This paper provides the first epidemiological evidence that IGF axis genes may be involved in the pathogenesis of advanced AMD and that IGF1R may predispose obese people to higher risk.
Abstract
Purpose.
To investigate whether insulin-like growth factor (IGF) axis genes, together with a novel dietary risk factor, the dietary glycemic index (dGI), and body mass index (BMI) affect the risk for age-related macular degeneration (AMD).
Methods.
This case–control study involved 962 subjects originally recruited through the Age-Related Eye Disease Study (AREDS) Genetic Repository. After those with missing covariates or invalid calorie intake (n = 23), diabetes (n = 59), and non-Caucasian race (n = 16) were excluded, 864 participants were used, including 209 AREDS category 1 participants (control group), 354 category 2 or 3 participants (drusen group), and 301 category 4 participants (advanced AMD group). A total of 25 single-nucleotide polymorphisms (SNPs) selected from IGF-1 (n = 9), IGF-2 (n = 1), IGF binding protein 1 (IGFBP1; n = 3), IGFBP3 (n = 3), acid-labile subunit of IGFBP (IGFALS; n = 2), IGF1 receptor (IGF1R; n = 4), and IGF2R (n = 3) were genotyped. SNP-AMD associations were measured with genotype, allele χ2 tests and Armitage's trend test. Odds ratios (OR), 95% confidence intervals (CIs), and SNP-exposure interactions were evaluated by multivariate logistic regression.
Results.
One SNP (rs2872060) in IGF1R revealed a significant association with advanced AMD (P-allele = 0.0009, P-trend = 0.0008; the significance level was set at 0.05/25 = 0.002 for multiple comparisons). The risk allele (G) in the heterozygous and homozygous states (OR, 1.67 and 2.93; 95% CI, 1.03–2.71 and 1.60–5.36, respectively) suggests susceptibility and an additive effect on AMD risk. Further stratification analysis remained significant for both neovascularization (OR, 1.49 and 2.61; 95% CI, 0.90–2.48 and 1.39–4.90, respectively) and geographic atrophy (OR, 2.57 and 4.52; 95% CI, 0.99–6.71 and 1.49–13.74, respectively). The G allele interaction analysis with BMI was significant for neovascularization (P = 0.042) but not for geographic atrophy (P = 0.47). No significant interaction was found with dGI.
Conclusions.
These data suggest a role of IGF1R on the risk for advanced AMD in this group of subjects.
Age-related macular degeneration (AMD) is the major cause of irreversible vision loss in the Western world,1 affecting approximately 15% of the elderly. At present, there is no widely practicable treatment for AMD. It is believed that AMD is a multifactorial disease, and the risk of AMD is determined by multiple genetic and environmental (including nutritional) factors.2,3 In recent studies, we observed a link between glycemic index (GI) and increased risk for AMD in two American cohorts: the Nutrition and Vision Project (NVP) substudy of the Nurses' Health Study (NHS) and the Age-Related Eye Diseases Study (AREDS).4–6 Both studies indicate that consuming diets that cause higher blood glucose loads (i.e., diets with higher glycemic index [GI]) is associated with higher risk for AMD in otherwise healthy, nondiabetic individuals. The findings were also replicated in the Blue Mountain Eye Study (BMES) cohort, Australia.7
The GI is a physiological measure of the “glycemic quality” of carbohydrate-containing foods.8 Intake of high-GI foods results in rapid elevation of postprandial blood glucose levels relative to low-GI foods. The clinical and public health implication of GI is that it can help people to choose foods. The dietary glycemic index (dGI) for each subject was calculated as Σ (GIi × Wi)/W: the weighted average of the GI values for each food item i (GIi) with the amount of carbohydrate consumed from each food item i as the weight Wi/W.9 Thus, dGI can be thought of as a description of a dietary pattern that describes the glycemic quality of a diet. Our recent evaluations indicated that the dGI is a better correlate of AMD than is total carbohydrate intake (quantity).4–6 Our analyses also found that the association with dGI is stronger in individuals with bilateral AMD progression (more susceptible to AMD progression).6 Since bilateral AMD progression suggests a stronger genetic influence, we hypothesized that genetic variants play a epistatic or modifier role in the novel association between dGI and AMD risk.6 Specifically, we speculate that gene–carbohydrate interactions are etiologic factors for AMD.3 In this study, we investigated roles for insulin-like growth factor (IGF)–related (often referred to as the IGF axis) genes because carbohydrate nutrition has been shown to affect postprandial circulating levels of IGFs and their responses10–12 and several mechanistic studies have related these genes to choroidal neovascularization, a late-stage manifestation of AMD.13–15 To test our hypothesis, we used a candidate gene approach in a association study, to evaluate whether genetic polymorphisms in specific IGF-axis genes alter the risk of AMD.
Material and Methods
Study Subjects
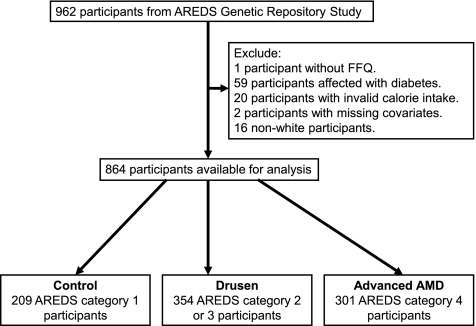
The subjects were participants from the AREDS Genetic Repository Study (n = 962; age range, 55–80 years; median, 69 years; 56% female) who had reliable dietary data and genomic DNA samples (n = 864; Fig. 1). To avoid potential bias from population stratification, we excluded nonwhite participants in our main analyses. After excluding those without dietary information, missing covariates or invalid calorie intake (n = 23), diabetes (n = 59), and non-Caucasian race (n = 16), the following remained in the sample: 209 AREDS category 1 participants (control group), 354 category 2 or 3 participants (drusen group), and 301 category 4 participants (advanced AMD group). The protocol complied with the Declaration of Helsinki.
Figure 1.
Exclusion criteria and eligible participants from the AREDS Genetic Repository Study.
Case and Control Definitions
The baseline AMD category was assessed according to AREDS AMD grading procedures.16,17 Persons in category 1 were free of AMD and had a total drusen area of less than five small drusen (<63 μm in diameter) and visual acuity (VA) of 20/32 or better in both eyes. Category 2 participants had mild age-related macular lesions (multiple small drusen, nonextensive (<20) intermediate drusen (63–124 μm in diameter), pigment abnormalities, or a combination thereof) in the most advanced eye and VA of 20/32 or better in both eyes. Category 3 required the absence of advanced AMD in both eyes and at least 1 eye with VA of 20/32 or better with at least one large druse (≥125 μm in diameter) and extensive (as measured by drusen area) intermediate drusen, or geographic atrophy that did not involve the center of the macula, or a combination thereof. In category 3a, both eyes met these criteria, whereas in category 3b, one eye either had reduced VA not resulting from AMD or a disqualifying ocular condition. Category 4 participants had VA of 20/32 or better and no advanced AMD (geographic atrophy involving the center of the macula or features of choroidal neovascularization) in the study eye, and the fellow eye had either lesions of advanced AMD (category 4a) or VA less than 20/32 and AMD abnormalities sufficient to explain reduced VA (category 4b), as determined by examination of photographs at the reading center. Persons aged 55 to 59 years were eligible for the study only if they met the criteria for categories 3 or 4.
Single-Nucleotide Polymorphism (SNP) Identifying and Genotyping
Genotyping data were derived from genomic DNA for the SNPs of interest. In addition to functional or previously studied polymorphisms,18–24 tagging SNPs were selected for each gene that included coverage of the promoter and 3′ flanking regions (Table 1). Tagging SNPs were identified from build 36 of the European-derived (CEU) HapMap database using a minor allele frequency of ≥20% and an r2 ≥ 0.80. A total of 25 SNPs selected from IGF-1 (n = 9), IGF-2 (n = 1), IGF binding protein 1 (IGFBP1; n = 3), IGFBP3 (n = 3), acid-labile subunit of IGFBP (IGFALS; n = 2), IGF-1 receptor (IGF1R; n = 4), and IGF2R (n = 3) were genotyped. Because of the large number of tagging SNPs necessary to evaluate IGF1R and IGF2R, only the polymorphisms previously studied were genotyped for these two genes.23,24 The National Center for Biotechnology Information (Bethesda, MD; PubMed web site (http://www.ncbi.nlm.nih.gov/pubmed) was used to search for candidate SNPs with keywords: “igf1r AND polymorphism” and “igf2r AND polymorphism.” SNPs with significant associations with diseases were preferred. Four SNPs were selected for IGF1R and three SNPs were selected for IGF2R. The search was performed at the end of 2007, when the study was initiated.
Table 1.
The 25 SNPs Selected from 7 IGF Axis Genes
| Gene | Functional or Significant SNPs | Tagging SNPs |
|---|---|---|
| IGF-1 | rs2162679, rs7965399 | rs6214, rs5742678, rs7956547, rs12821878, rs5742632, rs7136446, rs9989002 |
| IGF-2 | Not done | rs3213221 |
| IGFBP1 | rs1995051, rs3793344 | rs4619 |
| IBGBP3 | Not done | rs2453839, rs2471551, rs3110697 |
| IGFALS | rs3751893 | rs17559 |
| IGF1R | rs1319868, rs1567811, rs8041224, rs2872060 | Not done |
| IGF2R | rs1805075, rs8191754, rs629849 | Not done |
All SNPs were genotyped by 5′ exonuclease allele discrimination (Taqman, on a 7900HT with SDS 2.0 software; Applied Biosystems, Foster City, CA). Positive controls were used to assure plate-to-plate consistency of genotypes, and all genotype calls were conducted using the double-masked genotype assignments and discrepancies addressed using raw data.
Defining Potential Covariates
Data on possible risk factors for AMD were obtained from a baseline general physical and ophthalmic examination, a detailed questionnaire on basic characteristics and demographic data, and a validated food-frequency questionnaire (FFQ).25
The 90-item modified block FFQ collected information about usual dietary intakes over the previous year and classified them into nine possible response categories, ranging from never or less than once per month to two or more times per day. The daily total nutrient intake of an individual was calculated by summing the product of the frequency, serving size, and carbohydrate content per serving from individual food items derived from the nutrition database of the Nutrition Coordinating Center at the University of Minnesota.
The GI values for foods in the FFQ were either derived from published values with white bread used as the reference food, or imputed from GI values of comparable foods.26 Indigestible fiber content was subtracted from the carbohydrate content. The dGI was adjusted for total energy intake by using the residuals method.27
In addition, the following were considered as potential covariates in our analyses: age, sex, education level (college graduate, and high school or less), body mass index (BMI, computed from weight [in kilograms] divided by height [in meters squared]), smoking status (ever or never), sunlight exposure (hours per day),28 and hypertension history.
Statistical Analysis
We tested the hypothesis that genetic polymorphisms of IGF-axis genes and their interactions with dGI and BMI affect the risk for AMD in nondiabetic individuals from the AREDS. For each genotyped SNP (Table 1), we first examined whether the genotype frequencies were in Hardy-Weinberg equilibrium (P-HWE[b], exact P value for HWE) and evaluated the relationships with AMD status using a genotype case–control test (P-genotype),29 allele case–control test (P-allele),30 and Armitage's trend test (P-trend)31 (calculations by SAS/Genetics; SAS, Cary NC). The trend test and allele case–control test are most useful when there is an additive allele effect on the disease susceptibility. When HWE holds in the combined sample of cases and controls, these statistics are approximately equal and have an asymptotic χ12 distribution. However, if the assumption of HWE in the combined sample is violated, then the variance for the allele case–control statistic is incorrect; only the trend test remains valid under this violation. If dominance effects of alleles were also suspected to contribute to disease susceptibility, the genotype case–control test was used. The standard 2 × 3 contingency table analysis was used to form the χ22 statistic for the genotype case–control test, which tests for both additive and dominance (nonadditive) allelic effects.
For those with significant associations, we further calculated the multivariate-adjusted odds ratios (ORs) and 95% confidence intervals (95% CIs) estimated from logistic models. Age, sex, education level, BMI, smoking status, sunlight exposure, hypertension history, and dGI were included as covariates in multivariate models. The genotype with the lowest OR served as the referent group. Allele-specific ORs and 95% CIs were calculated in a similar way. The interactions between markers and dGI and BMI were evaluated by incorporating an interaction term in the model.32,33 We estimated ORs and CIs by logistic regression analysis (PROC LOGISTIC and PROC CASECONTROL; SAS).
Population Stratification
Population stratification occurs when there are allele frequency differences between cases and controls due to systematic ancestry differences that can cause spurious associations. To assess whether population stratification confounds the associations between the 25 SNPs and advanced AMD, we used a method based on principal component analysis (PCA).34 The PCA was performed using the data for the 25 SNPs from advanced AMD cases and controls in our sample (n = 485, all white; see Study Subjects section).
Results
Compared with the control group (Table 2), cases in the drusen group were significantly older, less educated, more likely to be a female, and more likely to be smokers. In addition to those characteristics, cases in the advanced AMD group, had higher BMI and dGI. These two factors were further evaluated in the interaction analysis.
Table 2.
Baseline Characteristics by AREDS AMD Categories*
| Control (n = 209) | Drusen (n = 354) | Advanced AMD (n = 301) | |
|---|---|---|---|
| Age, y, mean ± SE | 66.3 ± 0.29 | 68.6 ± 0.27 | 69.4 ± 0.30 |
| P < 0.0001† | P < 0.0001† | ||
| Education | |||
| Some high school or less | 47 (22.49) | 120 (33.90) | 143 (47.51) |
| Some college or higher | 162 (77.51) | 234 (66.10) | 158 (52.49) |
| P = 0.004‡ | P < 0.0001‡ | ||
| Female | 104 (49.76) | 213 (60.17) | 178 (59.14) |
| P = 0.02‡ | P = 0.04‡ | ||
| Smoking status | |||
| Never | 104 (49.76) | 146 (41.24) | 119 (39.53) |
| Current or ever | 105 (50.24) | 208 (58.76) | 182 (60.47) |
| P = 0.05‡ | P = 0.02‡ | ||
| Hypertension history | 69 (33.01) | 128 (36.16) | 113 (37.54) |
| P = 0.45‡ | P = 0.29‡ | ||
| Body mass index, kg/m2, mean ± SE | 27.1 ± 0.25 | 27.4 ± 0.31 | 28.3 ± 0.30 |
| P = 0.42† | P = 0.01† | ||
| Sunlight exposure, h/d, mean ± SE | 1.0 ± 0.07 | 1.2 ± 0.06 | 1.1 ± 0.07 |
| P = 0.13† | P = 0.62† | ||
| Dietary glycemic index, mean ± SE | 77.9 ± 0.35 | 77.8 ± 0.25 | 79.0 ± 0.27 |
| P = 0.85† | P = 0.01† |
Data are n (%) unless otherwise indicated.
Controls, AREDS category 1; Drusen cases, AREDS category 2 and 3; advanced AMD cases: AREDS category 4.
P values are for the distributional differences compared with control groups and were derived by t-tests.
P values are for the distributional differences compared with control groups and were derived by χ2-tests.
In our scanning of 25 SNPs one SNP (rs2872060) in IGF1R revealed a significant association with advanced AMD (P- allele = 0.0009, P-trend = 0.0008; α-level was set at 0.05/25 = 0.002 for multiple comparisons; Table 3). This result suggests that SNP rs2872060 may have an additive allele effect on the risk for advanced AMD. The other 24 SNPs (Table 1) showed no significant associations with either drusen or advanced AMD.
Table 3.
Association of SNP-rs2872060 in IGF1R with Risk for Advanced AMD and Drusen*
| Controls | Cases | P-HWE | P-Genotype | P-Allele | P-Trend | |
|---|---|---|---|---|---|---|
| Drusen | 201 | 345 | 0.73 | 0.12 | 0.047 | 0.045 |
| Advanced AMD | 201 | 284 | 0.72 | 0.004 | 0.0009* | 0.0008* |
“Determined” rate = 96.1%; P-HWE = 0.73.
Statistically significant, with α set at 0.05/25 = 0.002.
Further logistic analysis estimates that the risk G allele is associated with a 1.5-fold increased risk for advanced AMD whereas GG genotype confers threefold increased risk (OR, 1.67 and 2.93; 95% CI, 1.03–2.71 and 1.60–5.36, respectively; P-trend = 0.0005; Table 4). Further stratification analysis remained significant for both neovascularization (OR, 1.49 and 2.61; 95% CI, 0.90–2.48 and 1.39–4.90, respectively; P-trend = 0.003) and geographic atrophy (OR, 2.57 and 4.52; 95% CI, 0.99–6.71 and 1.49–13.74, respectively; P-trend = 0.007; Table 5).
Table 4.
Logistic Analysis of Association between SNP-rs2872060 in IGF1R and Advanced AMD
| Controls | Cases | OR | 95% CI | P | |
|---|---|---|---|---|---|
| Allele | |||||
| T | 65 | 62 | 1 | — | — |
| G | 136 | 222 | 1.97 | 1.24–3.12 | 0.004 |
| Genotype | |||||
| TT | 65 | 62 | 1 | — | — |
| GT | 103 | 144 | 1.67 | 1.03–2.71 | 0.038 |
| GG | 33 | 78 | 2.93 | 1.60–5.36 | 0.0005 |
| P-trend–logistic | 0.0005 |
Table 5.
Logistic Analysis of Associations between SNP-rs2872060 in IGF1R and Two Types of Advanced AMD
| Allele | Controls | Cases | OR | 95% CI | P |
|---|---|---|---|---|---|
| Neovascular AMD | |||||
| T | 65 | 55 | 1 | — | — |
| G | 136 | 182 | 1.76 | 1.09–2.86 | 0.02 |
| Genotype | |||||
| TT | 65 | 55 | 1 | — | — |
| GT | 103 | 119 | 1.49 | 0.90–2.48 | 0.12 |
| GG | 33 | 63 | 2.61 | 1.39–4.90 | 0.003 |
| P-trend–logistic | 0.003 | ||||
| Geographic AMD | |||||
| T | 65 | 7 | 1 | — | — |
| G | 136 | 40 | 2.97 | 1.17–7.52 | 0.02 |
| Genotype | |||||
| TT | 65 | 7 | 1 | — | — |
| GT | 103 | 25 | 2.57 | 0.99–6.71 | 0.05 |
| GG | 33 | 15 | 4.52 | 1.49–13.74 | 0.008 |
| P-trend–logistic | 0.007 |
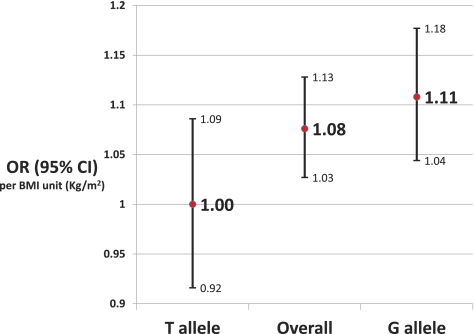
The G allele interaction analysis with BMI was significant for neovascularization (P = 0.049; Fig. 2) but not for geographic atrophy (P = 0.47). The OR per BMI unit (kilogram/meters squared) was higher in the subjects with the G allele than in the subjects with the T allele and in overall subjects as shown in Figure 2. To evaluate whether the interaction between G allele and BMI for neovascularization was modified by sex or smoking, we further stratified the analysis by sex and smoking. None of the P values was significant, probably because of the small sample size in each stratum. However, it appears that the interaction between the G allele and BMI was more dominant in the men (P = 0.07) than in the women (P = 0.24). The significance of the interaction did not differ between smokers (P = 0.10) and nonsmokers (P = 0.10).
Figure 2.
BMI-associated risk of neovascular AMD is highest in persons with the G allele in the rs2872060 polymorphism of IGF1R (P interaction = 0.042).
No significant interaction was found between the risk G allele and dGI.
In the analysis of population stratification, we plotted the first two principal components. The scatterplot shows no obvious cluster in our study sample, indicating no significant confounding effect from population stratification on our results.
Discussion
In this study on nondiabetic individuals from the AREDS, the SNP rs2872060 in IGF1R was significantly associated with the risk for advanced AMD and the association remained significant after stratification by the two types of the disease: neovascularization and geographic atrophy. The risk allele (G) showed an additive effect and a significant interaction with BMI on the risk for neovascularization, but not for geographic atrophy.
Genetic variants in IGF axis genes and differential expression of such genes have been related to diabetes or its complications,35,36 cardiovascular diseases,36 cancers,21,37–40 open angle glaucoma,41 and Alzheimer's disease,42,43 all of which share risk factors with AMD. To our knowledge, there has been no publication relating genetic polymorphisms of IGF axis genes to the risk for AMD. Because we had found that higher dGI is associated with increased risk for AMD,4–6,44–46 we hypothesized that the genetic polymorphisms of the IGF axis affects the risk for AMD and that this association may be modified by dGI. In this study, except for rs2872060 in IGF1R, we did not find any association between the 25 SNPs and early (drusen) or late AMD risk. We also did not find that the status of dGI modifies the association between rs2872060 and risk for advanced AMD. However, these results do not exclude the possibility that other genetic variants in the IGF axis may be responsible for the hypothesized associations.
The IGF axis is an important regulator of metabolic function and cellular development and growth that cells in almost every bodily organ use to communicate with the physiologic environment.10 This complex system consists of two cell-surface receptors (IGF1R and IGF2R), two growth factor ligands (IGF-1 and IGF-2), a family of six high-affinity IGF binding proteins (IGFBP 1–6), and associated IGFBP degrading enzymes, referred to collectively as IGFBP proteases.47,48
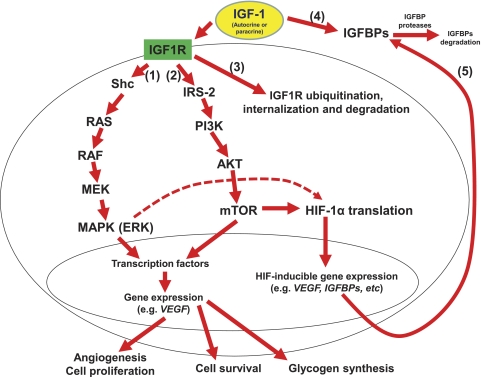
IGFs and IGFRs share high amino acid sequence homologies with insulin and insulin receptor (IR), respectively.49 Like insulin, the binding of IGF-1 to its receptors results in the activation of the intrinsic tyrosine kinase, and postreceptor phosphorylation of members of the insulin receptor substrate (IRS) family.50 The IR preferentially phosphorylates IRS-1, whereas IGF1R preferentially phosphorylates IRS-2.51 This factor may partly differentiate their activities: IGF-1 is a more potent mitogen with stronger antiapoptotic activity than insulin and plays a major role in regulating cell replication, differentiation, and survival,52 whereas insulin has stronger metabolic activity than does IGF-1.53 For example, ligand activation of IGF1R by IGF-1 was shown to result in activation of intracellular signaling pathways including Ras/mitogen-activated protein kinase (MAPK; a synonym for extracellular-signal-regulated kinase [ERK]) and phosphatidylinostiol-3 kinase (PI3K)/AKT, indicating an IGF-1-mediated cell proliferation (1 and 2 in Fig. 3),54,55 such as angiogenesis in diabetic retinopathy (DR) and AMD.56,57 Through different signal transduction mediators, ligand activation of IGF1R by IGF-1 may also lead to the ubiquitin-mediated degradation of IGF1R (3 in Fig. 3).58
Figure 3.
Signal transduction pathways demonstrate the different molecular consequences of ligand activation of IGF1R by IGF-1, including the activation of MAPK and PI3K/AKT cascades, which, in turn, induces expression of genes, such as VEGF, and ubiquitin-mediated IGF1R degradation. The activation of HIF-1 by mTOR in the PI3K/AKT cascade may result in the expression of IGFBPs and offer a feedback control on the IGF-1 activity. By binding IGF-1, IGFBPs can prevent proteolysis and extend the half-life of IGF-1, while reducing its bioavailability. Therefore, IGFBP proteases can regulate IGF half-life and play a critical role in modulating IGF availability at the cellular level.
Unlike insulin, which is mainly present in free form, nearly 99% of IGF-1 in the circulation is bound to one of the six IGFBPs (4 in Fig. 3).59 By binding IGF-1, IGFBPs can prevent the proteolysis and extend the half-life of IGF-1, while also reducing its bioavailability. Thus, IGFBP proteases may regulate the IGF-1 half-life and play a critical role in modulating IGF availability at the cellular level. IGFBPs also have biological activity independent of IGF-1.60 For example, IGFBP3 can directly bind to the ribonucleic acid polymerase II binding subunit 3, suggesting a possible role of IGFBP3 in directly regulating gene transcription and the cell cycle.61 However, the effects of IGFBP3 on the cell cycle are largely opposite those of IGF-1, since IGFBP3 is proapoptotic and antiproliferative.62
Although total circulating IGF-1 and IGFBP3 levels appear to show little or no intraindividual variation,63 there is extensive interindividual variation in levels of total IGF-1 and the IGFBPs.64 “Free” IGF-1 levels (about approximately 1% of IGF-1 in circulation) are also highly variable between individuals.65 Genetic factors may, at least partially, explain these interindividual variations. It is reasonable to assume that these interindividual differences could play a role in disease risk. Like some other hormones (e.g., estradiol and thyroid hormone), free IGF-1 has been proposed to be the main bioactive component of IGF-1 in circulation.66 However, unlike total IGF-1, free IGF-1 levels may vary significantly in the postprandial state, largely due to the regulation of IGFBP1 by insulin.67,68 Heritability studies have shown that genetic and environmental factors contribute equally to the variation in circulating IGF-1 levels.69–72
Nutrition is an important modifiable determinant of circulating IGF-1 levels.10,11 In a metabolic study, it was suggested that a high-GI diet is implicated in the risk of age-related diseases through modulating the IGF axis.12 The data implied that high-GI diets regulate IGF-1 levels through inhibiting IGFBP-3 but not directly through stimulating IGF-1 itself.
We recently proposed a novel hyperglycemic pathway: the hypoxia-inducible factor (HIF) pathway. In this model, in addition to hypoxia, hyperglycemia (e.g., the high postprandial hyperglycemia induced by high-GI diets) may also affect risk for DR, AMD, and other diseases through the activation of HIF-inducible genes, such as VEGF and VEGF receptor.46 Studies have also shown that IGF-1 can lead to HIF-1 activation and VEGF expression by a mechanism different from hypoxia.73–75 Interestingly, several IGF axis genes which are associated with the regulation of IGF-1, such as IGFBP1, 2, and 3, are also among the HIF-inducible genes.76 This appears to be part of the feedback control of HIF on the IGF axis (5 in Fig. 3).
Several lines of evidence from basic research suggest mechanistic explanations of the association between IGF axis genes and AMD risk, especially for neovascular AMD. IGF-1 protein and IGF1R appear to be colocalized in the retina. This suggests an autocrine function of IGF-1 in the normal human retina and may point toward a role for the IGF axis in the pathogenesis of neovascular AMD.15 It has also been shown that the IGF axis can contribute to angiogenesis directly by increasing proliferation of retinal endothelial cells,77,78 or indirectly through inducing vascular endothelial growth factor (VEGF) gene expression of cultured RPE cells.79 Furthermore, inhibitors of IGF1R, such as somatostatin and picropodophyllin,14,80 reduce IGF-1-dependent VEGF expression in human RPE cells and in vivo mouse model, respectively. VEGF is a potent inducer of postnatal neovascularization and angiogenesis and anti-VEGF (e.g., such as Lucentis and Avastin; Genentech, South San Francisco, CA; and Macugen, Pfizer, New York, NY) has been used in clinics to treat exudative AMD.81,82
Inflammation is associated with AMD. The IGF axis may also affect AMD risk through modulating inflammatory responses. Data suggest that intraocular IGF-I, but not systemic IGF-I, is sufficient to trigger processes leading to blood–retinal barrier breakdown and increased retinal vascular permeability.83–85 SNPs in IGF1R have also been implicated in differential immune response, such as lymphocyte recruitment and proliferation, resulting in difference in disease risk. For example, the TT genotype of rs2872060 was found to be associated with increased risk for bacteremia in sickle cell disease.23
Since SNP rs2872060 is located in the intron region of IGF1R, it is unlikely that this SNP directly affects the activity or stability of IGF1R protein. Instead, this SNP may affect the efficiency of transcription or RNA splicing for IGF1R and hence influence the risk for AMD.
Some epidemiologic studies have related higher BMI to increased risk for cancers, and it has been suggested that obesity-related tumorigenesis may be modulated by IGF1R.86 In Japanese breast cancer patients, it has been suggested that the molecular consequence of the increased BMI include increased expression of IGF1R, which, through modulation of the cell cycle and apoptosis, results in development and progression of postmenopausal breast cancer.87 In a case–control study of esophageal cancer, it was concluded that the common IGF1R gene polymorphism G1013A modulates the risk of obesity for esophageal cancer by influencing gene transcription or mRNA stability.86,88 Although BMI has long been recognized as a risk factor for AMD and,89 as discussed previously, the IGF axis has been implicated in the pathogenesis of AMD, our finding of an interaction between BMI and genetic polymorphism of IGF1R needs further confirmation and a mechanistic link underlying the interrelationship between obesity, the IGF axis, and AMD pathogenesis warrants further study.
There are several limitations in this study. First, to reduce cost, we used a candidate gene approach and subjectively selected 25 SNPs from 7 IGF axis genes for study. The coverage and information of our SNP list highly depended on the data from the HapMap project. Furthermore, other types of genetic polymorphisms, such as copy number variants,90 may be responsible for the underlying association with AMD risk. Therefore, false-negative findings are possible. On the other hand, because of the small sample size for geographic atrophy (Table 5), false-positive findings are also possible. In addition, we used Bonferroni's correction for our hypothesis testing on the 25 SNPs. Furthermore, multiple tests were performed for each SNP. This may also increase the likelihood of false-positive findings. Although rs2872060 is implicated in AMD risk, no study has related it to major systemic diseases or mortality. However, polymorphisms in IGF axis genes are related to risk for some diseases and aging, and this may create bias. If the relationship with the other diseases is stronger and subjects with these diseases had excluded from this study, it may have create false associations with AMD or attenuated existing ones. This survival bias could be assessed by comparing disease risk or survival time between different IGF axis gene genotypes. Because the primary interest of the AREDS is eye diseases and data regarding mortality are not available, it is difficult to assess the potential survival bias. Furthermore, it was found that survival bias results in no more than a 20% effect size erosion in cohorts with a mean age of <75 years,91 similar to our study sample. Finally, we included only white subjects in this study of the association between the 25 SNPs and advanced AMD. While previous genome-wide association studies (GWAS) in subsets of white participants from the AREDS showed inconsistencies in population stratification,92,93 we did not find obvious population stratification in our study sample. Unlike in GWAS, which must consider the confounding effects of population stratification on the associations between diseases and SNPs from across the whole genome, in this study of the 25 candidate SNPs we are not interested in the overall population stratification. Instead, we are interested in systematic ancestry differences in allele frequencies in the 25 SNPs between cases and controls. Although PCA can be applied to assess population stratification in GWAS and candidate gene association studies as well, its power depends on both the number of SNPS and the sample size.94 Based on the data from our advanced AMD case and controls (n = 485), we may have had inadequate power for detecting the underlying population stratification.
In summary, the findings in this study support an association between the IGF axis and risk for AMD in the AREDS cohort. More extensive screens of IGF axis genes, preferably in other cohorts, are needed to confirm the association. Elucidating the biochemical mechanisms by which the IGF axis affect AMD risk is also warranted. These insights will be helpful in designing intervention strategies and furthering our understanding of the underlying pathogenesis and would inform us about the designs of new therapeutics for AMD.
Footnotes
Supported by the U.S. Department of Agriculture under agreement, 1950-5100-060-01A (C-JC, AT); Grants R01-13250, R01 21212, and R03-EY014183-01A2 from the National Institutes of Health (NIH) (AT); grants from the Johnson and Johnson Focused Giving Program and American Health Assistance Foundation (AT), and the Ross Aging Initiative and NIH Grant R01 EY021826-01 (C-JC). The funding sources had no role in the design and conduct of the study; the collection, analysis, and interpretation of the data; or the preparation, review, and approval of the manuscript. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views or policies of the U.S. Department of Agriculture, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Disclosure: C.-J. Chiu, None; Y.P. Conley, None; M.B. Gorin, None; G. Gensler, None; C.-Q. Lai, None; F. Shang, None; A. Taylor, None
No reprints will be available.
References
- 1. Tomany SC, Wang JJ, van Leeuwen R, et al. Risk factors for incident age-related macular degeneration: pooled findings from 3 continents. Ophthalmology. 2004;111:1280–1287 [DOI] [PubMed] [Google Scholar]
- 2. Chiu CJ, Taylor A. Nutritional antioxidants and age-related cataract and maculopathy. Exp Eye Res. 2007;84:229–245 [DOI] [PubMed] [Google Scholar]
- 3. Haddad S, Chen CA, Santangelo SL, Seddon JM. The genetics of age-related macular degeneration: a review of progress to date. Surv Ophthalmol. 2006;51:316–363 [DOI] [PubMed] [Google Scholar]
- 4. Chiu CJ, Hubbard LD, Armstrong J, et al. Dietary glycemic index and carbohydrate in relation to early age-related macular degeneration. Am J Clin Nutr. 2006;83:880–886 [DOI] [PubMed] [Google Scholar]
- 5. Chiu CJ, Milton RC, Gensler G, Taylor A. Association between dietary glycemic index and age-related macular degeneration in the Age-Related Eye Disease Study. Am J Clin Nutr. 2007;86:180–188 [DOI] [PubMed] [Google Scholar]
- 6. Chiu CJ, Milton RC, Klein R, Gensler G, Taylor A. Dietary carbohydrate and progression of age-related macular degeneration, a prospective study from the Age-Related Eye Disease Study. Am J Clin Nutr. 2007;86:1210–1218 [DOI] [PubMed] [Google Scholar]
- 7. Kaushik S, Wang JJ, Flood V, et al. Dietary glycemic index and the risk of age-related macular degeneration. Am J Clin Nutr. 2008;88:1104–1110 [DOI] [PubMed] [Google Scholar]
- 8. Jenkins DJ, Wolever TM, Taylor RH. Glycemic index of foods: a physiological basis for carbohydrate exchange. Am J Clin Nutr. 1981;34:362–366 [DOI] [PubMed] [Google Scholar]
- 9. Wolever TM, Nguyen PM, Chiasson JL, et al. Determinants of diet glycemic index calculated retrospectively from diet records of 342 individuals with non-insulin-dependent diabetes mellitus. Am J Clin Nutr. 1994;59:1265–1269 [DOI] [PubMed] [Google Scholar]
- 10. Thissen JP, Ketelslegers JM, Underwood LE. Nutritional regulation of the insulin-like growth factors. Endocr Rev. 1994;15:80–101 [DOI] [PubMed] [Google Scholar]
- 11. Kaaks R, Lukanova A. Energy balance and cancer: the role of insulin and insulin-like growth factor-I. Proc Nutr Soc. 2001;60:91–106 [DOI] [PubMed] [Google Scholar]
- 12. Brand-Miller JC, Liu V, Petocz P, Baxter RC. The glycemic index of foods influences postprandial insulin-like growth factor-binding protein responses in lean young subjects. Am J Clin Nutr. 2005;82:350–354 [DOI] [PubMed] [Google Scholar]
- 13. Rosenthal R, Wohlleben H, Malek G, et al. Insulin-like growth factor-1 contributes to neovascularization in age-related macular degeneration. Biochem Biophys Res Commun. 2004;323:1203–1208 [DOI] [PubMed] [Google Scholar]
- 14. Sall JW, Klisovic DD, O'Dorisio MS, Katz SE. Somatostatin inhibits IGF-1 mediated induction of VEGF in human retinal pigment epithelial cells. Exp Eye Res. 2004;79:465–476 [DOI] [PubMed] [Google Scholar]
- 15. Lambooij AC, van Wely KH, Lindenbergh-Kortleve DJ, Kuijpers RW, Kliffen M, Mooy CM. Insulin-like growth factor-I and its receptor in neovascular age-related macular degeneration. Invest Ophthalmol Vis Sci. 2003;44:2192–2198 [DOI] [PubMed] [Google Scholar]
- 16. Age-Related Eye Disease Study Research Group The Age-Related Eye Disease Study system for classifying age-related macular degeneration from stereoscopic color fundus photographs: the Age-Related Eye Disease Study Report Number 6. Am J Ophthalmol. 2001;132:668–681 [DOI] [PubMed] [Google Scholar]
- 17. Age-Related Eye Disease Study Research Group The age-related eye disease study (AREDS) system for classifying cataracts from photographs: AREDS report no. 4. Am J Ophthalmol. 2001;131:167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aoi N, Nakayama T, Soma M, et al. Association of the insulin-like growth factor1 gene with myocardial infarction in Japanese subjects. Hereditas. 2010;147:215–224 [DOI] [PubMed] [Google Scholar]
- 19. Patel AV, Cheng I, Canzian F, et al. IGF-1, IGFBP-1, and IGFBP-3 polymorphisms predict circulating IGF levels but not breast cancer risk: findings from the Breast and Prostate Cancer Cohort Consortium (BPC3). PLoS One. 2008;3:e2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Taverne CW, Verheus M, McKay JD, et al. Common genetic variation of insulin-like growth factor-binding protein 1 (IGFBP-1), IGFBP-3, and acid labile subunit in relation to serum IGF-I levels and mammographic density. Breast Cancer Res Treat. 2010;123:843–855 [DOI] [PubMed] [Google Scholar]
- 21. Canzian F, McKay JD, Cleveland RJ, et al. Polymorphisms of genes coding for insulin-like growth factor 1 and its major binding proteins, circulating levels of IGF-I and IGFBP-3 and breast cancer risk: results from the EPIC study. Br J Cancer. 2006;94:299–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rosendahl AH, Hietala M, Henningson M, Olsson H, Jernström H. IGFBP1 and IGFBP3 polymorphisms predict circulating IGFBP-3 levels among women from high-risk breast cancer families. Breast Cancer Res Treat. 2011;127:785–794 [DOI] [PubMed] [Google Scholar]
- 23. Adewoye AH, Nolan VG, Ma Q, et al. Association of polymorphisms of IGF1R and genes in the transforming growth factor-beta/bone morphogenetic protein pathway with bacteremia in sickle cell anemia. Clin Infect Dis. 2006;43:593–598 [DOI] [PubMed] [Google Scholar]
- 24. Rezgui D, Williams C, Savage SA, et al. Structure and function of the human Gly1619Arg polymorphism of M6P/IGF2R domain 11 implicated in IGF2 dependent growth. J Mol Endocrinol. 2009;42:341–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kurinij N, Gensler G, Milton R, Age-Related Eye Disease Study (AREDS) Research Group Development and valuation of a food frequency questionnaire in a randomized trial of eye diseases. International Conference on Dietary Assessment Measures; May 7, 1998, Phoenix, AZ [Google Scholar]
- 26. Foster-Powell K, Holt SH, Brand-Miller JC. International table of glycemic index and glycemic load values: 2002. Am J Clin Nutr. 2002;76:5–56 [DOI] [PubMed] [Google Scholar]
- 27. Willett W, Stampfer MJ. Total energy intake: implications for epidemiologic analyses. Am J Epidemiol. 1986;124:17–27 [DOI] [PubMed] [Google Scholar]
- 28. McCarty CA, Lee SE, Livingston PM, Bissinella M, Taylor HR. Ocular exposure to UV-B in sunlight: the Melbourne visual impairment project model. Bull World Health Organ. 1996;74:353–360 [PMC free article] [PubMed] [Google Scholar]
- 29. Nielsen DM, Weir BS. A classical setting for associations between markers and loci affecting quantitative traits. Genet Res. 1999;74:271–277 [DOI] [PubMed] [Google Scholar]
- 30. Sasieni PD. From genotypes to genes: doubling the sample size. Biometrics. 1997;53:1253–1261 [PubMed] [Google Scholar]
- 31. Armitage P. Tests for linear trends in proportions and frequencies. Biometrics. 1955;11:375–386 [Google Scholar]
- 32. Chiu CJ, Chang ML, Chiang CP, Hahn LJ, Hsieh LL, Chen CJ. Interaction of collagen-related genes and susceptibility to betel quid-induced oral submucous fibrosis. Cancer Epidemiol Biomarkers Prev. 2002;11:646–653 [PubMed] [Google Scholar]
- 33. Chiu CJ, Chiang CP, Chang ML, et al. Association between genetic polymorphism of tumor necrosis factor-alpha and risk of oral submucous fibrosis, a pre-cancerous condition of oral cancer. J Dent Res. 2001;80:2055–2059 [DOI] [PubMed] [Google Scholar]
- 34. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909 [DOI] [PubMed] [Google Scholar]
- 35. Ukkola O, Sun G, Bouchard C. Insulin-like growth factor 2 (IGF2) and IGF-binding protein 1 (IGFBP1) gene variants are associated with overfeeding-induced metabolic changes. Diabetologia. 2001;44:2231–2236 [DOI] [PubMed] [Google Scholar]
- 36. Vaessen N, Heutink P, Janssen JA, et al. A polymorphism in the gene for IGF-I: functional properties and risk for type 2 diabetes and myocardial infarction. Diabetes. 2001;50:637–642 [DOI] [PubMed] [Google Scholar]
- 37. Miller BS, Yee D. Type I insulin-like growth factor receptor as a therapeutic target in cancer. Cancer Res. 2004;65:10123–10127 [DOI] [PubMed] [Google Scholar]
- 38. Cheng I, Stram DO, Penney KL, et al. Common genetic variation in IGF1 and prostate cancer risk in the Multiethnic Cohort. J Natl Cancer Inst. 2006;98:123–134 [DOI] [PubMed] [Google Scholar]
- 39. Le Marchand L, Kolonel LN, Henderson BE, Wilkens LR. Association of an exon 1 polymorphism in the IGFBP3 gene with circulating IGFBP-3 levels and colorectal cancer risk: the multiethnic cohort study. Cancer Epidemiol Biomarkers Prev. 2005;14:1319–1321 [DOI] [PubMed] [Google Scholar]
- 40. Zavras AI, Pitiphat W, Wu T, et al. Insulin-like growth factor II receptor gene-167 genotype increases the risk of oral squamous cell carcinoma in humans. Cancer Res. 2003;63:296–297 [PubMed] [Google Scholar]
- 41. Tsai FJ, Lin HJ, Chen WC, Chen HY, Fan SS. Insulin-like growth factor-II gene polymorphism is associated with primary open angle glaucoma. J Clin Lab Anal. 2003;17:259–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Messier C, Teutenberg K. The role of insulin, insulin growth factor, and insulin-degrading enzyme in brain aging and Alzheimer's disease. Neural Plast. 2005;12:311–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. de la Monte SM, Wands JR. Review of insulin and insulin-like growth factor expression, signaling, and malfunction in the central nervous system: relevance to Alzheimer's disease. J Alzheimers Dis. 2005;7:45–61 [DOI] [PubMed] [Google Scholar]
- 44. Chiu CJ, Klein R, Milton RC, Gensler G, Taylor A. Does eating particular diets alter risk of age-related macular degeneration in users of the Age-Related Eye Disease Study supplements? Br J Ophthalmol. 2009;93:1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chiu CJ, Milton RC, Klein R, Gensler G, Taylor A. Dietary compound score and risk of age-related macular degeneration in the Age-Related Eye Disease Study. Ophthalmology. 2009;116:939–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chiu CJ, Taylor A. Dietary hyperglycemia, glycemic index and metabolic retinal diseases. Prog Retin Eye Res. 2011;30:18–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yu H, Rohan T. Role of the insulin-like growth factor family in cancer development and progression. J Natl Cancer Inst. 2000;92:1472–1489 [DOI] [PubMed] [Google Scholar]
- 48. Furstenberger G, Senn HJ. Insulin-like growth factors and cancer. Lancet Oncol. 2002;3:298–302 [DOI] [PubMed] [Google Scholar]
- 49. Rajpathak SN, Gunter MJ, Wylie-Rosett J, et al. The role of insulin-like growth factor-I and its binding proteins in glucose homeostasis and type 2 diabetes. Diabetes Metab Res Rev. 2009;25:3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Holzenberger M, Kappeler L, De Magalhaes Filho C. IGF-1 signaling and aging. Exp Gerontol. 2004;39:1761–1764 [DOI] [PubMed] [Google Scholar]
- 51. Kim JJ, Accili D. Signalling through IGF-I and insulin receptors: where is the specificity? Growth Horm IGF Res. 2002;12:84–90 [DOI] [PubMed] [Google Scholar]
- 52. Zapf J, Schmid C, Froesch ER. Biological and immunological properties of insulin-like growth factors (IGF) I and II. Clin Endocrinol Metab. 1984;13:3–30 [DOI] [PubMed] [Google Scholar]
- 53. Guler HP, Zapf J, Froesch ER. Short-term metabolic effects of recombinant human insulin-like growth factor I in healthy adults. N Engl J Med. 1987;317:137–140 [DOI] [PubMed] [Google Scholar]
- 54. Larsson O, Girnita A, Girnita L. Role of insulin-like growth factor 1 receptor signalling in cancer. Br J Cancer. 2005;92:2097–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Samani AA, Yakar S, LeRoith D, Brodt P. The role of the IGF system in cancer growth and metastasis: overview and recent insights. Endocr Rev. 2007;28:20–47 [DOI] [PubMed] [Google Scholar]
- 56. Weng CY, Kothary PC, Verkade AJ, Reed DM, Del Monte MA. MAP kinase pathway is involved in IGF-1-stimulated proliferation of human retinal pigment epithelial cells (hRPE). Curr Eye Res. 2009;34:867–876 [DOI] [PubMed] [Google Scholar]
- 57. Díaz-Rodríguez E, Piñeiro A, Casanueva FF, Camiña JP. The bovine vitreous-derived lipid factor (bVLF) is a powerful inhibitor of retinal pigmented epithelial (hRPE) cell proliferation. FEBS Lett. 2005;579:4020–4030 [DOI] [PubMed] [Google Scholar]
- 58. Hupfeld CJ, Olefsky JM. Regulation of receptor tyrosine kinase signaling by GRKs and beta-arrestins. Annu Rev Physiol. 2007;69:561–577 [DOI] [PubMed] [Google Scholar]
- 59. Juul A. Serum levels of insulin-like growth factor I and its binding proteins in health and disease. Growth Horm IGF Res. 2003;13:113–170 [DOI] [PubMed] [Google Scholar]
- 60. Lee KW, Cohen P. Nuclear effects: unexpected intracellular actions of insulin-like growth factor binding protein-3. J Endocrinol. 2002;175:33–40 [DOI] [PubMed] [Google Scholar]
- 61. Oufattole M, Lin SW, Liu B, Mascarenhas D, Cohen P, Rodgers BD. Ribonucleic acid polymerase II binding subunit 3 (Rpb3), a potential nuclear target of insulin-like growth factor binding protein-3. Endocrinology. 2006;147:2138–2146 [DOI] [PubMed] [Google Scholar]
- 62. Firth SM, Baxter RC. Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev. 2002;23:824–854 [DOI] [PubMed] [Google Scholar]
- 63. Goodman-Gruen D, Barrett-Connor E. Epidemiology of insulin-like growth factor-I in elderly men and women: The Rancho Bernardo Study. Am J Epidemiol. 1997;145:970–976 [DOI] [PubMed] [Google Scholar]
- 64. Janssen JA, Stolk RP, Pols HA, Grobbee DE, de Jong FH, Lamberts SW. Serum free IGF-I, total IGF-I, IGFBP-1 and IGFBP-3 levels in an elderly population: relation to age and sex steroid levels. Clin Endocrinol (Oxf). 1998;48:471–478 [DOI] [PubMed] [Google Scholar]
- 65. Frystyk J. Free insulin-like growth factors: measurements and relationships to growth hormone secretion and glucose homeostasis. Growth Horm IGF Res. 2004;14:337–375 [DOI] [PubMed] [Google Scholar]
- 66. Chen JW, Hojlund K, Beck-Nielsen H, Sandahl Christiansen J, Orskov H, Frystyk J. Free rather than total circulating insulin-like growth factor-I determines the feedback on growth hormone release in normal subjects. J Clin Endocrinol Metab. 2005;90:366–371 [DOI] [PubMed] [Google Scholar]
- 67. Katz LE, DeLeon DD, Zhao H, Jawad AF. Free and total insulin-like growth factor (IGF)-I levels decline during fasting: relationships with insulin and IGF-binding protein-1. J Clin Endocrinol Metab. 2002;87:2978–2983 [DOI] [PubMed] [Google Scholar]
- 68. Nyomba BL, Berard L, Murphy LJ. Free insulin-like growth factor I (IGF-I) in healthy subjects: relationship with IGF-binding proteins and insulin sensitivity. J Clin Endocrinol Metab. 1997;82:2177–2181 [DOI] [PubMed] [Google Scholar]
- 69. Harrela M, Koistinen H, Kaprio J, et al. Genetic and environmental components of interindividual variation in circulating levels of IGF-I, IGF-II, IGFBP-1, and IGFBP-3. J Clin Invest. 1996;98:2612–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Verhaeghe J, Loos R, Vlietinck R, Herck EV, van Bree R, Schutter AM. C-peptide, insulin-like growth factors I and II, and insulin-like growth factor binding protein-1 in cord serum of twins: genetic versus environmental regulation. Am J Obstet Gynecol. 1996;175:1180–1188 [DOI] [PubMed] [Google Scholar]
- 71. Hong Y, Brismar K, Hall K, Pedersen NL, de Faire U. Associations between insulin-like growth factor-I (IGF-I), IGF-binding protein-1, insulin and other metabolic measures after controlling for genetic influences: results from middle-aged and elderly monozygotic twins. J Endocrinol. 1997;153:251–257 [DOI] [PubMed] [Google Scholar]
- 72. Hall K, Hilding A, Thoren M. Determinants of circulating insulin-like growth factor-I. J Endocrinol Invest. 1999;22(5 suppl):48–57 [PubMed] [Google Scholar]
- 73. Treins C, Murdaca J, Van Obberghen E, Giorgetti-Peraldi S. AMPK activation inhibits the expression of HIF-1alpha induced by insulin and IGF-1. Biochem Biophys Res Commun. 2006;342:1197–1202 [DOI] [PubMed] [Google Scholar]
- 74. Treins C, Giorgetti-Peraldi S, Murdaca J, Monthouël-Kartmann MN, Van Obberghen E. Regulation of hypoxia-inducible factor (HIF)-1 activity and expression of HIF hydroxylases in response to insulin-like growth factor I. Mol Endocrinol. 2005;19:1304–1317 [DOI] [PubMed] [Google Scholar]
- 75. Beppu K, Nakamura K, Linehan WM, Rapisarda A, Thiele CJ. Topotecan blocks hypoxia-inducible factor-1alpha and vascular endothelial growth factor expression induced by insulin-like growth factor-I in neuroblastoma cells. Cancer Res. 2005;65:4775–4781 [DOI] [PubMed] [Google Scholar]
- 76. Hong SS, Lee H, Kim KW. HIF-1alpha: a valid therapeutic target for tumor therapy. Cancer Res Treat. 2004;36:343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Grant MB, Caballero S, Millard WJ. Inhibition of IGF-I and b-FGF stimulated growth of human retinal endothelial cells by the somatostatin analogue, octreotide: a potential treatment for ocular neovascularization. Regul Pept. 1993;48:267–278 [DOI] [PubMed] [Google Scholar]
- 78. Spraul CW, Baldysiak-Figiel A, Lang GK, Lang GE. Octreotide inhibits growth factor-induced bovine choriocapillary endothelial cells in vitro. Graefes Arch Clin Exp Ophthalmol. 2002;240:227–231 [DOI] [PubMed] [Google Scholar]
- 79. Ulbig MW, Wolfensberger TJ, Hiscott P, Ationu A, Carter ND, Gregor ZJ. Insulin-like growth factor I (IGF-I) receptor/binding protein in human diabetic epiretinal membranes. Ger J Ophthalmol. 1995;4:264–268 [PubMed] [Google Scholar]
- 80. Economou MA, Wu J, Vasilcanu D, et al. Inhibition of VEGF secretion and experimental choroidal neovascularization by picropodophyllin (PPP), an inhibitor of the insulin-like growth factor-1 receptor. Invest Ophthalmol Vis Sci. 2008;49:2620–2626 [DOI] [PubMed] [Google Scholar]
- 81. Bressler NM. Antiangiogenic approaches to age-related macular degeneration today. Ophthalmology. 2009;116(10 suppl):S15–S23 [DOI] [PubMed] [Google Scholar]
- 82. Bressler SB. Introduction: Understanding the role of angiogenesis and antiangiogenic agents in age-related macular degeneration. Ophthalmology. 2009;116(10 Suppl):S1–S7 [DOI] [PubMed] [Google Scholar]
- 83. Ruberte J, Ayuso E, Navarro M, et al. Increased ocular levels of IGF-1 in transgenic mice lead to diabetes-like eye disease. J Clin Invest. 2004;113:1149–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Poulaki V, Joussen AM, Mitsiades N, Mitsiades CS, Iliaki EF, Adamis AP. Insulin-like growth factor-I plays a pathogenetic role in diabetic retinopathy. Am J Pathol. 2004;165:457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Haurigot V, Villacampa P, Ribera A, et al. Increased intraocular insulin-like growth factor-I triggers blood-retinal barrier breakdown. J Biol Chem. 2009;284:22961–22969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhao R, Macdonald K, Casson AG. Insulin-like growth factor type I receptor gene expression and obesity in esophageal adenocarcinoma. Mol Carcinog. 2009;48:982–988 [DOI] [PubMed] [Google Scholar]
- 87. Suga K, Imai K, Eguchi H, Hayashi S, Higashi Y, Nakachi K. Molecular significance of excess body weight in postmenopausal breast cancer patients, in relation to expression of insulin-like growth factor I receptor and insulin-like growth factor II genes. Jpn J Cancer Res. 2001;92:127–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. MacDonald K, Porter GA, Guernsey DL, Zhao R, Casson AG. A polymorphic variant of the insulin-like growth factor type I receptor gene modifies risk of obesity for esophageal adenocarcinoma. Cancer Epidemiol. 2009;33:37–40 [DOI] [PubMed] [Google Scholar]
- 89. Age-Related Eye Disease Study Research Group Risk factors for the incidence of Advanced Age-Related Macular Degeneration in the Age-Related Eye Disease Study (AREDS) AREDS report no. 19. Ophthalmology. 2005;112:533–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Cooper GM, Nickerson DA, Eichler EE. Mutational and selective effects on copy-number variants in the human genome. Nat Genet. 2007;39(7 suppl):S22–S29 [DOI] [PubMed] [Google Scholar]
- 91. Anderson CD, Nalls MA, Biffi A, et al. The effect of survival bias on case–control genetic association studies of highly lethal diseases. Circ Cardiovasc Genet. 2011;4:188–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ryu E, Fridley BL, Tosakulwong N, Bailey KR, Edwards AO. Genome-wide association analyses of genetic, phenotypic, and environmental risks in the age-related eye disease study. Mol Vis. 2010;16:2811–2821 [PMC free article] [PubMed] [Google Scholar]
- 94. Ma J, Amos CI. Theoretical formulation of principal components analysis to detect and correct for population stratification. PLoS One. 2010;5:pii: e12510 [DOI] [PMC free article] [PubMed] [Google Scholar]