Abstract
Select mutations in the human cytomegalovirus (HCMV) gene UL27 confer low-grade resistance to the HCMV UL97 kinase inhibitor maribavir (MBV). It has been reported that the 608-amino acid UL27 gene product (pUL27) normally localizes to cell nuclei and nucleoli, whereas its truncation at codon 415, as found in a MBV-resistant mutant, results in cytoplasmic localization. We now show that in the context of full-length pUL27, diverse single amino acid substitutions associated with MBV resistance result in loss of its nucleolar localization when visualized after transient transfection, whereas substitutions representing normal interstrain polymorphism had no such effect. The same differences in localization were observed during a complete infection cycle with recombinant HCMV strains over-expressing full-length fluorescent pUL27 variants. Nested UL27 C-terminal truncation expression plasmids showed that amino acids 596–599 were required for the nucleolar localization of pUL27. These results indicate that the loss of a nucleolar function of pUL27 may contribute to MBV resistance, and that the nucleolar localization of pUL27 during HCMV infection depends not only on a carboxy-terminal domain but also on a property of pUL27 that is affected by MBV-resistant mutations, such as an interaction with component(s) of the nucleolus.
Keywords: cytomegalovirus, maribavir, UL27, nucleolus, antiviral, drug resistance
1. Introduction
Human cytomegalovirus (HCMV) remains one of the most important infectious complications among neonates and after stem cell or solid organ transplantation (Boeckh and Geballe, 2011). Systemic agents currently approved by the Food and Drug Administration (FDA) for the treatment of HCMV infection are ganciclovir and its oral prodrug valganciclovir, foscarnet, and cidofovir. The use of these agents is limited by their adverse effects and by the development of resistance (Lurain and Chou, 2010). Thus, while the overall burden of HCMV-related disease has been reduced by modern antivirals, there remains a definite need for alternate, less toxic agents with novel viral targets or mechanisms of action (Prichard and Kern, 2011).
Maribavir (MBV) is a benzimidazole L-riboside that suppresses HCMV replication in vitro by inhibiting the activity of the viral UL97 serine/threonine kinase (pUL97) (Biron et al., 2002). Mutations in UL97 confer moderate-to-high level resistance (~9-fold to > 100-fold) to MBV (Biron et al., 2002; Chou, 2008; Chou, Van Wechel, and Marousek, 2007). Additionally, selection under MBV, or propagation of UL97-defective strains of HCMV without added drug, both result in mutations in the UL27 gene that confer low-level resistance (2–3 fold) to MBV (Chou, 2009; Chou et al., 2004; Komazin et al., 2003).
Mutations in UL27 that confer resistance to MBV include diverse single amino acid substitutions, premature stop codons, or frameshift mutations that result in a truncated protein (Chou, 2009; Chou et al., 2004; Komazin et al., 2003). This mutation pattern suggests that MBV resistance results from alteration or loss of pUL27 function, particularly that of its carboxy-terminal residues.
Wild type pUL27 (608 amino acids) localizes primarily to nuclei and nucleoli of human foreskin fibroblasts (HFFs) when expressed by transient transfection as a fluorescent fusion protein, while a MBV-resistant mutant truncated at codon 415 demonstrates primarily cytoplasmic localization (Chou et al., 2004), suggesting that altered subcellular localization of pUL27 may be a factor in MBV resistance. In this study, we investigated the effects of amino acid substitutions that confer resistance to MBV on pUL27 localization and the contribution of the carboxy terminus towards the normal trafficking of pUL27.
2. Materials and Methods
2.1 Cells
Human foreskin fibroblasts (HFFs) and human embryonic lung (HEL) fibroblasts were maintained at 37°C in a 5% CO2 atmosphere in Eagle’s minimal essential media supplemented with 10% Fetal Bovine Serum (FBS, Hyclone) during the growth phase and 3% FBS after reaching confluence.
2.2 Plasmids for transient expression of fluorescent fusion proteins
The plasmid SC127 (Chou et al., 2004) consists of the entire (608 codons) wild-type (wt; based on the HCMV AD169 sequence, Genbank accession no. X17403) UL27 open reading frame cloned in-frame with the enhanced green fluorescent protein (EGFP) coding sequence in the expression vector pEGFP-N1 (Clontech). Full length UL27 variants containing previously described amino acid substitutions (Chou, 2009; Chou et al., 2004) were PCR amplified using oligonucleotides 27UL1M (5′-CGCCGCAAGCTTCTTCTTCCTCTCAGTCATGA-3′) and 27UL1837M (5′-CGCCGGGATCCGATTGTGGCGTGACCTCCGA-3′), and the resulting PCR products were cloned into pEGFP-N1 using enzymes HindIII and BamHI, enabling the expression of UL27 variants as C-terminal EGFP fusion proteins (Table 1).
Table 1.
MBV resistance and nucleolar localization phenotypes of pUL27 variants
| Plasmid Name | UL27 Variant | MBV-R3 | Nucleolar Localization |
|---|---|---|---|
| SC127 | WT1 | No | Yes |
| SC514 | CS2 | No | Yes |
| SC518 | D361E | No | Yes |
| SC554 | K89N | No | Yes |
| SC493 | A269T | Yes | No |
| SC494 | R233S | Yes | No |
| SC506 | W153R | Yes | No |
| SC513 | L193F | Yes | No |
| SC523 | V353E | Yes | No |
| SC517 | W362R | Yes | No |
based on HCMV strain AD169 sequence
amino acid changes N289D+D298G+N300G+P307L+V310A+D351N+I367V compared to AD169
indicates resistance to MBV
Carboxy-terminal truncations of pUL27 were generated by PCR amplification from plasmid SC127 or HCMV strain AD169 genomic DNA using primers containing restriction sites and delimiting the desired UL27 codon range. PCR products were cloned in frame into the HindIII and BamHI sites of pEGFP-N1 to make the following plasmids comprising the indicated amino acids of pUL27: SC495 (1–559), SC496 (1–599), SC497 (1–595), and MH6 (1–565).
All new plasmid clones of PCR products were checked throughout the cloned segment for presence of the intended sequence variant and absence of errors, using standard dye deoxy sequencing (Applied Biosystems BigDye v3.1).
2.3 Viral clones and strains
The published bacterial artificial chromosome (BAC) BA1 was cloned from the AD169-derived wild type HCMV strain T2211 and contains a secreted alkaline phosphatase (SEAP) reporter gene under control of an ectopic CMV major immediate early (MIE) promoter adjacent to gene US3 (Chou, 2010; Chou et al., 2005). After transfection of BA1 into HFF, HCMV strain T3099 was recovered. BAC BA1 was modified to replace the native UL27 coding sequence (codons 10 to 598) with a bacterial galK selectable marker by conditional recombination in E. coli strain SW102 and galactose selection as described (Chou, 2010). The resulting BAC was purified by retransformation into E. coli strain SW105 and named BA101. The UL27 mutant BAC containing the K89N amino acid substitution was newly derived by generating a transfer vector incorporating K89N and an upstream Frt-flanked Kan selection marker, followed by recombination into BA101, kanamycin selection, and then Flp-induced removal of the Kan marker, using the same technical approach as described for generating UL97 mutants (Chou, 2010). The resulting BAC (BA153) was transfected into HFF cultures to yield the corresponding live HCMV mutant T3578. A wild type BAC (BA93) and live virus strain (T3386) containing the residual upstream Frt marker were generated as a control for phenotype comparisons. Using a previously reported co-transfection technique (Chou, 2009) not involving BAC clones, a recombinant HCMV mutant T3209 containing UL27 variant D361E was constructed and plaque purified.
To increase the level of expression of pUL27 in the context of a replicating HCMV genome, the SEAP coding sequence downstream of the ectopic MIE promoter in BAC BA1 was replaced with that of a UL27-EGFP fusion protein as follows. A 2-kb segment of BA1 DNA starting near the end of the MIE promoter, spanning the SEAP coding sequence and ending just inside the BeloBAC11 vector (Chou, 2010), was PCR-amplified with primers 5′-TCTGAGCGGTACCCGTTGCTGCCG-3′ and 5′-GAGACGTTGATCGGCACG-3′. The PCR product was digested with enzymes KpnI and NotI, and cloned into standard Bluescript vector pBS2KS+ (Stratagene) as SC528. The UL27-EGFP fusion protein coding sequence was PCR-amplified from plasmid SC514 (Table 1) using an upstream primer containing a BglII restriction site and digested with enzymes BglII and XbaI. A second PCR product containing the Frt-flanked Kan selection marker was amplified using flanking primers incorporating restriction sites for Bsu36I and BamHI, and then digested with these enzymes. The two digested PCR products were combined and ligated with plasmid SC528 that had been digested with Bsu36I and XbaI to release the SEAP coding sequence. This resulted in replacement of the SEAP sequence with that of Frt-Kan-Frt-UL27-EGFP, yielding transfer vector SC539. The SEAP coding sequence of the BAC BA1 was then replaced by UL27-EGFP by temperature-induced conditional recombination of a PCR product amplified from transfer vector SC539, followed by kanamycin selection and arabinose-induced removal of the Kan marker by Flp recombinase, as previously described (Chou, 2010), to yield BAC BA156. Transfection of BA156 into HFFs yielded strain T3580 (Table 2).
Table 2.
HCMV clones and strains over-expressing UL27 EGFP fusion proteins
| BAC Name | HCMV strain | UL27 Genotype
|
|
|---|---|---|---|
| Native UL27 locus | Over-expressed UL27/strain | ||
| BA156 | T3580 | wt AD169 | wt/clinical strain1 |
| BA166 | T3607 | galK2 | A269T/AD169 |
| BA169 | T3614 | galK2 | wt/clinical strain1 |
| BA186 | T3654 | galK2 | wt/AD169 |
| BA198 | T3675 | galK2 | R233S/clinical strain1 |
amino acid changes N289D+D298G+N300G+P307L+V310A+D351N+I367V compared to AD169
replacing most of native UL27 coding sequence
To make the additional UL27 over-expressing strains T3607, T3614, and T3654 (Table 2), the SEAP coding sequence of BAC BA101 (lacking most of the native UL27 sequence) was replaced with that of the UL27-EGFP fusion proteins derived from plasmids SC493 (BA166, T3607), SC514 (BA169, T3614), and SC127 (BA186, T3654), using a similar technique as described for T3580. To produce strain T3675 (Table 2) containing UL27 mutation R233S, “en passant” mutagenesis was used (Tischer, Smith, and Osterrieder, 2010). A plasmid clone containing an I-SceI recognition site upstream of a Kan cassette (SC556) was PCR amplified using primers 27UL659sF (5′-TGGACGGCACCCTGTGCCTGTTTCTGGAGCCCGAGGAGAGCGAACTCATC GGCCGCTGCCTCTAGCTAGGGATAACAG-3′) and 27UL738sR (5′-GCGGCACAGGGCCGCCGGCAGGCAGCGGCCGATGAGTTCGCTCTCCTCG GGCTCCAGAAAAAGTCAGCGTAATGCTCTGCCA-3′). The resulting PCR product containing flanking copies of the R233S sequence context were electroporated into strain GS1783 E. coli (a gift of Gregory Smith, Northwestern University) harboring BAC BA169 DNA. After kanamycin selection and arabinose induction of I-SceI activity in GS1783, a second round of heat-induced recombination produced the desired R233S mutant derivative (BA198). BA198 was transfected into HFFs to yield viral strain T3675.
All newly constructed HCMV recombinant viral strains were checked for the desired DNA sequence and absence of errors throughout the UL27 coding sequence and for the integrity of the in-frame EGFP fusion where applicable. BAC integrity and purity was monitored by HindIII restriction digests (Chou, 2010) and absence of parental sequence (galK or SEAP) by PCR.
2.4 DNA Transfection
Sub-confluent HFFs grown in multi-well chamber slides (Lab Tek II, Nalge Nunc International Corp) were transfected with 0.5–1.0 μg of plasmid DNA using Fugene 6 (Roche) according to the manufacturer’s instructions.
2.5 Fluorescence microscopy
For indirect immunofluorescence staining for nucleolin (C-23) (Santa Cruz Biotechnology, sc-55486, 1:100 dilution), and pUL44 (Virusys, CA006, 1:2500 dilution), HFFs were fixed for 30 minutes at room temperature (RT) in 4% paraformaldehyde in PBS and then permeabilized for 10 minutes in 0.1% Triton X-100 in PBS at RT. After blocking in PBS containing 5% BSA and 5% horse serum for one hour at RT or overnight at 4°C, the cells were incubated with primary antibody for 1 hour at RT, washed three times in PBS, then incubated with Texas Red-conjugated F(ab′)2 fragment goat anti-mouse secondary antibody (Santa Cruz Biotechnology, sc-3797, 1:100 dilution) for one hour at RT. Staining with 4′,6′-diamidino-2-phenylindole (DAPI; Sigma) was performed for 5 min at RT. The cells were then washed three times in PBS and the cell monolayer was covered with Fluoromount-G (Southern Biotech). Fluorescence microscopy was performed using a Leica DMI6000 microscope at 63X objective magnification. Pseudocoloring and image processing were performed using Adobe Photoshop 6.
2.6 Phenotypic assay of MBV sensitivity
SEAP yield reduction assays for MBV susceptibility of HCMV strains were performed as previously described (Chou, 2009; Chou et al., 2005; Chou, Van Wechel, and Marousek, 2006). Briefly, cell-free virus stock was inoculated onto 6-day-old HEL fibroblast monolayers in 24-well culture plates at a multiplicity of infection (MOI) of 0.01 to 0.02 and cultured for 6 days under a range of MBV concentrations (5 per assay) and a no-drug control. The drug concentration required to reduce the supernatant SEAP activity by 50% (EC50), assayed using a chemiluminescent substrate, was determined by curve fitting. Eleven or twelve assay replicates performed on at least four separate dates were used to calculate the mean EC50 and standard deviation for each strain.
3. Results
3.1 Amino acid substitutions conferring resistance to MBV are associated with exclusion of pUL27 from nucleoli
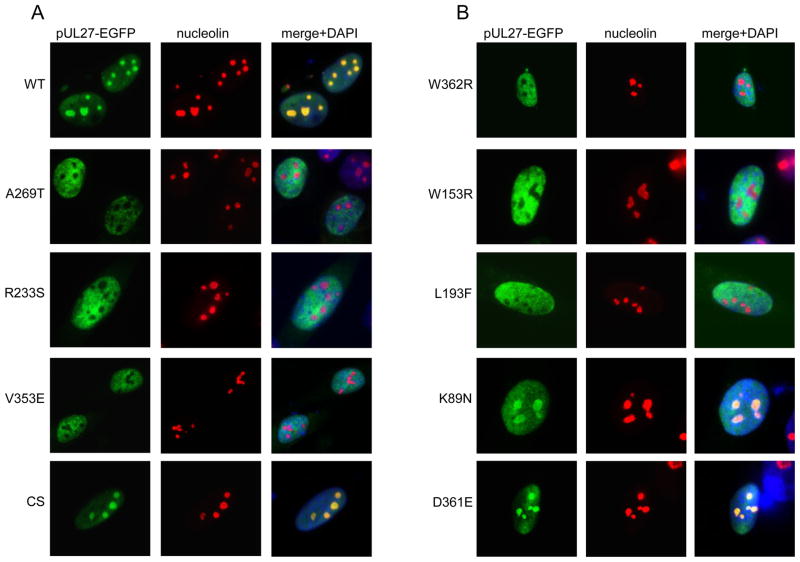
In order to determine the effect of individual MBV-resistant amino acid substitutions in pUL27 on the subcellular localization of pUL27, pUL27 variants (Chou, 2009; Chou et al., 2004) were cloned into pEGFP-N1 and their localization was examined by fluorescence microscopy after transfection of HFFs. Consistent with previous observations (Chou et al., 2004), full-length, wt pUL27 was observed primarily in nuclei and nucleoli (Figure 1A). Surprisingly, the MBV-resistant pUL27 mutants A269T, R233S, and V353E were present in nuclei but were uniformly excluded from nucleoli. To confirm that this observation was not simply due to an effect of normal UL27 sequence polymorphism, the localization of a MBV-susceptible (MBV-S) pUL27 variant from a clinical strain (CS) containing several common amino acid substitutions (N289D, D298G, N300G, P307L, V310A, D351N and I367V) (Chou, 2009) was examined. pUL27(CS) demonstrated nucleolar localization similar to that seen with wt pUL27. The localization of three additional MBV-resistant pUL27 mutants (W362R, W153R, L193F) was assessed, all of which were excluded from nucleoli (Figure 1B). In addition, two previously uncharacterized pUL27 variants, K89N (strain T3578) and D361E (strain T3209), were found to be MBV-susceptible (MBV-S), having MBV EC50s of 0.13 +/− 0.03 μM (n=12) and 0.08 +/− 0.02 μM (n=11), respectively, compared to EC50s of 0.10 μM for both baseline control MBV-S strains T2211 and T3386. Both of these MBV-S pUL27 variants, K89N and D361E, localized to nucleoli (Figure 1B). Thus, the amino acid substitutions resulting in resistance to MBV studied here are associated with the loss of nucleolar localization of pUL27 (Table 1).
Figure 1.
MBV-R amino acid substitutions result in exclusion of pUL27 from nucleoli. (A) HFFs were transfected with EGFP-fusion vectors expressing wild-type (WT) UL27, the MBV-S UL27 clinical strain (CS) variant, and the MBV-R variants A269T, R233S, and V353E, and (B) the MBV-R variants W362R, W153R, and L193F and the MBV-S variants D361E and K89N. 48–72 hours later, indirect immunofluorescence for nucleolin was performed, and the localization of each pUL27-EGFP fusion construct and nucleolin was assessed by fluorescence microscopy.
3.2 MBV-resistant pUL27 mutants are excluded from nucleoli throughout HCMV infection
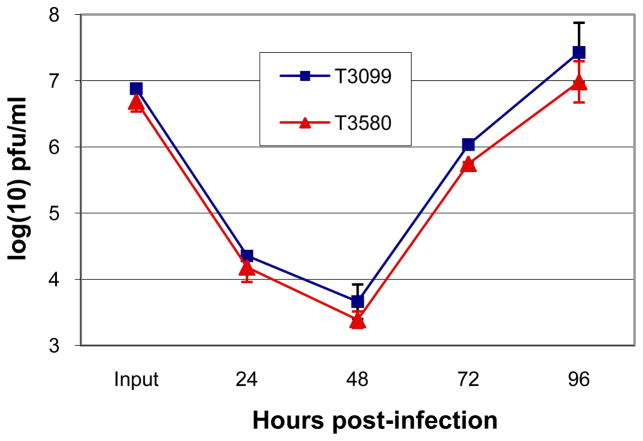
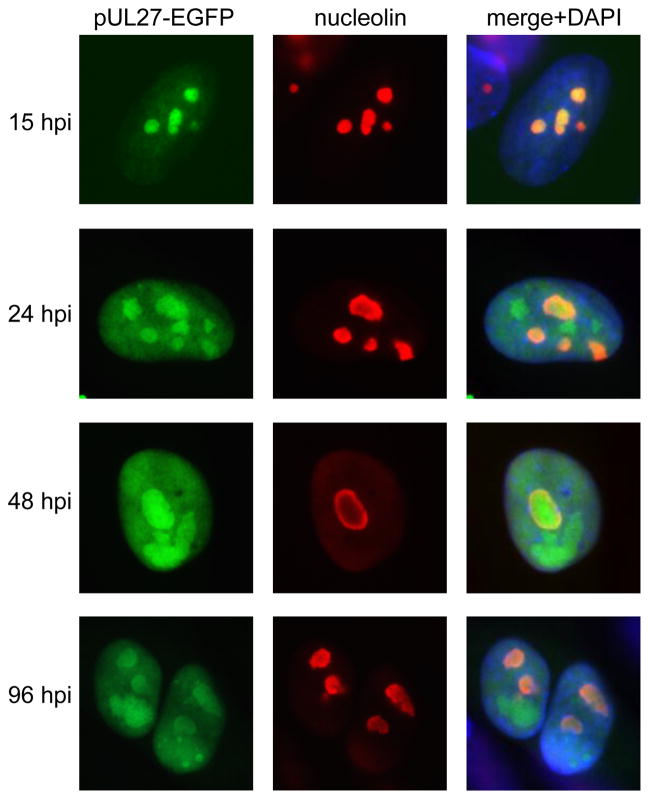
Initial efforts to visualize pUL27 trafficking in HCMV-infected cells by indirect immunofluorescence or by constructing recombinant viruses containing a pUL27 fluorescent fusion protein coding sequence were unsuccessful, likely because of the low-level expression of pUL27 from its native promoter during HCMV infection (data not shown). Therefore, recombinant HCMV strains were made in which UL27 expression was placed under control of an ectopic HCMV MIE promoter (Table 2). This strategy resulted in an approximately 10-fold and 100-fold increase in pUL27 levels, as determined by western blotting, by 24 hpi and 96 hpi, respectively (data not shown), and had little impact on viral replication as determined by single-step growth curves (Figure 2). To determine the subcellular trafficking of MBV-susceptible pUL27 during HCMV infection, HFFs were infected with T3580 and the localization of pUL27(CS)-EGFP was determined by fluorescence microscopy at various times post-infection. Co-localization with nucleolin at both early and late times during infection indicated that pUL27(CS)-EGFP was present in nuclei and nucleoli throughout the viral life cycle (Figure 3).
Figure 2.
Expression of pUL27 under control of an ectopic MIEP does not affect HCMV replication. HFFs were infected with T3099 (AD169 BAC; BA1) or T3580 (Table 2) at an MOI of ~3. Viral titers from supernatants collected at the indicated times post-infection were determined by plaque assay. The results shown represent duplicate infections with each strain.
Figure 3.
pUL27 localizes to nucleoli throughout HCMV infection. HFFs were infected with T3580 (Table 2). At the indicated times post-infection, indirect immunofluorescence for nucleolin was performed and the localization of pUL27-EGFP and nucleolin was determined by fluorescence microscopy.
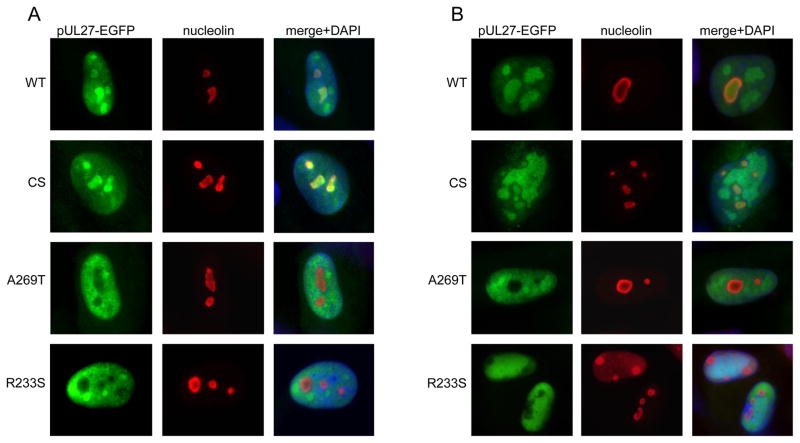
To determine the localization pattern of MBV-resistant pUL27 mutants during HCMV infection, and to control for the possibility that the native, untagged pUL27 present in T3580 could affect pUL27-EGFP localization, recombinant HCMV strains were constructed in which wt pUL27, pUL27(CS), and the MBV-resistant mutants pUL27(A269T) and pUL27(R233S) were expressed from the ectopic MIE promoter while the native UL27 open reading frame had been replaced by galK (Table 2). During infection of HFFs, the MBV-resistant pUL27 mutants pUL27(A269T) and pUL27(R233S) were excluded from nucleoli at 20 hpi (Figure 4A) and at 96 hpi (Figure 4B), whereas both wt pUL27 and pUL27(CS) were present in nucleoli throughout infection.
Figure 4.
The MBV-R mutants pUL27(A269T) and pUL27(R233S) are excluded from nucleoli throughout HCMV infection. HFFs were infected with T3607, T3614, T3654, and T3675 (Table 2). Indirect immunofluorescence for nucleolin was performed at 20 hpi (A) and 96 hpi (B), and the localization of each pUL27-EGFP construct and nucleolin was determined by fluorescence microscopy.
Beginning at 24–48 hpi, the accumulation of all four pUL27-EGFP fusion proteins in nuclear bodies distinct from nucleoli was observed (Figures 3 and 4B). Positive co-staining for HCMV pUL44 (Supplemental Figure 1) indicates that these represent replication centers (Ahn, Jang, and Hayward, 1999).
These results indicate that pUL27 is normally found predominantly in nuclei and nucleoli during HCMV infection. Amino acid substitutions conferring resistance to MBV result in exclusion of pUL27 from nucleoli throughout infection but do not affect nuclear localization or replication center accumulation.
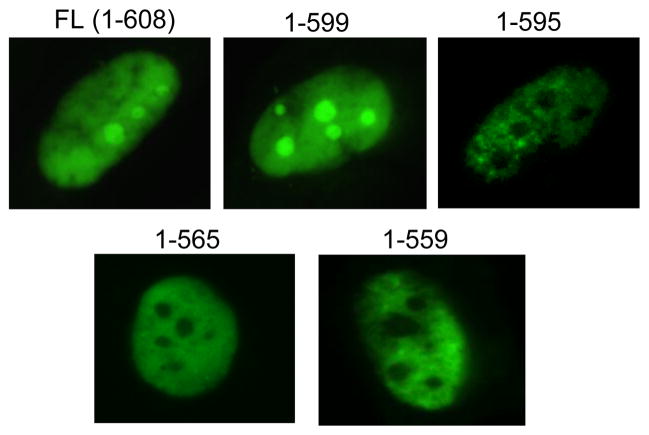
3.3 A carboxy-terminal domain in pUL27 is required for nucleolar localization
Many nucleolar proteins contain motifs that are necessary for localization of the native protein to the nucleolus (Emmott and Hiscox, 2009). These tend to be rich in arginine and lysine residues, may vary in size, and can be bipartite (Emmott and Hiscox, 2009). Analysis of pUL27 revealed the presence of two carboxy-terminal arginine-rich motifs, 560RCRRK564 and 596RRFR599. To evaluate the role of these regions in the nucleolar trafficking of pUL27, the localization of EGFP-fused truncations of pUL27 comprising amino acids 1–559, 1–565, 1–595, and 1–599, was examined by fluorescence microscopy after transient transfection of HFFs. pUL27(1–599) was present in the nucleus and nucleoli, similarly to full-length pUL27, while pUL27(1–559), pUL27(1–565), and pUL27(1–595) were excluded from nucleoli (Figure 5). Thus, the 596RRFR599 motif appears to be primarily responsible for the nucleolar localization of pUL27 in the absence of MBV-R amino acid substitutions.
Figure 5.
Amino acids 596–599 are necessary for localization of pUL27 to nucleoli. HFFs were transfected with EGFP-fusion vectors expressing the indicated amino acids of pUL27. 48–72 hours later, the localization of each pUL27-EGFP construct was determined by fluorescence microscopy.
4. Discussion
The role of pUL27 during HCMV infection and how genetic alterations in UL27 confer resistance to MBV are incompletely understood. pUL27 is dispensable for HCMV replication in cell culture and an in vivo model of infection (Chou, 2009; Prichard et al., 2006). Recent evidence indicates that pUL27 causes cell cycle arrest in G0/1 by promoting the degradation of the Tip60 acetyltransferase (Reitsma et al., 2011). Further, knock-down of Tip60 using RNA interference restored the ability of MBV to inhibit viral DNA synthesis during infection with HCMV deleted of UL27 (Reitsma et al., 2011). However, Tip60 degradation during HCMV infection is early and transient, while pUL27 levels do not decrease during the infection cycle (Reitsma et al., 2011). Therefore, it is likely that the role of pUL27 in MBV resistance is a dynamic one that changes over the course of HCMV infection, involving effects in addition to those related to Tip60.
Previous findings indicated that MBV resistance may be associated with abnormal subcellular localization of pUL27 (Chou et al., 2004). We found that single amino acid substitutions occurring throughout pUL27 that confer resistance to MBV result in the loss of nucleolar localization of pUL27 during HCMV infection, suggesting that the loss of a nucleolar function of pUL27 may contribute to MBV resistance.
Traditionally, the nucleolus has been thought of as the site for ribosome biogenesis, but it is now known that the nucleolus also participates in a wide array of cellular processes, such as control of the cell cycle and the response to cellular stressors such as DNA damage (Boisvert et al., 2007; Pederson and Tsai, 2009; Zhang and Lu, 2009). The nucleolus and nucleolar proteins play a similarly diverse role during infection with both RNA and DNA viruses, contributing to viral attachment and entry, replication, assembly, and egress (Greco, 2009). Numerous examples exist where abolishing the nucleolar localization of a viral protein alters viral replication (Boyne and Whitehouse, 2006; Boyne and Whitehouse, 2009; Cochrane, Perkins, and Rosen, 1990; Fazakerley et al., 2002; Michienzi et al., 2002).
Given that UL27 mutations arise when UL97 kinase function is disrupted, loss of pUL27 nucleolar function may affect a pathway directly or indirectly regulated by pUL97. Since deletion of UL27 has little effect on HCMV replication, the nucleolar function of pUL27 is unlikely to be important during normal infection. However, this function may be detrimental to HCMV replication when pUL97 activity is inhibited by MBV, resulting in mutations that exclude pUL27 from nucleoli. For example, it is plausible that because pUL27 is reported to interact with Tip60 (Reitsma et al, 2011), it may normally play a role in relocalizing Tip60 to the nucleolus, thereby contributing to its down-regulation early in the infection cycle.
Deletion of the motif 596RRFR599, along with the remaining nine carboxy-terminal amino acids of pUL27, abolished the nucleolar localization of pUL27. However, while this carboxy-terminal domain appears to be necessary for the nucleolar localization of pUL27, it is not sufficient, since single amino acid substitutions conferring resistance to MBV result in exclusion of full-length pUL27 from nucleoli.
The nucleolar trafficking of a protein that possesses a well-defined nucleolar localization signal may be regulated by additional properties of the protein (Meng, Yasumoto, and Tsai, 2006; Tsai and McKay, 2005). Our findings suggest that MBV-resistant amino acid substitutions occur at specific residues that are critical determinants of an additional property of pUL27 that governs nucleolar localization, such as a protein-protein interaction, protein-nucleic acid interaction, or post-translational modification (Carmo-Fonseca, Mendes-Soares, and Campos, 2000; Catez et al., 2002; Korgaonkar et al., 2005; Mekhail et al., 2004; Niedick et al., 2004).
In summary, we found that resistance to MBV is strongly associated with exclusion of pUL27 from nucleoli during HCMV infection, and that a carboxy-terminal motif is necessary, but not sufficient, for the nucleolar localization of pUL27. Further studies of the role of the interaction of pUL27 with the nucleolus in the development of MBV resistance are likely to contribute to our understanding of how HCMV compensates for lack of pUL97 activity.
Supplementary Material
Supplemental Figure 1. MBV-S and MBV-R pUL27 localize to replication centers. HFFs were infected with T3580 or T3607 (Table 2). At 96 hpi, indirect immunofluorescence for pUL44 was performed, and the localization of pUL27-EGFP and pUL44 was determined by fluorescence microscop
Highlights.
We studied the subcellular localization of the HCMV protein pUL27 in maribavir resistant mutants
UL27 mutations conferring maribavir resistance result in exclusion of pUL27 from nucleoli, an effect not seen with natural UL27 sequence polymorphisms
A carboxy-terminal domain of pUL27 comprising amino acids 596–599 is necessary but not sufficient for nucleolar localization.
Determining the function of pUL27 in nucleoli may provide insight into how HCMV compensates for lack of pUL97 kinase activity.
Acknowledgments
We wish to thank Gregory Smith (Northwestern University) for the gift of E. coli strain GS1783, and the National Cancer Institute Biological Resources Branch for E. coli strains SW102 and SW105. This work was supported by awards from the Medical Research Foundation of Oregon (MH), the Sunlin and Priscilla Chou Foundation (MH), NIH grant R01-AI39938 (SC), and Department of Veterans Affairs research funds (SC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn JH, Jang WJ, Hayward GS. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10) J Virol. 1999;73(12):10458–71. doi: 10.1128/jvi.73.12.10458-10471.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biron KK, Harvey RJ, Chamberlain SC, Good SS, Smith AA, 3rd, Davis MG, Talarico CL, Miller WH, Ferris R, Dornsife RE, Stanat SC, Drach JC, Townsend LB, Koszalka GW. Potent and selective inhibition of human cytomegalovirus replication by 1263W94, a benzimidazole L-riboside with a unique mode of action. Antimicrob Agents Chemother. 2002;46(8):2365–72. doi: 10.1128/AAC.46.8.2365-2372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckh M, Geballe AP. Cytomegalovirus: pathogen, paradigm, and puzzle. J Clin Invest. 2011;121(5):1673–1689. doi: 10.1172/JCI45449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8(7):574–85. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Boyne JR, Whitehouse A. Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc Natl Acad Sci U S A. 2006;103(41):15190–5. doi: 10.1073/pnas.0604890103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne JR, Whitehouse A. Nucleolar disruption impairs Kaposi’s sarcoma-associated herpesvirus ORF57-mediated nuclear export of intronless viral mRNAs. FEBS Lett. 2009;583(22):3549–56. doi: 10.1016/j.febslet.2009.10.040. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Mendes-Soares L, Campos I. To be or not to be in the nucleolus. Nat Cell Biol. 2000;2(6):E107–12. doi: 10.1038/35014078. [DOI] [PubMed] [Google Scholar]
- Catez F, Erard M, Schaerer-Uthurralt N, Kindbeiter K, Madjar JJ, Diaz JJ. Unique motif for nucleolar retention and nuclear export regulated by phosphorylation. Mol Cell Biol. 2002;22(4):1126–39. doi: 10.1128/MCB.22.4.1126-1139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Knutson E, Wang S, Martinez LA, Albrecht T. Stabilization of p53 in human cytomegalovirus-initiated cells is associated with sequestration of HDM2 and decreased p53 ubiquitination. J Biol Chem. 2007;282(40):29284–95. doi: 10.1074/jbc.M705349200. [DOI] [PubMed] [Google Scholar]
- Chou S. Cytomegalovirus UL97 mutations in the era of ganciclovir and maribavir. Rev Med Virol. 2008;18:233–46. doi: 10.1002/rmv.574. [DOI] [PubMed] [Google Scholar]
- Chou S. Diverse cytomegalovirus UL27 mutations adapt to loss of viral UL97 kinase activity under maribavir. Antimicrob Agents Chemother. 2009;53(1):81–5. doi: 10.1128/AAC.01177-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S. Recombinant phenotyping of cytomegalovirus UL97 kinase sequence variants for ganciclovir resistance. Antimicrob Agents Chemother. 2010;54(6):2371–8. doi: 10.1128/AAC.00186-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S, Marousek GI, Senters AE, Davis MG, Biron KK. Mutations in the human cytomegalovirus UL27 gene that confer resistance to maribavir. J Virol. 2004;78(13):7124–30. doi: 10.1128/JVI.78.13.7124-7130.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S, Van Wechel LC, Lichy HM, Marousek GI. Phenotyping of cytomegalovirus drug resistance mutations by using recombinant viruses incorporating a reporter gene. Antimicrob Agents Chemother. 2005;49(7):2710–5. doi: 10.1128/AAC.49.7.2710-2715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S, Van Wechel LC, Marousek GI. Effect of cell culture conditions on the anticytomegalovirus activity of maribavir. Antimicrob Agents Chemother. 2006;50(7):2557–9. doi: 10.1128/AAC.00207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S, Van Wechel LC, Marousek GI. Cytomegalovirus UL97 kinase mutations that confer maribavir resistance. J Infect Dis. 2007;196(1):91–4. doi: 10.1086/518514. [DOI] [PubMed] [Google Scholar]
- Cochrane AW, Perkins A, Rosen CA. Identification of sequences important in the nucleolar localization of human immunodeficiency virus Rev: relevance of nucleolar localization to function. J Virol. 1990;64(2):881–5. doi: 10.1128/jvi.64.2.881-885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmott E, Hiscox JA. Nucleolar targeting: the hub of the matter. EMBO Rep. 2009;10(3):231–8. doi: 10.1038/embor.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazakerley JK, Boyd A, Mikkola ML, Kaariainen L. A single amino acid change in the nuclear localization sequence of the nsP2 protein affects the neurovirulence of Semliki Forest virus. J Virol. 2002;76(1):392–6. doi: 10.1128/JVI.76.1.392-396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco A. Involvement of the nucleolus in replication of human viruses. Rev Med Virol. 2009;19(4):201–14. doi: 10.1002/rmv.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komazin G, Ptak RG, Emmer BT, Townsend LB, Drach JC. Resistance of human cytomegalovirus to the benzimidazole L-ribonucleoside maribavir maps to UL27. J Virol. 2003;77(21):11499–506. doi: 10.1128/JVI.77.21.11499-11506.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korgaonkar C, Hagen J, Tompkins V, Frazier AA, Allamargot C, Quelle FW, Quelle DE. Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol Cell Biol. 2005;25(4):1258–71. doi: 10.1128/MCB.25.4.1258-1271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurain NS, Chou S. Antiviral drug resistance of human cytomegalovirus. Clin Microbiol Rev. 2010;23(4):689–712. doi: 10.1128/CMR.00009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhail K, Gunaratnam L, Bonicalzi ME, Lee S. HIF activation by pH-dependent nucleolar sequestration of VHL. Nat Cell Biol. 2004;6(7):642–7. doi: 10.1038/ncb1144. [DOI] [PubMed] [Google Scholar]
- Meng L, Yasumoto H, Tsai RY. Multiple controls regulate nucleostemin partitioning between nucleolus and nucleoplasm. J Cell Sci. 2006;119(Pt 24):5124–36. doi: 10.1242/jcs.03292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michienzi A, Li S, Zaia JA, Rossi JJ. A nucleolar TAR decoy inhibitor of HIV-1 replication. Proc Natl Acad Sci U S A. 2002;99(22):14047–52. doi: 10.1073/pnas.212229599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedick I, Froese N, Oumard A, Mueller PP, Nourbakhsh M, Hauser H, Koster M. Nucleolar localization and mobility analysis of the NF-kappaB repressing factor NRF. J Cell Sci. 2004;117(Pt 16):3447–58. doi: 10.1242/jcs.01129. [DOI] [PubMed] [Google Scholar]
- Pederson T, Tsai RY. In search of nonribosomal nucleolar protein function and regulation. J Cell Biol. 2009;184(6):771–6. doi: 10.1083/jcb.200812014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard MN, Kern ER. The search for new therapies for human cytomegalovirus infections. Virus Res. 2011;157(2):212–21. doi: 10.1016/j.virusres.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard MN, Quenelle DC, Bidanset DJ, Komazin G, Chou S, Drach JC, Kern ER. Human cytomegalovirus UL27 is not required for viral replication in human tissue implanted in SCID mice. Virol J. 2006;3:18. doi: 10.1186/1743-422X-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitsma JM, Savaryn JP, Faust K, Sato H, Halligan BD, Terhune SS. Antiviral inhibition targeting the HCMV kinase pUL97 requires pUL27-dependent degradation of Tip60 acetyltransferase and cell-cycle arrest. Cell Host Microbe. 2011;9(2):103–14. doi: 10.1016/j.chom.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Tischer BK, Smith GA, Osterrieder N. En passant mutagenesis: a two step markerless red recombination system. Methods Mol Biol. 2010;634:421–30. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- Tsai RY, McKay RD. A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J Cell Biol. 2005;168(2):179–84. doi: 10.1083/jcb.200409053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16(5):369–77. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. MBV-S and MBV-R pUL27 localize to replication centers. HFFs were infected with T3580 or T3607 (Table 2). At 96 hpi, indirect immunofluorescence for pUL44 was performed, and the localization of pUL27-EGFP and pUL44 was determined by fluorescence microscop