CaBP5 interacts with Munc18–1 and myosin VI and stimulates neurite outgrowth and neurotransmitter release in PC12 cells.
Abstract
Purpose.
CaBP5 is a neuronal calmodulin-like Ca2+-binding protein that is expressed in the retina and in the cochlea. Although CaBP5 knockout mice displayed reduced sensitivity of retinal ganglion cell light responses, the function of CaBP5 in vivo is still unknown. To gain further insight into CaBP5 function, the authors screened for CaBP5-interacting partners.
Methods.
Potential retinal interacting partners for CaBP5 were identified using affinity chromatography followed by mass spectrometry and by yeast two-hybrid screening of a bovine retina cDNA library. Interacting partners were further analyzed using coimmunoprecipitation. Immunohistochemistry and subcellular fractionation were performed to determine their colocalization in the retina. The effect of CaBP5 on dopamine release and neurite outgrowth of PC12 cells was analyzed using ELISA and fluorescent labeling.
Results.
Using affinity chromatography, the authors identified Munc18–1 and myosin VI as interacting partners for CaBP5. Munc18–1 was also identified using the yeast two-hybrid system. Colocalization and coimmunoprecipitation of CaBP5 with these two proteins in retinal tissue further established their physiological interactions. Furthermore, CaBP5 expression in NGF-stimulated PC12 cells stimulates neurite outgrowth and dopamine exocytosis.
Conclusions.
This study shows that CaBP5 interacts with Munc18–1 and myosin VI, two proteins involved in the synaptic vesicle cycle. Together with the effect of CaBP5 in stimulating neurite outgrowth and vesicle exocytosis in PC12 cells, these results suggest that CaBP5 plays a role in neurotransmitter release.
Asubfamily of neuronal Ca2+-binding proteins is highly similar to calmodulin (CaBP1–8), and CaBP5 is a member of that subfamily.1–3 CaBP4 function has been well characterized. It is localized at the photoreceptor synaptic terminals and is essential for photoreceptor synaptic function through enhanced activation of Cav1.4 L-type voltage-gated Ca2+ channels and transmitter release.4,5 Mutations in the Cabp4 gene have been shown in patients with autosomal recessive incomplete congenital stationary night blindness and cone-rod synaptic disorder.6–8 In contrast, the specific function of CaBP5 in vivo has not yet been clearly established. In mice, CaBP5 is expressed in rod bipolar cells, in type 5 ON-cone bipolar cells, and in type 3 OFF-cone bipolar cells.3,9,10 In both human and monkey retina, CaBP5 is also expressed in rod bipolar cells and both ON and OFF cone bipolar cells.11 Like CaBP1, CaBP2, and CaBP4, CaBP5 has been observed in cochlear inner hair cells.12 We previously generated and characterized CaBP5 knockout mice to investigate the function of CaBP5 in the retina. No evidence of morphologic changes and no significant differences in the amplitude of the ERG responses were observed in CaBP5 knockout mice compared with wild-type mice. However, the sensitivity of retinal ganglion cell light responses was reduced by ∼50% in Cabp5−/− mice, suggestive of a role for CaBP5 in the normal transmission of light signals throughout the retinal circuitry. In transfected HEK293T cells, CaBP5 has a small effect in suppressing calcium-dependent inactivation of Cav1.2 and Cav1.3.12,13
To gain further insight into the function of CaBP5 in the retina, we investigated CaBP5 interaction with other retinal proteins using affinity chromatography and yeast two-hybrid screening of a retina cDNA library. We identified Munc18–1 and myosin VI, which are involved in synaptic vesicle cycling and trafficking. A physiological interaction between these proteins is corroborated by our findings that CaBP5 also stimulates dopamine vesicle exocytosis and neurite outgrowth of PC12 cells.
Materials and Methods
Antibodies
Commercially available antibodies were alkaline phosphatase-conjugated anti–mouse and anti–rabbit (Promega Corp., Madison, WI), rabbit anti–myosin VI (Proteus Biosciences, Inc, Ramona, CA), mouse anti–Munc18–1 (BD Biosciences, San Jose, CA; for immunohistochemistry and Western blot analysis experiments), rabbit anti–syntaxin-3 (Synaptic Systems, Göttingen, Germany), and Alexa Fluor 555 goat anti–mouse and Alexa Fluor 488 goat anti–rabbit (Invitrogen, Carlsbad, CA).
CaBP5 Affinity Chromatography
The full-length bovine 6His-tagged CaBP53 was coupled to CNBr-activated gel filtration media (Sepharose; GE Healthcare, Piscataway, NJ) according to the manufacturer's protocol. Bovine retinas obtained from In Vision BioResources (Seattle, WA) were homogenized in 50 mM Hepes, pH 7.4, 100 mM NaCl, 5 mM MgCl2,0.5 mM dithiothreitol (DTT), 10 mM dodecyl-β-maltoside, and a cocktail of inhibitor of proteases (Sigma, St. Louis, MO) with or without 1 mM CaCl2 using a glass-glass homogenizer. The homogenate was centrifuged at 40,000g for 20 minutes at 4°C. The supernatant was loaded on the CaBP5-gel filtration media (Sepharose; GE Healthcare), and the column was washed with the homogenization buffer containing 150 mM NaCl. Bound proteins were eluted with 0.1 M glycine buffer, pH 2.5.
Identification of CaBP5-Binding Proteins Using Mass Spectrometry
The identification of interacting partners for CaBP5 was carried out by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Briefly, proteins eluted from the affinity columns were separated on SDS-PAGE. After in-gel trypsinization of the excised bands, the proteolytic digests were sent to the mass spectrometry center of the School of Pharmacy at the University of Washington, where they were subjected to separation on a capillary LC coupled to the nano-spray (ESI) source of a tandem hybrid mass spectrometer (LTQ-FT, Q-TOF) (http://sop.washington.edu/medchem/mass-spectrometry-center/mass-spectrometry-center.html). The probable identity of the proteins was determined by the matching of sequence data obtained from the collisionally induced dissociation of selected peptide ions in the digest's chromatogram against those of candidate proteins found in the International Protein Index database. Proteins were selected using their calculated molecular weight and for a number of hits of at least five.
Yeast Two-Hybrid Screening of a Bovine Retinal Library
A bovine retina cDNA library14 (a kind gift of Wolfgang Baehr, Moran Eye Institute, University of Utah) was screened using the full-length bovine CaBP5 following a standard transformation protocol (Yeast Protocols Handbook; Clontech, Palo Alto, CA). Clones containing CaBP5-interacting proteins were isolated in the AH109 strain on synthetic dropout (SD) medium without tryptophan (Trp), leucine (Leu), histidine (His, reporter gene), adenine (Ade, reporter gene)–containing X-gal (reporter gene), and 10 mM 3-amino-1,2,4-triazole (3-AT; Sigma), which was also added to inhibit leaky expression of His3 proteins. Plasmids were recovered from yeast colonies by glass bead homogenization and phenol-chloroform extraction, transferred to Escherichia coli for amplification, and sequenced.
CaBP5 Interaction with Munc18-1 in Yeasts
Mouse Munc18–1 was cloned from mouse retina cDNA after PCR amplification with primers F816 (5′-CACCATGGCCCCCATTGGCCTCAAG-3′) and FH817 (5′-TCACTCCATTGTTGGAGCCTGATCC-3′). Domain I of Munc18–1,15 encoding amino acids 1 to 136, was subcloned by PCR amplification with F816 and F863 (5′-TCAAAACGCAATGTTGATTTCCGTC-3′). Domain II+III encoding amino acid 131-Stop of Munc18–1 was amplified with primer F867 (5′-CACCAACATTGCGTTTCTCCCCTATG-3′). The cDNA encoding the mouse CaBP53 was subcloned in fusion to the DNA-binding domain into the pGBKT7- BD vector (carrying the gene for tryptophan; Clontech). Full-length and fragments of Munc18–1 were subcloned in fusion to the Gal4-activation domain into the pGADT7 vector (carrying the gene for leucine; Clontech). AH109 yeasts were cotransformed with both plasmids and cotransformants selected on SD medium without tryptophan and leucine. To test for reporter gene expression, colonies were further streaked on selective SD medium without Trp, Leu, His + 10mM 3-AT or without Trp, Leu, His, Ade and with X-gal, and 10 mM 3-AT.
Preparation of Anti–Munc18-1 Polyclonal Antibodies
Full-length Munc18-1 was cloned in fusion to a 6His-tag in the pET30B vector, expressed, and purified from bacteria. Rabbit anti–Munc18-1 polyclonal antibodies (UW248) were raised in a New Zealand White rabbit by subcutaneous immunization with purified recombinant mouse Munc18-1 protein mixed with an equal volume of Freund's adjuvant (Cocalico Biologicals, Inc., Reamstown, PA). The rabbit anti–Munc18-1 antibody was used in coimmunoprecipitation assays.
Immunohistochemistry
Mouse retinas were obtained from C57Bl/6J mice treated according to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. All the procedures for the maintenance and use of animals were approved by the Institutional Animal Care and Use Committee of the University of Washington. Mouse eyecups were fixed in 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.4 (PB), for 1 hour. After fixation, tissues were incubated with increasing concentrations of sucrose to 20% sucrose in PB and then embedded in 33% OCT compound (Miles, Elkhart, NY) diluted with 20% sucrose in PB. Eye tissues were cut in 12-μm sections. To block nonspecific labeling, retinal sections were incubated with 3% normal goat serum in PBST buffer (10 mM sodium phosphate, 150 mM NaCl, 0.1% Triton X-100, pH 7.4) for 20 minutes at room temperature. Sections were incubated overnight at 4°C in a mix of diluted primary antibodies (mouse anti–CaBP5 1:400 + rabbit anti–myosin VI, 1:50, or rabbit anti–CaBP5 1:400 with mouse anti–Munc18-1, 1:500 [BD Biosciences]). A mix of Alexa Fluor 555-conjugated goat anti–mouse IgG and Alexa 488-conjugated goat anti–rabbit IgG or Alexa Fluor 488-conjugated goat anti–mouse IgG with Alexa Fluor 555-conjugated goat anti–rabbit IgG, respectively, was reacted with the sections for 1 hour at room temperature. Hoechst 33342 (Invitrogen) was added with the secondary antibodies when indicated. Then the sections were rinsed in PBST and mounted with antifade reagent (Prolong; Invitrogen). Sections were analyzed under a confocal microscope (LSM710; Carl Zeiss, Inc., Thornwood, NY). Immunofluorescent images were obtained with a 63×/1.4 oil objective (Plan Apochromat; Carl Zeiss, Inc.). Scans were performed sequentially by excitation at 488 nm and imaging with a 488/561 main dichroic beam splitter (MBS) and a 494- to 551-nm emission filter for Alexa 488, followed by excitation at 561 nm and imaged using a 458/561 MBS and a 565- to 697-nm emission filter for Alexa 555. Single confocal images were acquired at an optical thickness of 0.8 μm.
Fractionation of Bovine Retina
Bovine retinas were homogenized in extraction buffer (10 mM Hepes, pH 7.2, 100 mM NaCl, 1 mM CaCl2, 2 mM MgCl2) using a glass-glass homogenizer. The homogenate was divided into two equal fractions. EGTA was added to one of the fractions at a final concentration of 5 mM. Both fractions were further processed the same way but in the presence of CaCl2 or EGTA. Each homogenate was centrifuged at 18,000g for 50 minutes and was separated into supernatant S1 and pellet P1. The supernatant (S1) was centrifuged again at 100,000g for 40 minutes to isolate the soluble (S2) and insoluble (P2) fractions. P1 pellets were washed twice in extraction buffer containing 0.5% Triton X-100 and centrifuged for 50 minutes at 18,000g. P3 pellets were extracted with 10 mM Hepes, pH 7.2, containing 1 M NaCl, 1 mM DTT, and 1 mM ATP. Samples were centrifuged at 120,000g for 40 minutes to separate the S4 supernatant and the P4 pellet. Fractions were then analyzed by Western blotting for the presence of CaBP5, myosin VI, Munc18-1, and syntaxin-3.
Coimmunoprecipitation Assays
Five bovine retinas were homogenized in 12 mL of 20 mM Hepes, pH 7.2, 150 mM NaCl, 1 mM CaCl2, 2 mM MgCl2, 0.005% Triton X-100, and protease inhibitor. The homogenate was divided into two equal aliquots H1 and H2. EGTA was added to H2 at a final concentration of 5 mM. Lysates were centrifuged at 10,000g for 10 minutes at 4°C, and the supernatants were precleared by incubation with protein G magnetic beads. Beads were then discarded, and affinity-purified rabbit anti–CaBP5 (UW89) or anti–Munc18-1 (UW248) antibodies were added. After 1-hour incubation at 4°C with antibodies, protein G magnetic beads were added and incubation was carried out overnight. Protein G beads were then washed 10 times in homogenization buffer with or without EGTA. Proteins were eluted with 50 μL of 0.2 M glycine pH 2.8 and analyzed by Western blot.
Establishment of PC12 Cell Stably Expressing CaBP5-mCherry or Control mCherry
Rat pheochromocytoma-derived PC12 cells were obtained from ATCC (Manassas, VA). The stop codon of the mouse CaBP5 was first removed by PCR and cloned with a fragment encoding a (Gly)7-Leu-(Gly)7 linker fused to mCherry into the pcDNA3.1 vector. PC12 cells were transfected using Lipofectamine (Invitrogen) with pcDNA3.1-mCherry or pcDNA3.1-CaBP5-mCherry and were cultured for 4 weeks in RPMI medium containing 10% horse serum, 5% FBS, streptomycin/penicillin, and 400 μg/mL G418. G418-resistant colonies expressing mCherry were identified using an epifluorescence microscope and expanded in the same medium containing 200 μg/mL G418.
Analysis of Neurite Outgrowth
PC12 cells stably expressing CaBP5-mCherry or control mCherry alone were plated on poly-l-lysine–coated glass-bottom Petri dishes (Mattek, Ashland, MA), additionally coated with 10 μg/mL mouse collagen IV (BD Biosciences). Cells were cultured in RPMI medium containing 50 ng/mL of mouse nerve growth factor (NGF) from submaxillary glands (2.5s) (BD Biosciences). Randomly selected areas that contained dispersed cells with distinguishable neurites were analyzed by confocal microscopy before and after 2, 4, and 6 days of neurite outgrowth stimulation by NGF. Because both mCherry and CaBP5-mCherry are soluble and are present in the neurites, immunofluorescent images of live cells were obtained with a 40×/1.3NA (Plan-Neofluar; Carl Zeiss, Inc.) objective lens. Neurite length was determined by manually tracing the neurites and was measured using Zeiss (Zen 2009) software on images taken at day 4 after stimulation with NGF. Processes were measured from the soma to the tip of each branch. The number of roots is the number of roots starting from the soma. The number of extremities represents the number of branches.
Analysis of FM1-43 Uptake
PC12 cells stably expressing CaBP5-mCherry or control mCherry were cultured in collagen-coated, six-well plates for 3 days in RPMI medium containing 50 ng/mL NGF. To test basal endocytosis, the vesicles were labeled by exposure of the cells to 5 μM dye (FM1-43; Invitrogen) for 10 minutes in buffer A (5 mM Hepes, pH7.4, 5 mM glucose, 1 mM MgCl2, 3 mM CaCl2, 3 mM KCl, 147 mM NaCl). To test regulated endocytosis, cells were stimulated for 2 minutes with high-potassium buffer B (5 mM Hepes, pH 7.4, 5 mM glucose, 1 mM MgCl2, 3 mM CaCl2, 50 mM KCl, 100 mM NaCl) and labeled for another 10 minutes by incubation in buffer A containing 5 μM dye (FM1-43; Invitrogen). The dye in the exposed membrane was then washed away by multiple washes with buffer A for 10 minutes, followed by a 5-minute wash with 1 mM ADVASEP-716 (Sigma) in buffer A and washed again for another 10-minute wash in buffer A. Cells, typically 200,000 cells/well, were collected and analyzed in the next hour after staining. Fluorescence emission was recorded using a fluorometer (FluoroLog; Jobin Yvon, Horiba Scientific, Irvine, CA). Excitation wavelengths for FM1-43 and mCherry were 488 nm and 561 nm, whereas emissions were recorded at 560 nm and 610 nm, respectively. At 560 nm, there is no crosstalk from mCherry excitation using 488-nm excitation wavelength. The FM1-43 fluorescence was normalized to the emission of CaBP5-mCherry or mCherry. mCherry fluorescence at 610 nm had been estimated to be ∼3.3-fold that of CaBP5-mCherry by comparing the emission fluorescence at 610 nm to the cell number, using cell counting and Hoechst staining. The 3.3-fold factor was taken into consideration in normalizing the FM1-43 signal to that of mCherry. For confocal images, cells were processed as described but were cultured in glass-bottom Petri dishes, and KCl-stimulated FM1-43 uptake was analyzed under a confocal microscope.
Analysis of Dopamine Release by PC12 Stable Cell Lines
Cells were seeded in collagen-coated, 24-well plates and cultured for 3 days in RPMI containing 50 ng/mL NGF. The cells were washed and equilibrated for 30 minutes in buffer A. The buffer was then replaced with fresh buffer A or high KCl buffer B. After 2-minute incubation at 37°C, the supernatant was collected and the amount of secreted dopamine was analyzed using an ELISA kit obtained from Rocky Mountain Diagnostics (Colorado Springs, CO) according to the manufacturer's instructions. Although the same number of cells was seeded in the 24-well plates, we evaluated the number of cells after the assay because we observed a lower density of CaBP5-mCherry–expressing cells than mCherry-expressing cells. mCherry-expressing cells were approximately twice as abundant as CaBP5-mCherry–expressing cells, and this was taken into account in the dopamine release quantification.
Results
CaBP5-Interacting Proteins Identified Using Yeast Two-Hybrid Screening of a Bovine Retinal Library
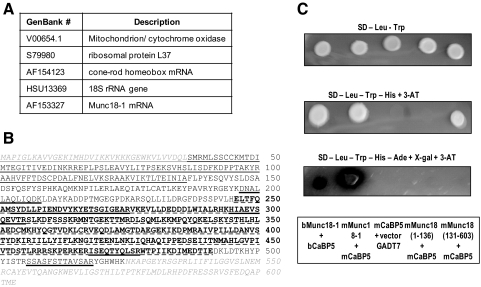
To identify CaBP5-interacting proteins, a bovine retina cDNA library was screened using the full-length bovine CaBP5. Clones containing CaBP5-interacting proteins were isolated in the AH109 strain on media depleted of leucine and tryptophan (selection of both plasmids) and of histidine and adenine and containing X-gal (selection of reporter genes requiring the proximity of the DNA-binding domain and the activation domain). Screening of 5 × 105 clones led to the identification of five clones (Fig. 1A). Although we screened a retina cDNA library using three reporter genes, we isolated cytochrome oxidase and ribosomal proteins that are commonly encountered as false positives in yeast two-hybrid screening and were thus not further investigated. Because of the location of CaBP5 in bipolar cells, the cone-rod homeobox mRNA was also eliminated. Munc18-1 is the only physiologically relevant interacting partner for CaBP5 identified in this two-hybrid screening. The Munc18-1 clone isolated from the bovine retina library and interacting with CaBP5 did not include the first 36 and the last 79 amino acids of the protein sequence (Fig. 1B).
Figure 1.
Yeast two-hybrid assay. (A) CaBP5-interacting proteins identified using a yeast two-hybrid screening of a bovine retinal library with the full-length bovine CaBP5. (B) Amino acid sequence of bovine Munc18–1. The clone isolated from the library did not include the first 36 and the last 79 amino acids of Munc18–1 (shown in italic gray letters). Domain I (1–135) of Munc18–1 is underlined, and domain III (246–490) is in bold letters and underlined with a dashed line. The other amino acids constitute domain II. The peptides identified by mass spectrometry analysis are underlined with a double line. (C) Analysis of CaBP5 interaction with full-length Munc18-1 and Munc18-1 domains I or domains II+III using the yeast two-hybrid assay. Replicates of yeast colonies initially selected on SD-Leu-Trp on SD-Leu-Trp, SD-Leu-Trp-His+3-AT (media selective for low-affinity protein interaction), or SD-Leu-Trp-His+3-AT-Ade+X-gal 9 (for selection of high-affinity protein interaction). Yeast colonies cotransformed with bovine CaBP5 (bCaBP5) and bovine Munc18-1 (amino acids 37-524, bMunc18-1), mouse CaBP5 (mCaBP5), and full-length mouse Munc18-1 (mMunc18-1), mouse CaBP5 and mouse Munc18-1 (amino acids 1-136, domain I), mouse CaBP5 and mouse Munc18-1 (amino acids 130-end, domains II + III), mouse CaBP5 and pGADT7 vector (negative control). Plates were photographed after 6-day incubation at 30°C.
Identification of CaBP5-Binding Proteins by Affinity Chromatography and Mass Spectrometry
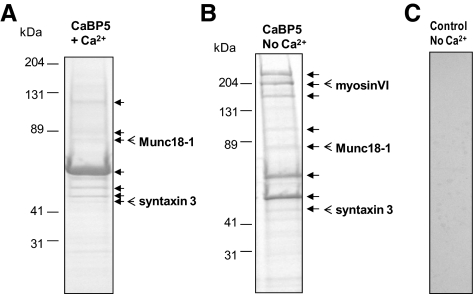
We next identified retinal proteins that interact with CaBP5 by affinity chromatography on immobilized CaBP5. Because CaBP5 is a Ca2+-binding protein, we probed CaBP5-interacting partners by affinity chromatography in the presence or absence of 1 mM CaCl2. The eluted proteins were separated by SDS-PAGE. The major bands, indicated by arrows in Figures 2A and 2B, were excised. After in-gel trypsin digestion, they were analyzed by LC-MS/MS. All proteins listed in Table 1 were identified after affinity chromatography without added Ca2+. Those identified in Ca2+ condition (Table 1, right column) did not include high molecular–weight proteins because they were not represented as major bands and were not analyzed by mass spectrometry. Although the array of major bands appeared different in the presence of Ca2+ (Fig. 2), the same proteins were identified by mass spectrometry in both conditions. Multiple proteins were identified in the analysis of each major band. Identified proteins included multiple cytoskeleton proteins and proteins associated with the cytoskeleton or involved in its assembly (Table 1). Chaperones, ion/proton transport ATPases, and proteins involved in metabolic pathways and in synaptic vesicle cycle were also isolated using this method. As a control for nonspecific binding of retinal proteins to the gel filtration media (Sepharose; GE Healthcare) resin, we incubated retina extract with the resin devoid of CaBP5. No major bands were detected after analysis of the proteins eluted from the control gel filtration media (Sepharose; GE Healthcare) (Fig. 2C).
Figure 2.
Identification of CaBP5-binding proteins by affinity chromatography. Affinity chromatography on CaBP5-gel filtration media (Sepharose) was performed using extracts of bovine retina prepared (A) with or (B) without 1 mM CaCl2. The eluted proteins were separated on SDS-PAGE. The major bands that were further processed for identification using LC-MS/MS are indicated with a black arrow. (C) A control experiment was carried out in parallel using a gel filtration media (Sepharose) column without CaBP5 and shows no major bands. The bands containing the proteins further analyzed in this study, i.e., myosin VI, munc18-1 and syntaxin-3, are indicated with an open arrow although multiple proteins were identified in all major bands.
Table 1.
CaBP5-Binding Proteins identified by Mass Spectrometry
| Description | Accession No. | MWt | Unique Peptides | Ca2+ |
|---|---|---|---|---|
| SPTAN1: spectrin, alpha, nonerythrocytic 1 | IPI:IPI00732001.3 | 285 | 45 | |
| SPTBN1: spectrin, beta, nonerythrocytic 1 isoform 1 | IPI:IPI00825390.3 | 274 | 29 | |
| SPTBN2: spectrin, beta, nonerythrocytic 2 | IPI:IPI00691115.3 | 262 | 2 | |
| MYO18A: similar to myosin 18A isoform 2 | IPI:IPI00712754.5 | 232 | 20 | |
| MYH10: myosin-10 | IPI:IPI00709219.1 | 229 | 84 | |
| MYH14: similar to myosin, heavy chain 14 isoform 4 | IPI:IPI00695094.3 | 229 | 80 | |
| MYH9: nonmuscle myosin heavy chain | IPI:IPI00696012.4 | 229 | 82 | |
| MYO5A: similar to myosin Va | IPI:IPI00691592.4 | 206 | 32 | |
| LOC514078: similar to myosin-Vb,LOC514078 134-kDa protein | IPI:IPI00709039.2 | 159 | 10 | |
| MYO6: similar to myosin-VI | IPI:IPI00707581.4 | 146 | 62 | |
| RBP3: retinol-binding protein 3 | IPI:IPI00687859.1 | 139 | 18 | |
| DCTN1: 142-kDa protein, DCTN1 DCTN1 protein | IPI:IPI00714222.4 | 127 | 4 | |
| NCAM1: Neural cell adhesion molecule 1 | IPI:IPI00710515.2 | 120 | 5 | |
| ATP1A3: ATPase, Na+/K+ transporting, alpha 3 polypeptide isoform 1 | IPI:IPI00732365.4 | 112 | 7 | |
| NEFH: similar to neurofilament heavy polypeptide (NF-H) | IPI:IPI00705914.4 | 112 | 9 | |
| AP2B1: AP-1 complex subunit beta-1 | IPI:IPI00696360.2 | 105 | 7 | + |
| NEF3; LOC787355: neurofilament triplet M protein | IPI:IPI00705032.3 | 103 | 24 | |
| HNRNPU: heterogeneous nuclear rihonucleoprotein U | IPI:IPI00688257.4 | 90 | 9 | + |
| GSN: gelsolin a | IPI:IPI00883474.1 | 85 | 37 | |
| PFKM: 6-phosphofructokinase, muscle type | IPI:IPI00685357.4 | 85 | 5 | + |
| HSP90AB1: heat shock protein HSP 90-beta | IPI:IPI00709435.3 | 83 | 4 | |
| SV2A: synaptic vesicle glycoprotein 2A | IPI:IPI00708267.1 | 82 | 3 | |
| SV2B: SV2B protein | IPI:IPI00845192.1 | 78 | 3 | |
| HSPA5: 78-kDa glucose-regulated protein | IPI:IPI00717234.3 | 78 | 5 | + |
| PPEF1: similar to Serine/threonine-protein phosphatase with EF-hands 1 | IPI:IPI00687922.2 | 75 | 4 | + |
| HSPA9: stress-70 protein, mitochondrial | IPI:IPI00692247.1 | 73 | 9 | + |
| HSPA8: heat shock cognate 71-kDa protein | IPI:IPI00708526.2 | 71 | 3 | + |
| HSPA12A: similar to heat shock 70-kDa protein 12A | IPI:IPI00699608.4 | 70 | 4 | + |
| STXBP1: syntaxin-binding protein 1, also known as MUNC18–1 | IPI:IPI00690962.2 | 68 | 5 | + |
| ATP6V1A: ATPase, H+ transporting, lysosomal V1 subunit A | IPI:IPI00826624.2 | 68 | 24 | + |
| NEFL: neurofilament light polypeptide | IPI:IPI00717196.2 | 62 | 45 | + |
| ATP5A1: ATP synthase subunit alpha, mitochondrial | IPI:IPI00694295.1 | 60 | 7 | + |
| PHGDH: D-3 phosphoglycerate dehydrogenase | IPI:IPI00699717.2 | 57 | 3 | + |
| ATP5B: ATP synthase subunit beta, mitochondrial | IPI:IPI00717884.1 | 56 | 5 | |
| ATP6V: 1H isoform alpha of V-type proton ATPase subunit H | IPI:IPI00700783.2 | 55 | 7 | + |
| ALDH9A1: 4-trimethylaminobutyraldehyde dehydrogenase | IPI:IPI00703131.5 | 54 | 2 | + |
| VIM: vimentin | IPI:IPI00689228.3 | 53 | 18 | + |
| TUBA3E: tubulin alpha-3 chain | IPI:IPI00795426.1 | 50 | 13 | + |
| TUBB2A: tubulin beta-2B chain | IPI:IPI00697107.2 | 50 | 3 | + |
| TUBA4A: tubulin alpha-4A chain | IPI:IPI00704353.2 | 50 | 10 | + |
| TUBB2C: tubulin beta-2C chain | IPI:IPI00706594.1 | 50 | 4 | + |
| GFAP: glial fibrillary acidic protein | IPI:IPI00716718.2 | 49 | 19 | + |
| CKB: creatine kinase B-type | IPI:IPI00716827.1 | 43 | 3 | + |
| ACTB: actin, cytoplasmic 1 | IPI:IPI00698900.1 | 41 | 15 | + |
| SAG: isoform 8 of S-arrestin | IPI:IPI00702970.2 | 41 | 8 | + |
| ALDOA: fructose-bisphosphate aldolase | IPI:IPI00867095.1 | 39 | 4 | + |
| ALDOC: fructose-bisphosphate aldolase | IPI:IPI00852561.1 | 39 | 6 | + |
| STX3: syntaxin 3 | IPI:IPI00867107.1 | 34 | 6 | + |
Proteins isolated by affinity chromatography and constituting the major bands are indicated by arrows in Figures 2A and 2B. Those bands were excised and analyzed by LC-MS/MS after in-gel trypsin digestion. The data output from the LC-MS/MS was searched against the International Protein Index database. The number of unique peptides identified is listed. Proteins are sorted in descending order according to their molecular weight (MWt). All listed proteins were identified after affinity chromatography without added Ca2+. Those identified in Ca2+ conditions are indicated in the right column and did not include high molecular–weight proteins because they did not constitute the major bands.
Because we could not validate all the proteins identified by affinity chromatography, we selected some of them for further analysis based on their properties. Among identified cytoskeleton proteins, myosin VI was chosen because it has been shown to colocalize with CaBP5 in inner ear cilia.12,17 Munc18-1 was our strongest candidate because it was identified as a CaBP5-interacting partner using the two complementary methods. Munc18-1 is also expressed in the cochlea and is a neuronal protein interacting with syntaxin-1.15,18 Although syntaxin-1 was not identified as a binding partner for CaBP5 using affinity chromatography, we identified syntaxin-3 instead. Syntaxin-3 has been reported as a Munc18-1–interacting protein and is expressed in retinal bipolar cell synapses.19–23 Therefore, syntaxin-3 was further analyzed for its interaction with CaBP5 or Munc18-1 in a complex with CaBP5.
Colocalization of CaBP5 with Myosin VI and Munc18-1 in the Retina
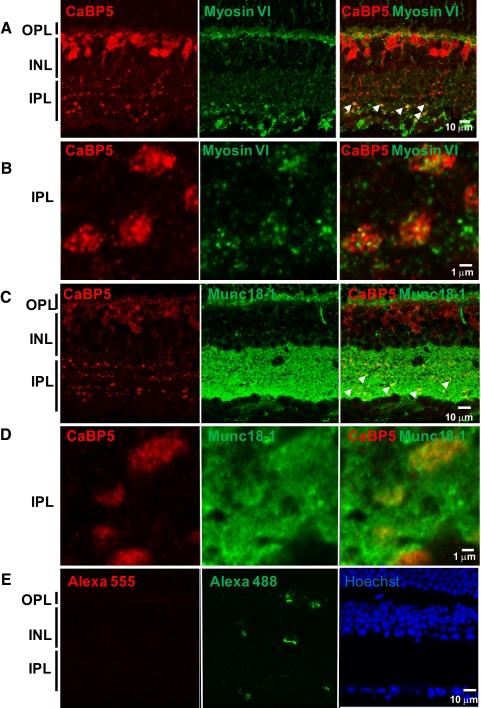
The full-length Munc18-1 and a C-terminal domain of myosin VI were amplified by PCR from mouse retina cDNA as a first confirmation of their expression in the retina (data not shown). Given that Munc18-1 cDNA from the retina cDNA library and the Munc18-1 peptides identified by mass spectrometry are shared by Munc18-1 and Munc18-2, we tried to amplify Munc18-2 by PCR. However, no PCR products were obtained with Munc18-2–specific primers using retina cDNA as template (data not shown). The full-length Munc18-1 was fused to a 6His-tag and was expressed in bacteria and used to generate anti–Munc18-1 antibody. These antibodies were characterized by immunoblotting on retinal extract and recombinant Munc18-1 protein and were shown to recognize specifically Munc18-1 (Supplementary Fig. S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-8246/-/DCSupplemental). We then analyzed the colocalization of Munc18-1 and CaBP5 in the retina using immunohistochemistry. As previously reported,3 CaBP5 localizes in bipolar cells (Fig. 3). Myosin VI colocalizes with CaBP5 in the inner and outer plexiform layers (Figs. 3A, 3B). Munc18-1 is observed in the outer plexiform layer and in the inner plexiform layer, wherein it colocalizes with CaBP5 (Figs. 3C, 3D).
Figure 3.
Single confocal images of mouse retina sections double labeled with anti–CaBP5, (A, B) anti– myosin VI, and (C, D) anti–Munc18-1. (A, C) Myosin VI and Munc18-1 colocalize with CaBP5 in the inner plexiform layer. Arrows in the merged images (right) point to some of the yellow colocalization signals. (B, D) Higher magnification of cross-sections through the inner plexiform layer at spots equivalent to those shown with arrows in (A) and (C). (E) Negative control in which the primary antibody was omitted. Nonspecific labeling of blood vessels only is observed in the negative control. Control sections were stained with Hoechst dye to visualize the retinal cell layers. OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer.
Distribution of CaBP5 with Munc18-1 and Myosin VI in Subcellular Retinal Fractions
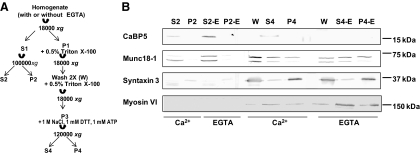
In vivo interaction of CaBP5 with Munc18-1 and myosin VI would require their presence in the same subcellular compartments. Although their subcellular colocalization at the bipolar axon terminals was first revealed through our immunohistochemistry approach, we further characterized their colocalization in native tissue using biochemical fractionation of bovine retina in the presence or absence of calcium (Fig. 4). Soluble cytoplasmic (S) and nonsoluble membrane (P) fractions were analyzed by Western blot for the presence of CaBP5, Munc18-1, myosin VI, and syntaxin-3. As expected, the transmembrane syntaxin-3 is present in the nonsoluble fractions (P and Triton X-100–extracted W fractions). Munc18-1, which is a soluble protein that binds to syntaxin, is distributed between soluble and insoluble fractions. We found similar distribution patterns of Munc18-1 and CaBP5 in the soluble fractions S2 and S2-E and the high salt extraction fraction S4. As shown in Figure 4B, myosin VI, an actin-based motor protein, accordingly distributes in actin-bound protein fractions S4 and P4. Ca2+-bound CaBP5 does enrich with myosin VI in the actin-associated protein fraction extracted with high salts (S4). However, in the absence of Ca2+, CaBP5 does not enrich with myosin VI in this fraction (S4E). This is also consistent with our observation of higher solubility of CaBP5 in its Ca2+-free form.
Figure 4.
Distribution of CaBP5, Munc18-1, myosin VI, and syntaxin-3 in subcellular retinal fractions. (A) Diagram of the fractionation steps used to generate the retinal soluble, insoluble, and actin-based protein fractions. (B) Western blot analysis of retinal subfractions probed with anti–CaBP5, anti–myosin VI, anti–Munc18-1, and anti–syntaxin-3. CaBP5 and Munc18-1 are both enriched in the cytosolic fraction. In the presence of Ca2+, CaBP5 is also present with myosin VI in the actin-associated protein fraction. Syntaxin-3 is isolated mostly in the nonsoluble fractions.
CaBP5 Interacts with Munc18-1 and Myosin VI in the Retina
We carried out coimmunoprecipitation experiments as an independent method to investigate the interaction of CaBP5 with Munc18-1 and myosin VI using physiological concentrations of both proteins in a retina extract.
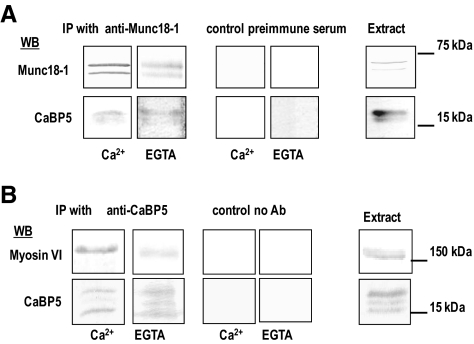
We performed coimmunoprecipitation with anti–Munc18-1 in bovine retina homogenates in the presence or absence of Ca2+. A protein with a molecular weight of ∼70 kDa, corresponding to Munc18-1, was immunoprecipitated with the anti–Munc18-1 antibody but not with preimmune serum IgG (Fig. 5A). As shown in Figure 5A, CaBP5 is coimmunoprecipitated with Munc18-1 in either the presence or the absence of Ca2+. CaBP5 was not detected in the control experiment with the preimmune serum. The reciprocal experiment in which we tried to coimmunoprecipitate Munc18-1 from retinal homogenate using anti–CaBP5 antibodies was unsuccessful. It is possible that the CaBP5 antibodies mask the interaction domain of CaBP5 with Munc18-1.
Figure 5.
Interaction of CaBP5 with Munc18-1 and myosin VI in the retina. (A) CaBP5 coimmunoprecipitates with Munc18-1 in the retina. Munc18-1 was immunoprecipitated from bovine retina extract using an anti–Munc18-1 polyclonal antibody or with the preimmune serum as a control and in the presence or the absence of Ca2+. The coimmunoprecipitated proteins were analyzed by Western blotting using anti–Munc18-1 (upper lane) or anti– CaBP5 (lower lane). Right: initial extract probed for those proteins. (B) Myosin VI coimmunoprecipitates with CaBP5 in the retina. Coimmunoprecipitation was carried using bovine retina extract with or without an anti–CaBP5 polyclonal antibody and in the presence or the absence of Ca2+. The coimmunoprecipitated proteins were analyzed by Western blotting using anti–myosin VI (upper lane) or anti–CaBP5 (lower lane) antibodies. Right: initial extract.
We also analyzed the interaction of myosin VI with CaBP5 by coimmunoprecipitation from bovine retina lysates using anti–CaBP5 antibodies. Two proteins with molecular weights of ∼17 and ∼150 kDa, corresponding to CaBP5 and myosin VI, were coimmunoprecipitated with anti–CaBP5 antibody. Neither CaBP5 nor myosin VI was detected in control experiments with protein G beads alone (Fig. 5B). This interaction appeared to be Ca2+ independent. Because syntaxin-3 was also identified by affinity chromatography and was previously reported to interact with Munc18-1, we also tested for its coimmunoprecipitation with CaBP5. Syntaxin-3 was not immunoprecipitated with CaBP5 (data not shown).
CaBP5 Interacts with Full-Length Munc18-1 in Yeast
We next tested whether CaBP5 interacts with a specific domain of Munc18-1 in the yeast two-hybrid assay. Fusion proteins consisting of the full-length mouse Munc18-1, the N-terminal peptide (domain I, amino acids 1-13615), and C-terminal peptide (domain II+III, amino acids 130-603) (Fig. 1B) were fused to the Gal4-activation domain (AD) by subcloning into the yeast expression vector (pGADT7-AD), and the full-length mouse CaBP5 cDNA was fused to the DNA-binding domain in pGBKT7 vector. The reporter yeast strain AH109 was cotransformed with mouse CaBP5 and Munc18-1 fusion proteins. As a negative control, CaBP5 was cotransformed with the pGADT7 vector. The bovine CaBP5 interaction with the Munc18-1 clone of the retinal library was used as a positive control. Cotransformants were first selected on SD-Leu-Trp (plasmid selection). For low-affinity interaction, restreaking of colonies would result in growth on SD-Leu-Trp-His+3-AT, and stronger interaction can be selected on SD-Leu-Trp-His-Ade+ X-gal. Yeast growth on highly selective medium was observed after the interaction of CaBP5 with the full-length Munc18-1 (Fig. 1C). Low-affinity interaction of CaBP5 was detected with domain II+III only. Control experiments in which yeast was cotransformed with pGBKT7-CaBP5 and pGADT7 vector did not show growth on interaction selective media. These data suggest that the N-terminal domain I is not absolutely required for CaBP5 interaction with Munc18-1. Together with the deletions in domains I and II observed in the Munc18-1 plasmid isolated from the retina cDNA library, these results suggest that CaBP5 interacts primarily with domain III of Munc18-1 (Fig. 1B). We could not confirm these interactions in vitro using pull-down assays with purified recombinant proteins because the Munc18-1 protein fragments were sticky and gave high signals with the negative controls even in more stringent salt conditions.
CaBP5 Expression in PC12 Cells Increases NGF-Induced Neurite Outgrowth
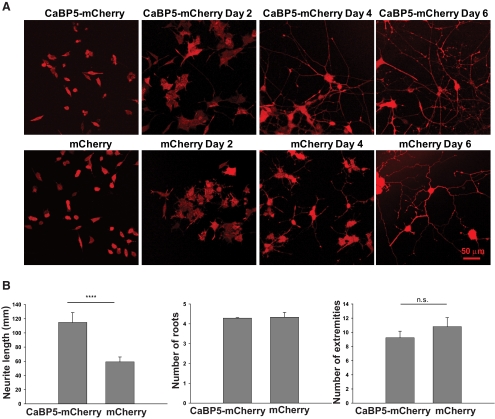
Because of the role of Munc18-1 in synaptic vesicle docking and fusion, we next investigated the effect of CaBP5 expression on neurotransmitter release in PC12 cells, which have been used as a model to investigate exocytosis.24,25 They resemble neurons on NGF-stimulated differentiation and express the proteins involved in synaptic vesicle exocytosis.26,27 Because of the low efficiency of transfection of PC12 cells, we chose to establish a stable PC12 cell line expressing CaBP5-mCherry or mCherry as a negative control. CaBP5 was fused to mCherry to select stably expressing CaBP5 clones and to use the emission fluorescence of mCherry for normalization to the cell number in quantification of FM1-43 dye uptake and dopamine release. The mCherry emission fluorescence also allowed us to visualize the neurites of PC12 cells because CaBP5-mCherry is a soluble protein that localizes throughout the cell. Although the cell lines were initially created to analyze synaptic vesicle cycling, we first noticed a change in the neurite outgrowth of CaBP5-mCherry expressing cells after stimulation with NGF compared with those stably expressing mCherry. CaBP5-mCherry–expressing cells showed increased neurite extension compared with cells expressing mCherry only (Fig. 6). The change in neurite outgrowth was more prominent 3 days after NGF stimulation. Two independent cell lines were analyzed for CaBP5-mCherry and mCherry. Both CaBP5-mCherry PC12 cell lines displayed increased neurite growth rates compared with mCherry PC12 cell lines. The number of neurite roots at the soma was similar in mCherry and CaBP5-mCherry expressing cells (Fig. 6B, middle). Although the number of neurite branches showed no statistically significant difference between mCherry and CaBP5-mCherry cells (Fig. 6B, right), the cells expressing CaBP5 often appeared to form a less elaborate network than control mCherry cells.
Figure 6.
CaBP5 expression in PC12 cells increases NGF-induced neurite outgrowth. (A) Confocal images of PC12 cells stably expressing CaBP5-mCherry or mCherry grown on collagen IV–coated plates before (left) and 2, 4, or 6 days after the addition of 50 ng/mL NGF. (B) Quantification of neurite growth in stable PC12 cell lines expressing mCherry (n = 21) or CaBP5-mCherry (n = 21) after treatment with NGF for 4 days. Left: mean neurite length (****P < 0.0001, Student's t-test); center: mean neurite roots (no significant difference); right: mean neurite extremities number (P > 0.05, no significant difference). Bars are presented as mean ± SEM.
CaBP5 Stimulates Both Constitutive and KCl-Induced Endocytosis in PC12 Cells
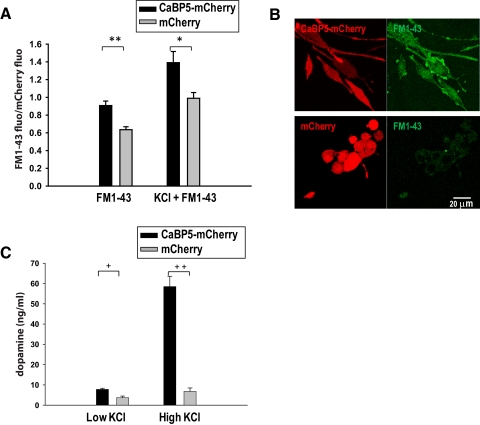
We observed that CaBP5 interacts with Munc18-1 and promotes neurite outgrowth; therefore, we analyzed whether CaBP5 stimulates endocytosis. Because neurotransmitter vesicle endocytosis follows vesicles exocytosis, the uptake of FM1-43 dye in vesicles after the stimulation of PC12 cells provides an indirect measurement of neurotransmitter vesicle exocytosis. FM dyes have been used to measure synaptic vesicle exocytosis/endocytosis.28 FM dye emission increased by >2 orders of magnitude on partition into membranes and constituted almost the entire signal uptake by the cells. We took advantage of this property and measured the uptake of FM1-43 by PC12 cells to determine whether dense-core vesicle formation was different in CaBP5-mCherry–expressing than in mCherry cells. As shown in Figure 7B, FM1-43 fluorescence originated from internalized dye because FM1-43 was largely absent in the plasma membrane after the cells were washed. FM1-43 that endocytosed in PC12 cells was measured in a fluorometer using an average of 200,000 cells per data point. We noticed that CaBP5 stimulates the basal rate of FM1-43 uptake. As shown in Figure 7A, FM1-43 uptake by CaBP5-mCherry–expressing PC12 cells was higher than that of mCherry-expressing cells, even without stimulation. CaBP5 also stimulated the uptake of FM1-43 after stimulation with 50 mM KCl compared with mCherry-expressing cells (Figs. 7A, 7B).
Figure 7.
CaBP5 stimulates exocytosis and endocytosis in PC12 cells. (A) CaBP5 stimulates both basal and KCl-induced endocytosis in PC12 cells. PC12 cells stably expressing CaBP5-mCherry or mCherry and cultured for 3 days with 50 ng/mL NGF were loaded with FM1-43 for 10 minutes without stimulation (left) or after stimulation for 2 minutes with high KCl buffer (right). After a 30-minute wash, including a 5-minute wash with 1 mM ADVASEP-7, the cells were collected and their fluorescence was measured in a fluorometer. FM1-43 fluorescence at 560 nm was normalized to the mCherry fluorescence at 610 nm. *P < 0.02, **P < 0.002, Student's t-test. (B) Uptake of FM1-43 dye in PC12 cells expressing CaBP5-mCherry or mCherry after 2-minute KCl stimulation. Representative confocal images of PC12 cells stably expressing CaBP5-mCherry or mCherry (3 days after stimulation with 50 ng/mL NGF) loaded with FM1-43 after stimulation for 2 minutes with high K+ buffer and washed with 1 mM ADVASEP-7. Images were taken on the same day and with the same laser power. PC12 cells expressing CaBP5-mCherry uptake more FM1-43 dye than cells expressing mCherry. (C) Increased dopamine release by PC12 stable cell lines expressing CaBP5-mCherry. PC12 cells expressing CaBP5-mCherry or mCherry were cultured for 3 days with 50 ng/mL NGF. The PC12 cell supernatant was then analyzed by ELISA for dopamine content after a 2-minute incubation with 3 mM KCl or 50 mM KCl buffer. The bars are presented as mean ± SEM. For each bar, n = 3. *P < 0.05, **P < 0.001, Student's t-test.
CaBP5 Stimulates Dopamine Release in PC12 Cells
Another way to analyze whether CaBP5 has an effect on neurotransmitter vesicle exocytosis is to measure dopamine secretion in PC12 cells using an ELISA kit. As seen in Figure 7C, at low KCl concentrations, cells expressing CaBP5-mCherry released twice as much dopamine as mCherry cells. In addition, after stimulation with high KCl buffer, CaBP5-mCherry–expressing cells secreted eight times more dopamine than mCherry cells.
Discussion
We showed previously that CaBP5 is required for normal transmission of the light signal throughout the retina.13 The sensitivity of retinal ganglion cells light responses were reduced by ∼50% in CaBP5 knockout mice. Using a candidate approach, we showed that CaBP5 modestly suppressed calcium-dependent inactivation of Cav1.2 and Cav1.3 in transfected HEK293T cells.12,13 Although Cav1 channels might be a physiological target for CaBP5, we chose to pursue the study of CaBP5 function by identifying CaBP5-binding partners using affinity chromatography coupled to mass spectrometry and screening of a bovine retina cDNA library using the yeast two-hybrid system. In this study, we identified Munc18-1 and myosin VI as physiological partners for CaBP5. Although CaBP5 is a Ca2+-binding protein, we have not observed a Ca2+-dependent interaction with myosin VI and Munc18-1. Ca2+-independent interactions of other members of the CaBP subfamily, CaBP1 and CaBP4, with their L-type, voltage-gated calcium channel targets, were previously reported, though CaBPs bind their targets more effectively in the presence of Ca2+.5,29 We also present evidence that CaBP5 stimulates neurite outgrowth and neurotransmitter vesicle exocytosis in PC12 cells.
Using affinity chromatography, we identified many CaBP5-binding proteins involved in different pathways. Munc18-1 was selected as our strongest candidate as a CaBP5 physiological interacting partner because it was identified using two independent methods. Here we showed that CaBP5 partially colocalizes with Munc18-1 and myosin VI at the synaptic terminals of retinal bipolar cells. We confirmed the physiological interactions of Munc18-1 and myosin VI with CaBP5 and showed their partitions in the same subcellular fractions.
Myosins are involved in actin-based motility and have been involved in many forms of cytoplasmic transport, including synaptic vesicle transport. Compared with other myosins, myosin VI is unique in that it acts toward the minus end of actin filaments—that is, in a reverse directionality—and is thus localized to endocytic vesicles (see Refs. 30, 31 for review). Mutations in its gene are associated with human deafness.32,33 Although we focused on myosin VI interaction with CaBP5, multiple nonmuscular myosins were captured by affinity chromatography on immobilized CaBP5. One of them is myosin V, which has been identified as vesicle-associated protein and mediates F-actin–dependent synaptic vesicle conveyance to the membrane.34 Myosin V has been shown to play a role in exocytic activity in retinal neurons.35 Binding of syntaxin-1 to the neck domain of myosin V, which also binds calmodulin, has been shown to be involved in exocytosis.36 Unfortunately, we could not confirm the myosin V/CaBP5 interaction because of a lack of good commercial anti–myosin V antibodies, but we validated its expression in the retina using RT-PCR (data not shown). CaBP1, a member of the calmodulin-like protein subfamily, has been shown to interact with myosin I. CaBP1 binds to its IQ motifs, which are known to bind calmodulin. CaBP1 also binds to a domain outside the defined IQ motifs, and this might explain CaBP1 binding to myosin I without displacing calmodulin in the absence of Ca2+.37 However, in the presence of Ca2+, CaBP1 competes with calmodulin for binding to the IQ motifs. Similarly, calmodulin binds to myosin VI, not only to its IQ motif but also to a domain located between the IQ motif and the converter region.38 It is possible that CaBP5 interacts in a similar way with myosins. The association of CaBP5 with myosin might affect its motility and, consequently, the traffic of synaptic proteins. It is also possible that CaBP5 interactions with myosins might compete with binding of other synaptic proteins. Further studies investigating the mechanism of interaction of CaBP5 with myosins will be required to answer those questions.
Munc18-1 was identified not only using affinity chromatography on immobilized CaBP5 but also using yeast two-hybrid screening of a retina library, indicating that CaBP5 interacts directly with Munc18-1. Because of its interaction with Munc18-1, we hypothesized that CaBP5 might have a role in synaptic vesicle cycling. Munc18-1 (also called Sec1, SM, and STXBP1 for syntaxin-binding protein 1) plays a role in multiple steps of the synaptic vesicle exocytosis.39,40 Munc18-1, together with the soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptors (SNARE proteins), syntaxin, SNAP25, and synaptobrevin/VAMP 2, all of which have been shown to be present in the retina ribbon synapses,22,23 are required for synaptic vesicle fusion with the membrane.27,39–41 Although Munc18-1 was first identified as important for synaptic vesicle docking, its role in synaptic vesicle fusion has been revealed by the absence of synaptic transmission in Munc18-1 knockout mice.42 Munc18-1 inhibits synaptic vesicle exocytosis in binding to the close conformation of syntaxin-1, but it is also necessary for synaptic vesicle fusion in binding to the N terminus of open syntaxin in complex with the other SNARE proteins. The present observation that CaBP5 interacts with Munc18-1 and stimulates basal and regulated secretion in PC12 cells is suggestive of a role of CaBP5 in vesicle cycling. It is also consistent with our previous study13 showing that the absence of CaBP5 reduces the sensitivity of rod-mediated responses. The change of ganglion cell responses in CaBP5 knockout mice might reflect a change in vesicle release at the bipolar synapse.
Although our previous study13 had shown that CaBP5 can directly interact, mostly in a Ca2+-dependent manner, with a cytoplasmic fragment of Cav1.2 α1, we did not identify Cav1.2 α1 as an interacting partner using affinity chromatography coupled with mass spectrometry. In the presence of Ca2+, high molecular–weight proteins that would include the Cav1.2 α1 did not constitute the major bands. This result is probably due to the low level of Cav1.2 in the retina. In addition, it is possible that, in the presence of Ca2+, the endogenous CaBP5 forms a tight complex with the Cav1.2 α1, limiting its retention on the column. In the absence of Ca2+, myosins were identified in high molecular–weight bands. In addition to the low level of Cav1.2, the interaction of CaBP5 with more abundant proteins, such as myosins and Munc18-1, could also mask the interaction with Cav1.2. In any case, our previous study and the present findings indicate that CaBP5 interacts with players involved in neurotransmitter release. CaBP5 might affect vesicle release indirectly by modulating Ca2+ entry through voltage-gated calcium channels and, more directly, through binding to Munc18-1. Precisely how CaBP5 modulates neurotransmitter cycling was not determined. It is also possible that CaBP5 might interfere with Munc18-1 interaction with syntaxin or Mint, Doc2, or SNARE complex proteins, which also bind to Munc18-1 and are involved in synaptic vesicle cycle.43–45
We tried to identify a specific domain of Munc18-1 responsible for its interaction with CaBP5 by expressing the domain I or II+III15 of Munc18-1 with CaBP5 in the yeast two-hybrid assay. No binding of CaBP5 to Munc18-1 domain I was detected, but weak interaction was observed between CaBP5 and Munc18-1 domain II+III. This is also in agreement with the observation that the Munc18-1 clone isolated in the yeast two-hybrid screening was missing 36 amino-acids at the N terminus. This clone was also deleted from the 79 C-terminal amino acids that constitute part of domain II of Munc18-1. Altogether, these data suggest that domain 3 is predominantly important for the interaction between CaBP5 and Munc18-1. Munc18-1 domain 3 has been shown to interact with the SNARE complex. Mutations in domain 3 impair binding to the SNARE complex and, consequently, inhibit the release kinetics of exocytosis.46–48 However, mutations in domain 3 do not affect the interaction with syntaxin. Domain I of Munc18-1, instead, is important for the interaction between Munc18-1 and syntaxin in its closed and open conformations.15,41,45 In the closed conformation of syntaxin, Munc18-1 domain 3 also binds to syntaxin.15,49 We would like to speculate that binding of CaBP5 to domain 3 of Munc18-1 might compete with binding of this domain to the closed syntaxin and that it thereby facilitates the conformational change of syntaxin from a closed to an open conformation prone to interact with the SNARE proteins. In this case, CaBP5 would have a role in priming of the docked vesicles. Such role in synaptic vesicle priming has been shown for Munc13-1 that accelerates opening of syntaxin-1 by interaction with the syntaxin-1 SNARE motif.50 Primed vesicles also spontaneously fuse with the plasma membrane, even in the absence of Ca2+. This is in agreement with our observation that CaBP5 stimulates both basal and regulated exocytosis. No significant changes in the number of synaptic vesicles or docked vesicles were observed in CaBP5 knockout mice bipolar terminals compared with wild-type mice (data not shown). These observations are suggestive of a role of CaBP5 in priming rather than in docking or fusion of synaptic vesicles.
Munc18-1 has also been shown to regulate early neurite outgrowth.51 Plasma membrane expansion in the growing neurite occurs through a mechanism of regulated exocytosis involving the components of the exocytosis machinery (see Ref. 52 for review). Although SNARE-dependent synaptic vesicle fusion has been shown to contribute to the supply of lipids and proteins necessary for developing neurites, Munc18-1 knockout mice with abolished synaptic vesicle release develop long neurites.42,51 However, a 42% decrease in outgrowth speed was observed for Munc18-1–deficient neurons compared with wild-type neurons.51 Therefore, we can speculate that through interaction with Munc18-1, CaBP5 could stimulate neurite outgrowth. In fact, we have observed in this study that CaBP5-mCherry expression in PC12 cells stimulates neurite outgrowth compared with PC12 cells expressing mCherry. It is possible that the enhanced neurite outgrowth in CaBP5-expressing cells also results from increased basal and regulated dense core vesicle cycling in PC12 cells.
In summary, we report in this study that CaBP5 interacts with Munc18-1 and myosin VI, two proteins involved in the synaptic vesicle cycle. Our results showing that CaBP5 stimulates neurite outgrowth and vesicle endocytosis and exocytosis in PC12 cells agree with the physiological interaction of CaBP5 with those proteins. Taken together, the present findings suggest a role for CaBP5 in fine-tuning vesicle cycling that might be important for the physiological function of CaBP5-expressing bipolar cells.
Supplementary Material
Acknowledgments
The authors gratefully thank Sharona Gordon and the members of her group for helpful discussions and the use of laboratory equipment.
Footnotes
Supported by National Institutes of Health/National Eye Institute Grant R21-EY017310 (FH) and in part by Grant R01-EY020850 (FH) and Vision CORE Grant EY01730.
Disclosure: I. Sokal, None; F. Haeseleer, None
References
- 1. Haeseleer F, Imanishi Y, Sokal I, Filipek S, Palczewski K. Calcium-binding proteins: Intracellular sensors from the calmodulin superfamily. Biochem Biophys Res Commun. 2002;290:615–623 [DOI] [PubMed] [Google Scholar]
- 2. Haeseleer F, Paczewski K. Calmodulin and Ca2+-binding proteins (CaBPs): variations on a theme. Photoreceptors Calcium. 2002:303–317 [DOI] [PubMed] [Google Scholar]
- 3. Haeseleer F, Sokal I, Verlinde C, et al. Five members of a novel Ca2+-binding protein (CABP) subfamily with similarity to calmodulin. J Biol Chem. 2000;275:1247–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haeseleer F. Interaction and colocalization of CaBP4 and Unc119 (MRG4) in photoreceptors. Invest Ophthalmol Vis Sci. 2008;49:2366–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haeseleer F, Imanishi Y, Maeda T, et al. Essential role of Ca2+-binding protein 4, a Ca(v)1.4 channel regulator, in photoreceptor synaptic function. Nat Neurosci. 2004;7:1079–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aldahmesh MA, Al-Owain M, Alqahtani F, Hazzaa S, Alkuraya FS. A null mutation in CABP4 causes Leber's congenital amaurosis-like phenotype. Mol Vis. 2010;16:207–212 [PMC free article] [PubMed] [Google Scholar]
- 7. Littink KW, van Genderen MM, Collin RWJ, et al. A novel homozygous nonsense mutation in cabp4 causes congenital cone-rod synaptic disorder. Invest Ophthalmol Vis Sci. 2009;50:2344–2350 [DOI] [PubMed] [Google Scholar]
- 8. Zeitz C, Labs S, Lorenz B, et al. Genotyping microarray for CSNB-associated genes. Invest Ophthalmol Vis Sci. 2009;50:5919–5926 [DOI] [PubMed] [Google Scholar]
- 9. Ghosh KK, Bujan S, Haverkamp S, Feigenspan A, Wässle H. Types of bipolar cells in the mouse retina. J Comp Neurol. 2004;476:202–203 [DOI] [PubMed] [Google Scholar]
- 10. Haverkamp S, Ghosh KK, Hirano AA, Wässle H. Immunocytochemical description of five bipolar cell types of the mouse retina. J Comp Neurol. 2003;455:463–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haverkamp S, Haeseleer F, Hendrickson A. A comparison of immunocytochemical markers to identify bipolar cell types in human and monkey retina. Vis Neurosci. 2003;20:589–600 [DOI] [PubMed] [Google Scholar]
- 12. Cui G, Meyer A, Calin-Jageman I, et al. Ca2+-binding proteins tune Ca2+-feedback to Cav1.3 Ca 2+ channels in auditory hair cells. J Physiol. 2007;585:791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rieke F, Lee A, Haeseleer F. Characterization of Ca2+-binding protein 5 knockout mouse retina. Invest Ophthalmol Vis Sci. 2008;49:5126–5135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang HB, Liu XH, Zhang K, et al. Photoreceptor cGMP phosphodiesterase delta subunit (PDE delta) functions as a prenyl-binding protein. J Biol Chem. 2004;279:407–413 [DOI] [PubMed] [Google Scholar]
- 15. Misura KMS, Scheller RH, Weis WI. Three-dimensional structure of the neuronal-Sec1-syntaxin-1a complex. Nature. 2000;404:355–362 [DOI] [PubMed] [Google Scholar]
- 16. Kay AR, Alfonso A, Alford S, et al. Imaging synaptic activity in intact brain and slices with FM1-43 in C. elegans, lamprey, and rat. Neuron. 1999;24:809–817 [DOI] [PubMed] [Google Scholar]
- 17. Avraham KB, Hasson T, Steel KP, et al. The mouse Snell's waltzer deafness gene encodes an unconventional myosin required for structural integrity of inner-ear hair-cells. Nat Genet. 1995;11:369–375 [DOI] [PubMed] [Google Scholar]
- 18. Uthaiah RC, Hudspeth AJ. Molecular anatomy of the hair cell's ribbon synapse. J Neurosci. 2010;30:12387–12399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ibaraki K, Horikawa HPM, Morita T, et al. Identification of 4 different forms of syntaxin-3. Biochem Biophys Res Commun. 1995;211:997–1005 [DOI] [PubMed] [Google Scholar]
- 20. Pevsner J, Hsu SC, Scheller RH. N-SEC1, a neural-specific syntaxin-binding protein. Proc Natl Acad Sci U S A. 1994;91:1445–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sherry DM, Mitchell R, Standifer KM, du Plessis B. Distribution of plasma membrane-associated syntaxins 1 through 4 indicates distinct trafficking functions in the synaptic layers of the mouse retina. BMC Neurosci. 2006;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morgans CW, Brandstätter JH, Kellerman J, Betz H, Wässle H. A SNARE complex containing syntaxin-3 is present in ribbon synapses of the retina. J Neuroscience. 1996;16:6713–6721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ullrich B, Südhof TC. Distribution of synaptic markers in the retina—implications for synaptic vesicle traffic in ribbon synapses. J Physiol. 1994;88:249–257 [DOI] [PubMed] [Google Scholar]
- 24. Westerink RHS, Ewing AG. The PC12 cell as model for neurosecretion. Acta Physiol. 2008;192:273–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Banerjee A, Kowalchyk JA, DasGupta BR, Martin TFJ. SNAP-25 is required for a late postdocking step in Ca2+-dependent exocytosis. J Biol Chem. 1996;271:20227–20230 [DOI] [PubMed] [Google Scholar]
- 26. Han L, Jiang TD, Han GA, et al. Rescue of Munc18-1 and-2 double knockdown reveals the essential functions of interaction between Munc18 and closed syntaxin in PC12 cells. Mol Biol Cell. 2009;20:4962–4975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bajohrs M, Darios F, Peak-Chew SY, Davletov B. Promiscuous interaction of SNAP-25 with all plasma membrane syntaxins in a neuroendocrine cell. Biochem J. 2005;392:283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gaffield MA, Betz WJ. Imaging synaptic vesicle exocytosis and endocytosis with FM dyes. Nat Protoc. 2006;1:2916–2921 [DOI] [PubMed] [Google Scholar]
- 29. Zhou H, Kim SA, Kirk EA, et al. Ca2+-binding protein-1 facilitates and forms a postsynaptic complex with Ca(v)1.2 (L-Type) Ca2+ channels. J Neurosci. 2004;24:4698–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sweeney HL, Houdusse A. What can myosin VI do in cells? Curr Opin Cell Biol. 2007;19:57–66 [DOI] [PubMed] [Google Scholar]
- 31. Sweeney HL, Houdusse A. Myosin VI rewrites the rules for myosin motors. Cell. 2010;141:573–582 [DOI] [PubMed] [Google Scholar]
- 32. Ahmed ZM, Morell RJ, Riazuddin S, et al. Mutations of MYO6 are associated with recessive deafness, DFNB37. Am J Hum Genet. 2003;72:1315–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Melchionda S, Ahituv N, Bisceglia L, et al. MYO6, the human homologue of the gene responsible for deafness in Snell's waltzer mice, is mutated in autosomal dominant nonsyndromic hearing loss. Am J Hum Genet. 2001;69:635–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prekeris R, Terrian DM. Brain myosin V is a synaptic vesicle-associated motor protein: evidence for a Ca2+-dependent interaction with the synaptobrevin-synaptophysin complex. J Cell Biol. 1997;137:1589–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Libby RT, Lillo C, Kitamoto J, Williams DS, Steel KP. Myosin Va is required for normal photoreceptor synaptic activity. J Cell Sci. 2004;117:4509–4515 [DOI] [PubMed] [Google Scholar]
- 36. Watanabe M, Nomura K, Ohyama A, et al. Myosin-Va regulates exocytosis through the submicromolar Ca2+-dependent binding of syntaxin-1A. Mol Biol Cell. 2005;16:4519–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tang N, Lin TM, Yang J, Foskett JK, Ostap EM. CIB1 and CaBP1 bind to the myo1c regulatory domain. J Muscle Res Cell Motil. 2007;28:285–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bahloul A, Chevreux G, Wells AL, et al. The unique insert in myosin VI is a structural calcium-calmodulin binding site. Proc Natl Acad Sci U S A. 2004;101:4787–4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rizo J, Südhof TC. SNAREs and Munc18 in synaptic vesicle fusion. Nat Rev Neurosci. 2002;3:641–653 [DOI] [PubMed] [Google Scholar]
- 40. Jahn R. Sec1/Munc18 proteins: mediators of membrane fusion moving to center stage. Neuron. 2000;27:201–204 [DOI] [PubMed] [Google Scholar]
- 41. Hata Y, Slaughter CA, Sudhof TC. Synaptic vesicle fusion complex contains unc-18 homolog bound to syntaxin. Nature. 1993;366:347–351 [DOI] [PubMed] [Google Scholar]
- 42. Verhage M, Maia AS, Plomp JJ, et al. Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science. 2000;287:864–869 [DOI] [PubMed] [Google Scholar]
- 43. Verhage M, deVries KJ, Røshol H, Burbach JPH, Gispen WH, Südhof TC. DOC2 proteins in rat brain: complementary distribution and proposed function as vesicular adapter proteins in early stages of secretion. Neuron. 1997;18:453–461 [DOI] [PubMed] [Google Scholar]
- 44. Okamoto M, Südhof TC. Mints, Munc18-interacting proteins in synaptic vesicle exocytosis. J Biol Chem. 1997;272:31459–31464 [DOI] [PubMed] [Google Scholar]
- 45. Dulubova I, Khvotchev M, Liu SQ, Huryeva I, Südhof TC, Rizo J. Munc18–1 binds directly to the neuronal SNARE complex. Proc Natl Acad Sci U S A. 2007;104:2697–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Boyd A, Ciufo LF, Barclay JW, et al. A random mutagenesis approach to isolate dominant-negative yeast Sec1 mutants reveals a functional role for domain 3a in yeast and mammalian Sec1/Munc18 proteins. Genetics. 2008;180:165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Graham ME, Prescott GR, Johnson JR, et al. Structure-function study of mammalian Munc18–1 and C. elegans UNC-18 implicates domain 3b in the regulation of exocytosis. PLoS ONE. 2011;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hu SH, Christie MP, Saez NJ, et al. Possible roles for Munc18-1 domain 3a and Syntaxin-1 N-peptide and C-terminal anchor in SNARE complex formation. Proc Natl Acad Sci U S A. 2011;108:1040–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Han GA, Malintan NT, Collins BM, Meunier FA, Sugita S. Munc18-1 as a key regulator of neurosecretion. J Neurochem. 2010;115: 1–10 [DOI] [PubMed] [Google Scholar]
- 50. Ma C, Li W, Xu Y, Rizo J. Munc13 mediates the transition from the closed syntaxin-Munc18 complex to the SNARE complex. Nat Struct Mol Biol. 2011;18:542–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Broeke JHP, Roelandse M, Luteijn MJ, et al. Munc18 and Munc13 regulate early neurite outgrowth. Biol Cell. 2010;102:479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pfenninger KH. Plasma membrane expansion: a neuron's Herculean task. Nat Rev Neurosci. 2009;10:251–261 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.