhou L, Mi QS, Huang S, She JX, Dong Z. MicroRNA-34a is induced via p53 during cisplatin nephrotoxicity and contributes to cell survival. Mol Med. 2010;16(9–10):409–416. doi: 10.2119/molmed.2010.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han WK, Bailly V, Abichandani R, Thadhani R, Bonventre JV. Kidney Injury Molecule-1 (KIM-1): a novel biomarker for human renal proximal tubule injury. Kidney Int. 2002;62(1):237–244. doi: 10.1046/j.1523-1755.2002.00433.x. [DOI] [PubMed] [Google Scholar]
- 43.Rosen S, Heyman S. Concerns about KIM-1 as a urinary biomarker for acute tubular necrosis (ATN) Kidney Int. 2003;63(5):1955. doi: 10.1046/j.1523-1755.2003.00944.x. [DOI] [PubMed] [Google Scholar]
- 44.Dyrskjot L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R, et al. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69(11):4851–4860. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- 45.Waters MD, Fostel JM. Toxicogenomics and systems toxicology: aims and prospects. Nat Rev Genet. 2004;5(12):936–948. doi: 10.1038/nrg1493. [DOI] [PubMed] [Google Scholar]
- 46.Colle A, Tavera C, Laurent P, Leung-Tack J, Girolami JP. Direct radioimmunoassay of rat cystatin C: increased urinary excretion of this cysteine proteases inhibitor during chromate nephropathy. J Immunoassay. 1990;11(2):199–214. doi: 10.1080/01971529008053269. [DOI] [PubMed] [Google Scholar]
- 47.Conti M, Moutereau S, Zater M, Lallali K, Durrbach A, Manivet P, et al. Urinary cystatin C as a specific marker of tubular dysfunction. Clin Chem Lab Med. 2006;44(3):288–291. doi: 10.1515/CCLM.2006.050. [DOI] [PubMed] [Google Scholar]
- 48.Dieterle F, Perentes E, Cordier A, Roth DR, Verdes P, Grenet O, et al. Urinary clusterin, cystatin C, beta2-microglobulin and total protein as markers to detect drug-induced kidney injury. Nat Biotechnol. 2010;28(5):463–469. doi: 10.1038/nbt.1622. [DOI] [PubMed] [Google Scholar]
- 49.Schaefer L, Gilge U, Heidland A, Schaefer RM. Urinary excretion of cathepsin B and cystatins as parameters of tubular damage. Kidney Int Suppl. 1994;47:S64–S67. [PubMed] [Google Scholar]
- 50.Vaidya VS, Waikar SS, Ferguson MA, Collings FB, Sunderland K, Gioules C, et al. Urinary biomarkers for sensitive and specific detection of acute kidney injury in humans. Clin Transl Sci. 2008;1(3):200–208. doi: 10.1111/j.1752-8062.2008.00053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ichimura T, Asseldonk EJ, Humphreys BD, Gunaratnam L, Duffield JS, Bonventre JV. Kidney injury molecule-1 is a phosphatidylserine receptor that confers a phagocytic phenotype on epithelial cells. J Clin Invest. 2008;118(5):1657–1668. doi: 10.1172/JCI34487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liangos O, Perianayagam MC, Vaidya VS, Han WK, Wald R, Tighiouart H, et al. Urinary N-acetyl-beta-(d)-glucosaminidase activity and kidney injury molecule-1 level are associated with adverse outcomes in acute renal failure. J Am Soc Nephrol. 2007;18(3):904–912. doi: 10.1681/ASN.2006030221. [DOI] [PubMed] [Google Scholar]
- 53.Prozialeck WC, Vaidya VS, Liu J, Waalkes MP, Edwards JR, Lamar PC, et al. Kidney injury molecule-1 is an early biomarker of cadmium nephrotoxicity. Kidney Int. 2007;72(8):985–993. doi: 10.1038/sj.ki.5002467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sieber M, Hoffmann D, Adler M, Vaidya VS, Clement M, Bonventre JV, et al. Comparative analysis of novel noninvasive renal biomarkers and metabonomic changes in a rat model of gentamicin nephrotoxicity. Toxicol Sci. 2009;109(2):336–349. doi: 10.1093/toxsci/kfp070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Timmeren MM, Vaidya VS, van Ree RM, Oterdoom LH, de Vries AP, Gans RO, et al. High urinary excretion of kidney injury molecule-1 is an independent predictor of graft loss in renal transplant recipients. Transplantation. 2007;84(12):1625–1630. doi: 10.1097/01.tp.0000295982.78039.ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang EJ, Snyder RD, Fielden MR, Smith RJ, Gu YZ. Validation of putative genomic biomarkers of nephrotoxicity in rats. Toxicology. 2008;246(2–3):91–100. doi: 10.1016/j.tox.2007.12.031. [DOI] [PubMed] [Google Scholar]
- 57.Zhou Y, Vaidya VS, Brown RP, Zhang J, Rosenzweig BA, Thompson KL, et al. Comparison of kidney injury molecule-1 and other nephrotoxicity biomarkers in urine and kidney following acute exposure to gentamicin, mercury, and chromium. Toxicol Sci. 2008;101(1):159–170. doi: 10.1093/toxsci/kfm260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen N, Aleksa K, Woodland C, Rieder M, Koren G. N-Acetylcysteine prevents ifosfamide-induced nephrotoxicity in rats. Br J Pharmacol. 2008;153(7):1364–1372. doi: 10.1038/bjp.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gatanaga H, Tachikawa N, Kikuchi Y, Teruya K, Genka I, Honda M, et al. Urinary beta2-microglobulin as a possible sensitive marker for renal injury caused by tenofovir disoproxil fumarate. AIDS Res Hum Retroviruses. 2006;22(8):744–748. doi: 10.1089/aid.2006.22.744. [DOI] [PubMed] [Google Scholar]
- 60.Hofstra JM, Deegens JK, Willems HL, Wetzels JF. Beta-2-microglobulin is superior to N-acetyl-beta-glucosaminidase in predicting prognosis in idiopathic membranous nephropathy. Nephrol Dial Transplant. 2008;23(8):2546–2551. doi: 10.1093/ndt/gfn007. [DOI] [PubMed] [Google Scholar]
- 61.Morel G, Ban M, Bonnet P, Zissu D, Brondeau MT. Effect of beta-naphthoflavone and phenobarbital on the nephrotoxicity of chlorotrifluoroethylene and 1,1-dichloro-2,2-difluoroethylene in the rat. J Appl Toxicol. 2005;25(2):153–165. doi: 10.1002/jat.1048. [DOI] [PubMed] [Google Scholar]
- 62.Schaub S, Wilkins JA, Antonovici M, Krokhin O, Weiler T, Rush D, et al. Proteomic-based identification of cleaved urinary beta2-microglobulin as a potential marker for acute tubular injury in renal allografts. Am J Transplant. 2005;5(4 Pt 1):729–738. doi: 10.1111/j.1600-6143.2005.00766.x. [DOI] [PubMed] [Google Scholar]
- 63.Zhu G, Xiang X, Chen X, Wang L, Hu H, Weng S. Renal dysfunction induced by long-term exposure to depleted uranium in rats. Arch Toxicol. 2009;83(1):37–46. doi: 10.1007/s00204-008-0326-6. [DOI] [PubMed] [Google Scholar]
- 64.Kern W, Braess J, Kaufmann CC, Wilde S, Schleyer E, Hiddemann W. Microalbuminuria during cisplatin therapy: relation with pharmacokinetics and implications for nephroprotection. Anticancer Res. 2000;20(5C):3679–3688. [PubMed] [Google Scholar]
- 65.Vallon V. The proximal tubule in the pathophysiology of the diabetic kidney. Am J Physiol Regul Integr Comp Physiol. 2011;300(5):R1009–R1022. doi: 10.1152/ajpregu.00809.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arici M, Chana R, Lewington A, Brown J, Brunskill NJ. Stimulation of proximal tubular cell apoptosis by albumin-bound fatty acids mediated by peroxisome proliferator activated receptor-gamma. J Am Soc Nephrol. 2003;14(1):17–27. doi: 10.1097/01.ASN.0000042167.66685.EA. [DOI] [PubMed] [Google Scholar]
- 67.Russo LM, Sandoval RM, McKee M, Osicka TM, Collins AB, Brown D, et al. The normal kidney filters nephrotic levels of albumin retrieved by proximal tubule cells: retrieval is disrupted in nephrotic states. Kidney Int. 2007;71(6):504–513. doi: 10.1038/sj.ki.5002041. [DOI] [PubMed] [Google Scholar]
- 68.Gautier JC, Riefke B, Walter J, Kurth P, Mylecraine L, Guilpin V, et al. Evaluation of novel biomarkers of nephrotoxicity in two strains of rat treated with Cisplatin. Toxicol Pathol. 2010;38(6):943–956. doi: 10.1177/0192623310379139. [DOI] [PubMed] [Google Scholar]
- 69.Yu Y, Jin H, Holder D, Ozer JS, Villarreal S, Shughrue P, et al. Urinary biomarkers trefoil factor 3 and albumin enable early detection of kidney tubular injury. Nat Biotechnol. 2010;28(5):470–477. doi: 10.1038/nbt.1624. [DOI] [PubMed] [Google Scholar]
- 70.Venkat KK. Proteinuria and microalbuminuria in adults: significance, evaluation, and treatment. South Med J. 2004;97(10):969–979. doi: 10.1097/01.SMJ.0000140833.51533.46. [DOI] [PubMed] [Google Scholar]
- 71.Peterson PA, Evrin PE, Berggard I. Differentiation of glomerular, tubular, and normal proteinuria: determinations of urinary excretion of beta-2-macroglobulin, albumin, and total protein. J Clin Invest. 1969;48(7):1189–1198. doi: 10.1172/JCI106083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shihabi ZK, Konen JC, O’Connor ML. Albuminuria vs urinary total protein for detecting chronic renal disorders. Clin Chem. 1991;37(5):621–624. [PubMed] [Google Scholar]
- 73.Madsen J, Nielsen O, Tornoe I, Thim L, Holmskov U. Tissue localization of human trefoil factors 1, 2, and 3. J Histochem Cytochem. 2007;55(5):505–513. doi: 10.1369/jhc.6A7100.2007. [DOI] [PubMed] [Google Scholar]
- 74.Taupin D, Podolsky DK. Trefoil factors: initiators of mucosal healing. Nat Rev Mol Cell Biol. 2003;4(9):721–732. doi: 10.1038/nrm1203. [DOI] [PubMed] [Google Scholar]
- 75.Debata PR, Panda H, Supakar PC. Altered expression of trefoil factor 3 and cathepsin l gene in rat kidney during aging. Biogerontology. 2007;8(1):25–30. doi: 10.1007/s10522-006-9032-z. [DOI] [PubMed] [Google Scholar]
- 76.Dvergsten J, Manivel JC, Correa-Rotter R, Rosenberg ME. Expression of clusterin in human renal diseases. Kidney Int. 1994;45(3):828–835. doi: 10.1038/ki.1994.109. [DOI] [PubMed] [Google Scholar]
- 77.Hidaka S, Kranzlin B, Gretz N, Witzgall R. Urinary clusterin levels in the rat correlate with the severity of tubular damage and may help to differentiate between glomerular and tubular injuries. Cell Tissue Res. 2002;310(3):289–296. doi: 10.1007/s00441-002-0629-5. [DOI] [PubMed] [Google Scholar]
- 78.Ishii A, Sakai Y, Nakamura A. Molecular pathological evaluation of clusterin in a rat model of unilateral ureteral obstruction as a possible biomarker of nephrotoxicity. Toxicol Pathol. 2007;35(3):376–382. doi: 10.1080/01926230701230320. [DOI] [PubMed] [Google Scholar]
- 79.Shannan B, Seifert M, Boothman DA, Tilgen W, Reichrath J. Clusterin and DNA repair: a new function in cancer for a key player in apoptosis and cell cycle control. J Mol Histol. 2006;37(5–7):183–188. doi: 10.1007/s10735-006-9052-7. [DOI] [PubMed] [Google Scholar]
- 80.Price SA, Davies D, Rowlinson R, Copley CG, Roche A, Falkenberg FW, et al. Characterization of renal papillary antigen 1 (RPA-1), a biomarker of renal papillary necrosis. Toxicol Pathol. 2010;38(3):346–358. doi: 10.1177/0192623310362246. [DOI] [PubMed] [Google Scholar]
- 81.Hildebrand H, Rinke M, Schluter G, Bomhard E, Falkenberg FW. Urinary antigens as markers of papillary toxicity. II: Application of monoclonal antibodies for the determination of papillary antigens in rat urine. Arch Toxicol. 1999;73(4–5):233–245. doi: 10.1007/s002040050612. [DOI] [PubMed] [Google Scholar]
- 82.Shaw M. Cell-specific biomarkers in renal medicine and research. Methods Mol Biol. 2010;641:271–302. doi: 10.1007/978-1-60761-711-2_16. [DOI] [PubMed] [Google Scholar]
- 83.Lu Q, Lu C, Zhou GP, Zhang W, Xiao H, Wang XR. MicroRNA-221 silencing predisposed human bladder cancer cells to undergo apoptosis induced by TRAIL. Urol Oncol. 2009;28(6):635–641. doi: 10.1016/j.urolonc.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 84.Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Uchida Y, Kawahara K et al. (2011) Functional role of LASP1 in cell viability and its regulation by microRNAs in bladder cancer. Urol Oncol (in press) [DOI] [PubMed]
- 85.Cao Y, Yu SL, Wang Y, Guo GY, Ding Q, An RH. MicroRNA-dependent regulation of PTEN after arsenic trioxide treatment in bladder cancer cell line T24. Tumour Biol. 2010;32(1):179–188. doi: 10.1007/s13277-010-0111-z. [DOI] [PubMed] [Google Scholar]
- 86.Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125(2):345–352. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 87.Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Korner H, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7(16):2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 88.Hanke M, Hoefig K, Merz H, Feller AC, Kausch I, Jocham D, et al. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol Oncol. 2009;28(6):655–661. doi: 10.1016/j.urolonc.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 89.Catto JW, Miah S, Owen HC, Bryant H, Myers K, Dudziec E, et al. Distinct microRNA alterations characterize high- and low-grade bladder cancer. Cancer Res. 2009;69(21):8472–8481. doi: 10.1158/0008-5472.CAN-09-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, et al. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69(6):2623–2629. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- 91.Huang Q, Dunn RT, 2nd, Jayadev S, DiSorbo O, Pack FD, Farr SB, et al. Assessment of cisplatin-induced nephrotoxicity by microarray technology. Toxicol Sci. 2001;63(2):196–207. doi: 10.1093/toxsci/63.2.196. [DOI] [PubMed] [Google Scholar]
- 92.Ostenfeld MS, Bramsen JB, Lamy P, Villadsen SB, Fristrup N, Sorensen KD, et al. miR-145 induces caspase-dependent and -independent cell death in urothelial cancer cell lines with targeting of an expression signature present in Ta bladder tumors. Oncogene. 2010;29(7):1073–1084. doi: 10.1038/onc.2009.395. [DOI] [PubMed] [Google Scholar]
- 93.Wiklund ED, Bramsen JB, Hulf T, Dyrskjot L, Ramanathan R, Hansen TB, et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int J Cancer. 2011;128(6):1327–1334. doi: 10.1002/ijc.25461. [DOI] [PubMed] [Google Scholar]
- 94.Kenney PA, Wszolek MF, Rieger-Christ KM, Neto BS, Gould JJ, Harty NJ, et al. Novel ZEB1 expression in bladder tumorigenesis. BJU Int. 2010;107(4):656–663. doi: 10.1111/j.1464-410X.2010.09489.x. [DOI] [PubMed] [Google Scholar]
- 95.Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15(16):5060–5072. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu W, Zabirnyk O, Wang H, Shiao YH, Nickerson ML, Khalil S, et al. miR-23b targets proline oxidase, a novel tumor suppressor protein in renal cancer. Oncogene. 2010;29(35):4914–4924. doi: 10.1038/onc.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kort EJ, Farber L, Tretiakova M, Petillo D, Furge KA, Yang XJ, et al. The E2F3-Oncomir-1 axis is activated in Wilms’ tumor. Cancer Res. 2008;68(11):4034–4038. doi: 10.1158/0008-5472.CAN-08-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 99.Godwin JG, Ge X, Stephan K, Jurisch A, Tullius SG, Iacomini J. Identification of a microRNA signature of renal ischemia reperfusion injury. Proc Natl Acad Sci U S A. 2010;107(32):14339–14344. doi: 10.1073/pnas.0912701107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Agrawal R, Tran U, Wessely O. The miR-30 miRNA family regulates Xenopus pronephros development and targets the transcription factor Xlim1/Lhx1. Development. 2009;136(23):3927–3936. doi: 10.1242/dev.037432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, et al. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A. 2007;104(9):3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kato M, Putta S, Wang M, Yuan H, Lanting L, Nair I, et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol. 2009;11(7):881–889. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang Q, Wang Y, Minto AW, Wang J, Shi Q, Li X, et al. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J. 2008;22(12):4126–4135. doi: 10.1096/fj.08-112326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sun H, Li QW, Lv XY, Ai JZ, Yang QT, Duan JJ, et al. MicroRNA-17 post-transcriptionally regulates polycystic kidney disease-2 gene and promotes cell proliferation. Mol Biol Rep. 2010;37(6):2951–2958. doi: 10.1007/s11033-009-9861-3. [DOI] [PubMed] [Google Scholar]
- 105.Lee SO, Masyuk T, Splinter P, Banales JM, Masyuk A, Stroope A, et al. MicroRNA15a modulates expression of the cell-cycle regulator Cdc25A and affects hepatic cystogenesis in a rat model of polycystic kidney disease. J Clin Invest. 2008;118(11):3714–3724. doi: 10.1172/JCI34922. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview of RIFLE and AKIN processes for assessing DIKI/AKI. Patients who receive renal replacement therapy (RRT) are classified as stage 3 in the AKIN schema (Cr serum creatinine, GFR glomerular filtration rate, AKI acute kidney injury; JPEG 25 kb)
The overlap of human, mouse, and rat miRNAs identified in renal tissue shows the conservation of miRNA over different species. The total number of miRNAs determined per species is shown under the name of species. Out of these, a number of 73 miRNAs were detectable in all three species. Adapted from (44) (JPEG 9 kb)
The five kidney damage stages discerned by the GFR (DOC 29 kb)
An overview of international projects focused on the identification of biomarkers for the detection of nephrotoxicity using novel technologies like toxicogenomic and proteomics (DOC 32 kb)