Summary
Colorectal carcinoma continues to be a leading cause of cancer morbidity and mortality despite widespread adoption of screening methods. Targeted detection and therapy using recent advances in our knowledge of in vivo cancer biomarkers promise to significantly improve methods for early detection, risk stratification, and therapeutic intervention. The behavior of molecular targets in transformed tissues is being comprehensively assessed using new techniques of gene expression profiling and high throughput analyses. The identification of promising targets is stimulating the development of novel molecular probes, including significant progress in the field of activatable and peptide probes. These probes are being evaluated in small animal models of colorectal neoplasia and recently in the clinic. Furthermore, innovations in optical imaging instrumentation are resulting in the scaling down of size for endoscope compatibility. Advances in target identification, probe development, and novel instruments are progressing rapidly, and the integration of these technologies has a promising future in molecular medicine.
Keywords: colon, adenocarcinoma, targeted, molecular imaging, biomarker, endoscopy
Introduction
Colorectal cancer is the second leading cause of cancer related deaths in the western world, leading to approximately 148,800 new cases and 50,000 deaths annually in the US [1]. The morbidity and mortality associated with this disease may be significantly improved by developing better methods of early detection, risk stratification, and therapeutic monitoring [2,3]. Currently, early detection is performed using standard white light endoscopy to identify morphological changes, such as polyps, in the mucosa to guide tissue biopsy. However, the polyp miss rate, as determined by tandem colonoscopy, can be as high as 20% [4], and there is evidence that up to 10% of spontaneously occurring adenomas arise from sporadically occurring flat or depressed lesions that are not seen on standard endoscopy [5]. These lesions have a greater association with progression to carcinoma compared to that of polypoid neoplasms irrespective of size [6,7]. Furthermore, the presence of flat dysplasia in the setting of chronic ulcerative colitis presents a significantly increased risk for the development of colorectal cancer [8,9]. As a result, current screening guidelines recommend that random biopsies be collected every 10 cm, an approach that is limited by sampling error and requires a significant increase time, cost, and risk of the procedure [10]. Risk stratification is needed to provide an accurate prediction of the clinical and biological behavior of colorectal neoplasms. Currently risk is determined by factors related to the patient, including age, personal history, family history, and symptoms, and by those associated with the tumor, including depth of invasion, extent of differentiation, lymphatic and vascular invasion, and genetic features. Despite knowledge of all of these parameters, our ability to predict future progression is inadequate in most cases. Finally, therapeutic monitoring is important for assessing tumor response to chemotherapy. While the primary treatment for colorectal cancer is surgery, adjuvant chemotherapy can reduce the risk of developing recurrent or metastatic disease [11,12]. Improved diagnostic information is needed to determine how a tumor will respond to therapy and which therapies will be most effective. Patients who would not benefit from chemotherapy could be spared from associated toxicities.
Consequently, greater knowledge and better use of in vivo cancer biomarkers is greatly needed to improve outcomes. Tremendous progress in our understanding of the molecular mechanisms of cancer transformation has been made in recent years which can provide new insights into achieving this aim. There are several genetic pathways that are known to lead to colorectal cancer development. First, the adenoma-carcinoma sequence is a widely accepted model for cancer transformation in which normal mucosa progress to malignancy through a pre-malignant (dysplastic) phase that results from chromosomal instability or loss of heterozygosity, accounting for ∼80% of all sporadic colon cancers [13,14]. These tumors are characterized by mutations in specific genes, including the oncogenes k-ras, c-erb2, and c-myc, and the tumor suppressor genes APC and p53 [15]. Also, microsatellite instability may result from mutations in mismatch repair genes that lead to replication errors that occur during DNA synthesis [16,17]. These defects accumulate, resulting in cancer due to abnormal cell cycle regulation or apoptosis. Finally, the CpG island methylation phenotype (CIMP) occurs when aberrant methylation occurs in this promoter region and is associated with transcriptional inactivation of tumor suppressor genes [18,19]. Methylation affects genes that regulate the growth and differentiation of colonocytes, and stimulates hyperproliferative states that precede the development of colorectal cancer. Moreover, technological advances in high throughput gene expression profiles and a diverse library of antibodies for molecular markers have provided new approaches for evaluating the human genome to perform colon cancer staging and risk stratification [20-22].
In addition to human studies, the development of pre-clinical (mouse) models plays an important role in advancing our understanding of the molecular biology, environmental factors, and novel therapies for colorectal cancer. These animal models can be studied in a longitudinal fashion to reveal evolving details about key biological mechanisms involved in tumorigenesis over a shortened natural history. A number of spontaneous tumor models have been genetically engineered to mimic cancer syndromes in humans, such as hereditary non-polyposis colon cancer (HNPCC) and familial adenomatous polyposis (FAP) [23,24]. Mouse models are now being developed using conditional knockouts, regulated oncogenes, and tumor suppressor genes to more faithfully reproduce the process of sporadic tumor formation [25]. In these mice, somatic mutations are induced in a tissue-specific and time-controlled fashion [26] to provide new strategies for studying the role of various genes in the initiation, progression and treatment of colorectal cancer. The combination of gene knockouts of tumor suppressors have resulted in a number of models that cover the range from dysplastic lesions to invasive cancers and they correlate, in part, with common genetic alterations in sporadic cancers [27]. In addition, other transgenic mouse strains have modeled colon cancer development in the setting of inflammatory bowel disease [28]. These and other emerging models can be used to better understand the biological and environmental factors and to test the efficacy of novel therapeutic agents.
Molecular Biomarkers
Cancer biomarkers are expressed by transformed tissues rather than by normal tissues and not only reflect tumor phenotype but can also predict how the cancer will behave [29,30]. Recent advances in biotechnology have allowed for the development on new molecular biomarkers that enhance diagnostic accuracy, better predict patient outcome, accurately measure disease progression, and assess risk of recurrence. For example, gene expression profiling provides a systematic approach to examining potential biomarkers from a tumor specimen by providing a simultaneous analysis of a large number of genes for level of over-expression [31,32]. This method provides a comprehensive assessment of the molecular events involved in tumor development and progression, enabling profiles from a group of tumors to be analyzed and used to predict the natural history of the disease. Molecular changes in biomarker expression can also be detected much earlier than morphological differences in tissue histology. In addition, these measurements are quantitative and thus less prone to inter-observer variation than interpretation of histologic features. Biomarkers may also be used to determine the most effective type of therapy and to predict patient response to treatment. Currently, the use of single targeted biomarkers, such as vascular endothelial growth factor (VEGF) and epidermal growth factor (EGF) are promising; however, multiple biomarkers will likely to be needed for clinical use because cancer arises from many accumulated mutations.
Vascular Endothelial Growth Factor (VEGF)
VEGF stimulates angiogenesis and enhances the development of neoplasia by binding to tyrosine kinase receptors on the plasma membrane of cells [33,34]. VEGF plays an important role in the proliferation and migration of vascular endothelial cells which in turn feed the growth of tumors. Furthermore, both primary and metastatic colorectal neoplasms have been found to express high levels of VEGF and its receptor (VEGFR. The level of expression not only correlates with more advanced stages of cancer but with metastatic disease. [35]. Moreover, in one study, 30% of 121 colorectal cancer specimens were found to stain positive for VEGF, and the recurrence rate in patients with VEGF positive tumors was significantly higher than that for VEGF negative tumors (50% vs. 11%, p < 0.001). Higher levels of VEGF expression are also associated with increased recurrence, cancer-related mortality, and lymph node metastasis [36]. These findings have led the development of Bevacizumab (trade name Avastin), a monoclonal antibody therapy that targets VEGF in colorectal neoplasia. A number of clinical studies using anti-VEGF therapy in colorectal cancer have demonstrated clinical benefit [37].
Epidermal growth factor receptor (EGFR)
EGF also binds to a tyrosine kinase receptor on the plasma membrane and activates an intracellular signaling cascade that promotes cell adhesion, proliferation, differentiation, apoptosis, and metastasis [38]. EGFR is expressed in between 60 to 80% of all human colorectal cancers, and is activated by transforming growth factor α (TGFα) as well as EGF [39]. EGFR provides prognostic information about cancer staging, as demonstrated by 35% of 134 colorectal cancer specimens staining positive. Moreover, the Kaplan–Meier curve for recurrence demonstrated that this level of expression was associated with an increased risk of recurrence (p = 0.04), and multivariate analysis has indicated that high EGFR staining was an independent negative predictor of survival (p = 0.01) [40]. In another study, resected specimens from 126 patients who were EGFR positive had a >10 fold risk of cancer-related death in comparison to those which were EGFR negative, and this difference was maintained after multivariate analysis [41]. These findings have led the development of Cetuximab (trade name Erbitux), an FDA-approved chimeric monoclonal (IgG1) antibody, as therapy that binds to the extracellular domain of EGFR to inhibit activation by EGF and TGFα, resulting in the blockage of downstream signaling and impaired cell growth and proliferation [42,43]. Panitumumab (trade name Vectibix) is a human monoclonal IgG2 antibody also targeting EGFR and approved by the FDA. Clinical studies with both anti-EGFR therapies are being conducted to evaluate their efficacy in the treatment of colorectal cancer [44].
Gene Expression Profiling
Gene expression profiling measures the activity of the entire human genome within a tissue specimen at once, creating a global picture of tumor function. These profiles can identify biomarkers that are either upregulated downregulated in neoplastic tissues and assess response to therapy. The use of gene expression profiling for target identification, risk stratification, and therapeutic monitoring has the potential to overcome the heterogeneity of gene expression among cancer cells by performing a comprehensive analysis of the genome in order to elucidate the different mechanisms involved in tumor development [45]. The first study of gene expression profiling in colon cancer used the U133a GeneChip (Affymetrix, Santa Clara, CA) to evaluate ∼22,000 genes in tumors from 74 patients with Dukes' B cancer [46]. A panel of 23 genes was found to predict recurrence in a validation set of 36 patients with an overall performance accuracy of 78%. Correct predictions were made in 13 of 18 patients that relapsed and 15 of 18 patients that were disease free after treatment, resulting in an odds ratio of 13 (95% CI, 2.6 to 65; p = 0.003). These results demonstrated the clinical value of gene expression profiling to identify patients with an increased risk of relapse on adjuvant chemotherapy. More recent studies have expanded the capabilities of this approach to more comprehensive datasets, including identifying a separate 43 gene signature with a 32,000 gene microarray to predict survival from 78 colon cancer specimens [47]. This gene cluster was able to separate patients into a good prognostic group (survival > 36 months) and a poor prognostic group (survival < 36 months). For all stages of tumors, a prognostic accuracy of 90% was found and for the individual stages B, C, and D, the results were 87%, 90%, and 91%, respectively. This gene set did a better job of risk stratifying patients into good and poor prognostic groups than did the conventional Dukes' B and C staging (p = 0.04).
In Vivo Molecular Imaging Instruments
Small Animal Molecular Imaging Systems
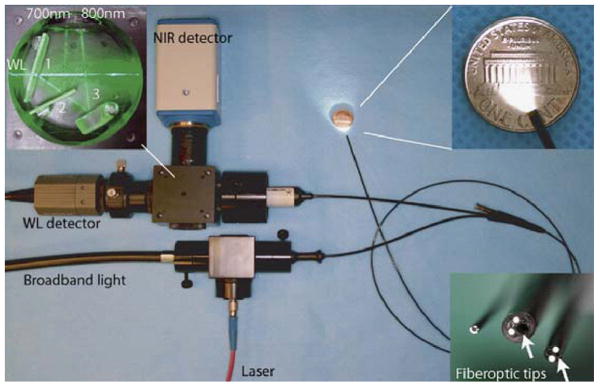
Small animal molecular imaging systems are useful for observing the onset and progression of neoplastic transformation after a prolonged period of latency, and endoscopic methods can be used to guide the collection of tissue specimens. The ability of small animal endoscopy to detect mucosal changes without requiring animal sacrifice can reduce the numbers needed to perform a statistically significant study by accurately timing the onset of disease and by using each animal as its own control, allowing for robust longitudinal studies related to chemoprevention and therapeutic intervention. Endoscopic instruments have been developed to image the colon in genetically engineered mice over organ level surface areas to guide tissue biopsy and to perform sequential in vivo studies [48-50]. In addition, small animal molecular imaging systems that use near-infrared (NIR) light have been developed to reduce tissue autofluorescence and to increase image contrast. For example, a flexible endoscope has been adapted to collected NIR fluorescence in two channels [48]. Broadband illumination is provided by a 300 W xenon lamp, and fluorescence excitation is generated by a 736 nm GaAs laser diode. Both beams are focused into the illumination port of the endoscope. A modified fiber optic angioscope (1.4 mm diameter) collects both the white light and NIR fluorescence images, as shown in Fig. 1. The schematic for spectral processing is shown in the top left inset. The first dichroic mirror transmits white light from the endoscope and reflects the broad band NIR fluorescence. The second dichroic transmits and reflects fluorescence in the upper (750 to 800 nm) and lower (675 to 725 nm) NIR bands, respectively. The lower right inset shows several different angioscopes with diameters of 0.8, 1.6, and 2.4 mm. A color video camera (WL) and a NIR sensitive CCD detector are used to collect the reflected white light and NIR fluorescence, respectively.
Fig. 1. Near-Infrared Small Animal Endoscope.
A modified fiber optic angioscope (1.4 mm diameter) collects both the white light and near-infrared (NIR) fluorescence images. The schematic for spectral processing is shown in the top left inset. The first dichroic mirror transmits white light from the endoscope and reflects the broad band near-infrared fluorescence. The second dichroic transmits and reflects fluorescence in the upper (750 to 800 nm) and lower (675 to 725 nm) NIR bands, respectively. The lower right inset shows several different angioscopes with diameters of 0.8, 1.6, and 2.4 mm.
Clinical Molecular Imaging Systems
Clinical systems for molecular imaging of the colon have become available as a result of several recent technological advancements in fluorescence detection that are compatible with medical endoscopes and are sensitive to FDA-approved fluorescence contrast agents. Endoscopes are available that image in several modes, including white light and fluorescence, to observe macroscopic surface areas in real time [51-53]. In white light mode, the full visible spectrum (400 to 700 nm) from a xenon lamp is delivered through the two light guides onto the tissue and the reflected light is collected by a CCD detector, as shown in Fig 2. In the fluorescence mode, a filter wheel flips into the illumination path and provides fluorescence excitation in the 395 to 475 nm spectral band [51]. In addition, illumination from 525 to 575 nm provides reflected light in the green wavelengths modulated by the absorption features of tissue hemoglobin. Fluorescence images are collected by a second CCD detector located on the distal tip of the endoscope that has a 490–625 nm band pass filter for blocking excitation light. Autofluorescence from normal mucosa appears as green, and that from neoplasia appears as magenta. Furthermore, hemoglobin absorbs both the fluorescence and excitation light, thus pre-malignant mucosa, which contains more hemoglobin due to increased vascularization, will appear with reduced green intensity.
Fig 2. Multi-modal in vivo imaging system.
A prototype medical endoscope provides standard wide area imaging with white light and is also sensitive to fluorescence for localization of probe binding. A confocal microscope can pass through the instrument channel of this endoscope to visualize below the mucosal surface to perform validation of probe binding.
Techniques of confocal microscopy have also been scaled down in size for compatibility with medical endoscopes so that optical sectioning can be performed below the tissue surface, as shown in Fig 2. This approach uses an optical fiber as a pinhole, placed in between the objective lens and the detector, to allow only the light that originates from within a tiny volume below the mucosal surface to be collected [54]. All other sources of scattered light do not have the correct path to be detected, and thus become ‘spatially filtered.’ These images can be collected at sufficiently fast frame rates using high speed scanning mechanisms to observe biological behavior with minimal disturbance from motion artifacts caused by patient peristalsis. The Cellvizio®-GI confocal imaging system(Mauna Kea Technologies, Paris, France) is one such system that consists of a flexible (1.5 mm diameter) miniprobe, control unit, and processing software [55]. A 488 nm (peak absorption of fluorescein) semiconductor laser delivers the excitation beam to a 4 kHz oscillating mirror for horizontal scanning (lines) and then to a 12 Hz galvanometer mirror for vertical scanning (frames) [56]. The mirrors raster scan the beam across the proximal face of an imaging fiber bundle that contains ∼30,000 optical fibers (1.9 μm core diameter, 3.3 μm average intercore spacing). A gradient index (GRIN) microlens is located at the distal end to focus the beam. The miniprobes have zero or 50 μm working distance with a corresponding lateral resolution of either 5 or 2.5 μm and an axial resolution of either 15 or 20 μm, respectively. Images are collected in a horizontal plane (en face) at 12 frames per second with a field of view of either 600×500 or 240×200 μm2. Fluorescence is collected by the same lens, and refocused back into the illumination fiber. The cores of the fiber act as collection pinholes for rejecting out of focus light to perform optical sectioning. A long pass filter rejects the excitation light, and fluorescence is detected with an avalanche photodiode. Image processing performed includes subtraction of fiber autofluorescence and calibration of individual fiber transmission efficiencies.
In Vivo Molecular Probes
New methods for the early detection of cancer can be greatly enhanced by the use of molecular probes that are specific to cancer biomarkers in vivo. A wide variety of different strategies for developing in vivo molecular probes are being pursued. For example, monoclonal antibodies and antibody fragments have been developed to target tumor-associated biomarkers for the detection of solid tumors and tumor-associated angiogenesis [57-59]. However, these approaches have been met with limited success for a variety of reasons, including insufficient probe delivery, limited target-to-background specificity, and immunogenicity. New methods are being developed to increase probe localization and improve binding, including activatable probes and tumor specific peptides.
Activatable Probes
There have been a number of fluorescent probes developed that produce near-infrared light to maximize tissue imaging depth and to minimize interference from autofluorescence in vivo in small animal models [60-62]. These near-infrared labeled molecular beacons are activated by proteases that cleave lysine bonds, resulting in signal amplification of 6 to 20 fold. Cathepsin B is an example of a protease that cleaves (activates) this probe in vivo. Various versions of this probe have been used in animal models for cancer detection and imaging of inflammatory response. A non-activatable probe used for control purposes consists of a magnetofluorescent nanoparticle labeled with Cy5.5, a monofunctional dye located adjacent to lysine cleavage sites on a macromolecular assembly, and acts in the intravascular space with a plasma half-life of ∼10 hours. The assembly consists of a synthetic graft copolymer containing partially pegylated poly-L-lysine. The injection dose of 2 nmol per animal and time of imaging after injection has been optimized in non–APC min mice. The use of this activatable probe has been demonstrated ex vivo in Apcmin/+ mice, where cathepsin B was found to be over-expressed in adenomas with a target-to-background ratio of 2.2±1 on NIR images collected, and lesions as small as 50 μm were detected [63]. This study demonstrates the potential for use of cathepsin B as an in vivo cancer biomarker.
Peptide Probes
Peptides have tremendous advantages for performing targeted detection and therapy in the colon of small animal models and human subjects because of their high diversity, rapid binding kinetics, and potential for deep diffusion in diseased mucosa [64-66]. In addition, peptides can be labeled easily, are generally non-toxic, and not immunogenic. Peptide probes are particularly attractive for clinical use in the digestive tract where the luminal surface can be easily accessed by medical endoscopes and methods of topical administration can be used safely. These probes have been developed using techniques of phage display, a powerful combinatorial method that uses recombinant DNA technology to generate a library of peptides that bind preferentially to the cell surface. The protein coat of bacteriophage, such as the filamentous M13 or T7, is genetically engineered to express a very large number (>109) of different peptides with unique sequences. Selection of sequences with affinity binding is then performed by biopanning the library against cultured cells that over-express desired targets. The DNA sequences are then recovered and used to synthesize the candidate peptides. Techniques of phage display have been successfully used to identify peptides that bind preferentially to dysplastic colonic mucosa and not to normal mucosa by employing a biopanning strategy that uses cultured cells and freshly excised normal and dysplastic tissue. First, non-specific binding phage are removed from the library by biopanning against ‘normal’ (non-transformed) human columnar intestinal cells. These cells exhibit morphology similar to that of normal colonic epithelium, and express a large number of non-specific cell surface antigens. Over 97% of the phage can be removed after three rounds of biopanning. The unbound phage was collected and then biopanned against a total of ten consecutive specimens of human colonic adenoma immediately upon excision, and yielded numerous candidate phage.
In Vivo Imaging Studies
Activatable Probes
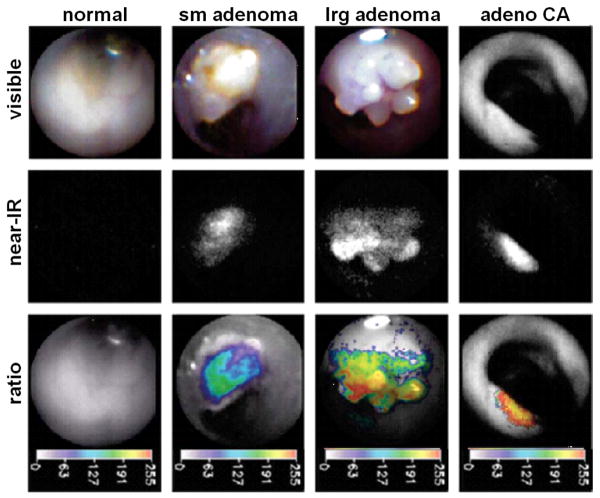
Molecular imaging has been performed using intravenously delivered activatable probes to evaluate colon tumors with real-time endoscopy in mouse models. In Fig. 3, in vivo white light images of normal mucosa, small adenoma, large adenoma, and adenocarcinoma collected with a 1.4 mm diameter angioscope in the colon of APCmin/+ mice are shown in the first row,. In the second row, NIR images from the same mucosal regions are collected after i.v. injection of the protease-activatable probe and reveal increased fluorescence intensity. In the third row, the ratio of the NIR images collected after injection of protease-activatable and non-activatable probes are shown in pseudocolor. This ratio corrects for differences in object distance, collection angle, tissue reflectance and probe delivery. The colons were resected after animal sacrifice and these regions of tissue were found on immunohistochemistry to demonstrate increased levels of cathepsin B expression in a progressive manner from normal colonic mucosa to dysplasia to adenocarcinoma. Furthermore, tumors in non-injected animals were not detectable on NIR fluorescence.
Fig. 3. Endoscopic images from APCmin/+ mouse.
In the first row, in vivo white light images of normal mucosa, small adenoma, large adenoma, and adenocarcinoma collected with a 1.4 mm diameter angioscope in the colon of APCmin/+ mice are shown. In the second row, NIR images from the same mucosal regions are collected after i.v. injection of the protease-activatable probe and reveal increased fluorescence intensity. In the third row, the ratio of the NIR images collected after injection of protease-activatable and non-activatable probes are shown in pseudocolor. This ratio corrects for differences in object distance, collection angle, tissue reflectance and probe delivery. Increased intensity on the ratio images was found to correlated to cathepsin B expression on immunohistochemistry.
Peptide Probes
Molecular imaging has also been performed using topically administered peptide probes using genetically engineered mice that develop adenomas in the distal colon. Mutations in the adenomatous polyposis coli (APC) gene are important in colorectal tumorigenesis; however, existing mouse intestinal tumor models display mainly small intestinal lesions and carcinomas are rare. A new mouse model of colon cancer has been developed that can express dysplastic polyps in the distal colon that can be easily imaged with a small animal endoscope. These mice that carry transgenes regulated by 9.5 kb fragment containing human CDX2 homeobox gene promoter and upstream flanking elements (CDX2P9.5), and have been shown to tightly restrict transgene expression in the mucosa of the distal small intestine, cecum, and colon of adult mice [67,68]. As a result, a CDX2P9.5-regulated Cre transgene with a long mononucleotide tract altering the reading frame can lead to colonic polyposis and tumor formation after about 30 weeks of age. The CDX2 sequences confer preferential transgene expression in colonic epithelium in the adult mouse, and mice carrying a CDX2P-NLS-Cre recombinase transgene and a loxP-targeted Apc allele developed mainly colon tumors, with carcinomas seen in 6 of 36 (17%) mice after 300 days. Like human colorectal lesions, the mouse tumors showed bi-allelic Apc inactivation, β-catenin dysregulation, global DNA hypomethylation, and chromosomal instability.
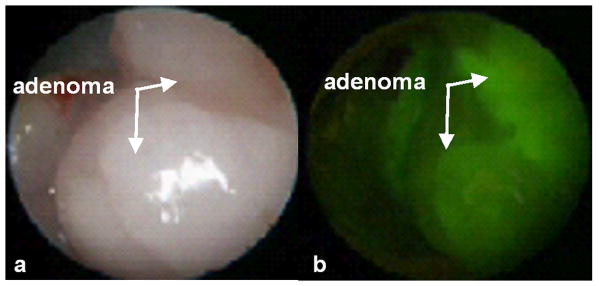
Molecular imaging has been performed using topically administered peptide probes to target pre-malignant colonic mucosa with real-time endoscopy. The predominantly distal distribution of tumors in affected CDX2 mice implies that somatic defects promoting clonal outgrowth of epithelial cells with a single Apc-mutant allele occur in a non-uniform fashion in the colon. The distal expression of colonic neoplasia in this genetically engineered mouse model allows for validation of peptide binding to colonic adenomas in vivo. Numerous adenomas that range in size from 2 to 5 mm can be found in the distal colon, and additional tumors can be seen in the small bowel. A small animal endoscope for imaging the mouse colon has been used with visible light, and consists of a 9.5 Fr (3 mm) diameter rigid Hopkins II 0 deg telescope with a 11.5 cm working length and a 3 Fr (1 mm) diameter instrument channel for performing tissue biopsy (Karl Storz Veterinary Endoscopy, Goleta, CA). Fluorescence excitation is produced with a 450 to 475 nm passband filter that can be manually switched into the optical path of a xenon (175 W) light source, and is delivered to the endoscope via a fluid light cable. Fluorescence images are collected with 510 nm barrier filter to block the excitation light, detected with a CCD camera, and digitized as a real time video. The distal colon and rectum of the mouse was first cleaned of stool and debris by performing tap water lavage, and then the distal end of the small animal endoscope was lubricated and inserted into the rectum of the mouse. Once adenomas were identified on white light, shown in Fig. 4a, a 1 ml solution of FITC-labeled target peptide “VRPMPLQ” at a concentration of 10 μM was applied through the instrument channel with a syringe, allowed to incubate for ∼10 minutes, and the unbound peptide was gently rinsed off with tap water. The corresponding fluorescence image, shown in Fig. 4b, demonstrates peptide binding to the adenomas. The rectum was then rinsed vigorously with PBS. Complete removal of the peptide from the rectal mucosa was observed on fluorescence. FITC-labeled scrambled (control) peptide was administered at the same concentration (10 μM) and did not reveal peptide binding. These results demonstrate the feasibility of performing in vivo targeted imaging in small animal models of colon cancer.
Fig. 4. Targeted in vivo images from adenoma in distal colon.
a) White light endoscopy shows spontaneous adenomas in the distal colon. b) Fluorescence image demonstrates increased intensity from binding with target peptide.
Macroscopic Peptide Imaging
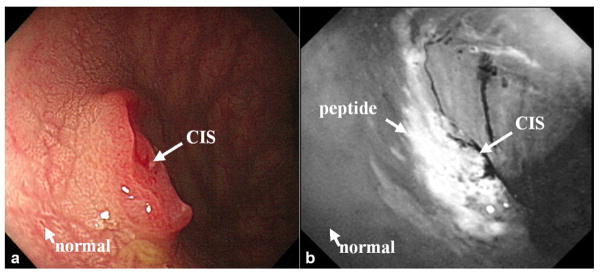
Fluorescence-labeled peptides can also be used for localizing pre-malignant mucosa in the colon that is difficult to visualize on macroscopic imaging in the clinic. These approaches are needed to screen large surface areas during routine endoscopy for further evaluation, such as by confocal microscopy. A study that included adult patients previously scheduled for elective outpatient screening colonoscopy has been performed to demonstrate the use of fluorescence-labeled target peptides. Each subject recruited into the study was required to obtain pre- and post-procedure blood tests to monitor for potential peptide toxicity. In Fig. 5a, a standard white light image shows a sessile mass approximately 10 mm in diameter, later found on histology to be carcinoma-in-situ (CIS). The lesion dimensions were assessed using the span of an opened biopsy forceps, and validated with a ruler after resection. The peptide targeted image, shown in Fig. 5b, and reveals increased fluorescence intensity at the site of the lesion compared to that of the adjacent normal mucosa. An average signal-to-noise ratio of 12±5 and a mean target-to-background ratio of 1.5±0.5 were found for n = 7 adenomas. The detection criterion for each site was compared to a threshold intensity, and designated positive if greater (negative if less). A sensitivity of 71% and specificity of 89% for detection was found using an intensity threshold of 1.20. Furthermore, no toxicities associated with peptide administration were observed in any of the patients based on follow up blood test results, defined by parameters > 20% above baseline, and by results of patient interviews.
Fig 5. Targeted macroscopic imaging in vivo.
Endoscopic images collected in vivo of carcinoma-in-situ (CIS) on a) white-light and b) fluorescence after target peptide was topically administered reveal increased fluorescence intensity at the site of the lesion with an average target-to-background ratio of 1.5.
Microscopic Peptide Imaging
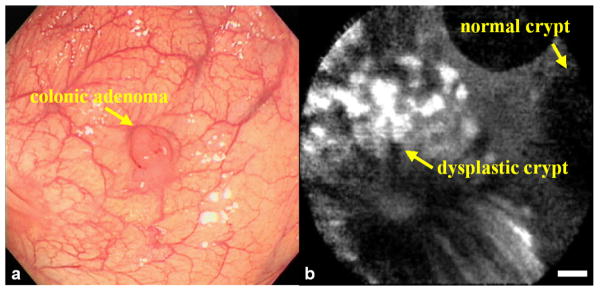
Binding of fluorescent-labeled peptides to colonic dysplasia in vivo on the microscopic scale with confocal microscopy has also been demonstrated. As before, adult patients who are already scheduled for elective outpatient screening colonoscopy were recruited for the study. Upon detection of a suspicious lesion, the confocal microscope was passed through the instrument channel of a standard colonoscope, and fluorescence videos were collected. In Fig. 6a, the conventional white light endoscopic image of the colonic adenoma shows a raised lesion, and in Fig. 6b, the confocal fluorescence image shows significant peptide binding to colonocytes in the dysplastic but not in the normal crypts. The target-to-background ratio for peptide binding from dysplastic and adjacent normal crypts was measured from the confocal images that met the following criteria: 1) minimum motion artifact, 2) lack of stool, debris, or excess mucus obscuring the image, and 3) the presence of crypt morphology. A mean target-to-background ratio of 18±4 was calculated for the target peptide “VRPMPLQ” from a total of n = 18 adenomas. The detection criterion for each site was compared to a threshold intensity and designated positive if greater (negative if less), and a sensitivity of 81% and specificity of 82% were achieved. As with the previous study, no toxicity associated with peptide administration was observed in any patients. This study demonstrated the feasibility of using topically administered peptides to target pre-malignant mucosa in the colon in vivo. The use of multiple peptides that bind to independent targets has potential to achieve even better results.
Fig 6. Targeted microscopic imaging in vivo.
a) Conventional white light endoscopic image of colonic adenoma, and b) in vivo confocal fluorescence image collected after topical administration of FITC-labeled target peptide shows preferential binding to dysplastic colonocytes. The dysplasia:normal border shows an average target-to-background ratio of 21, scale bars 20 μm.
Future Directions
In summary, a number of promising cancer biomarkers have been identified using new discoveries in biotechnology and novel techniques of molecular imaging have been developed for in vivo surveillance of neoplasia in the colon. Targeted detection and therapy in patients at increased risk of developing cancer represents an exciting new direction in the field of medicine. Before widespread use of these techniques can have an impact in clinical practice, greater progress is needed in the understanding of in vivo cancer biomarkers, development of molecular probes, and maturation of imaging technologies. In particular, the relationship between the expression of in vivo biomarkers and the natural history of colonic neoplasia needs to be better worked out so that risk stratification can be performed. Moreover, probes that bind to molecular targets with high specificity, low background, and minimal toxicity require further development, and delivery mechanisms that are simple, efficient, and effective are needed. Moreover, multi-modal imaging technologies that combine wide area surveillance to rapidly evaluate large surface areas for probe localization with high resolution techniques to observe sub-cellular features for validating probe binding are needed. Thereafter, well-designed multi-center, randomized controlled clinical trials are needed to validate and standardize these integrated imaging strategies. Thus, the use of in vivo cancer biomarkers has great potential to improve methods for the early detection of neoplasia, increase the efficacy of surveillance, and ultimately to improve patient outcomes.
References
- 1.American Cancer Society. Cancer Facts & Figures 2008. Atlanta: American Cancer Society; 2008. [Google Scholar]
- 2.Vogelaar I, van Ballegooijen M, Schrag D, et al. How much can current interventions reduce colorectal cancer mortality in the U.S.? Mortality projections for scenarios of risk-factor modification, screening, and treatment. Cancer. 2006;107:1624–33. doi: 10.1002/cncr.22115. [DOI] [PubMed] [Google Scholar]
- 3.O'brien MJ, Winawer SJ, Zauber AG, et al. National Polyp Study Workgroup. Flat adenomas in the National Polyp Study: is there increased risk for high-grade dysplasia initially or during surveillance? Clin Gastroenterol Hepatol. 2004;2:905–11. doi: 10.1016/s1542-3565(04)00392-1. [DOI] [PubMed] [Google Scholar]
- 4.van Rijn JC, Reitsma JB, Stoker J, Bossuyt PM, van Deventer SJ, Dekker E. Polyp miss rate determined by tandem colonoscopy: a systematic review. Am J Gastroenterol. 2006;101:343–50. doi: 10.1111/j.1572-0241.2006.00390.x. [DOI] [PubMed] [Google Scholar]
- 5.Soetikno RM, Kaltenbach T, Rouse RV, Park W, Maheshwari A, Sato T, Matsui S, Friedland S. Prevalence of nonpolypoid (flat and depressed) colorectal neoplasms in asymptomatic and symptomatic adults. JAMA. 2008;299:1027–35. doi: 10.1001/jama.299.9.1027. [DOI] [PubMed] [Google Scholar]
- 6.Hurlstone DP, Cross SS, Adam I, Shorthouse AJ, Brown S, Sanders DS, Lobo AJ. A prospective clinicopathological and endoscopic evaluation of flat and depressed colorectal lesions in the United Kingdom. Am J Gastroenterol. 2003;98:2543–9. doi: 10.1111/j.1572-0241.2003.07679.x. [DOI] [PubMed] [Google Scholar]
- 7.O'brien MJ, Winawer SJ, Zauber AG, Bushey MT, Sternberg SS, Gottlieb LS, Bond JH, Waye JD, Schapiro M National Polyp Study Workgroup. Flat adenomas in the National Polyp Study: is there increased risk for high-grade dysplasia initially or during surveillance? Clin Gastroenterol Hepatol. 2004;2:905–11. doi: 10.1016/s1542-3565(04)00392-1. [DOI] [PubMed] [Google Scholar]
- 8.Jaramillo E, Watanabe M, Befrits R, Ponce de Leon E, Rubio C, Slezak P. Small, flat colorectal neoplasias in long-standing ulcerative colitis detected by high-resolution electronic video endoscopy. Gastrointest Endosc. 1996;44:15–22. doi: 10.1016/s0016-5107(96)70223-7. [DOI] [PubMed] [Google Scholar]
- 9.von Roon AC, Reese G, Teare J, Constantinides V, Darzi AW, Tekkis PP. The risk of cancer in patients with Crohn's disease. Dis Colon Rectum. 2007;50:839–55. doi: 10.1007/s10350-006-0848-z. [DOI] [PubMed] [Google Scholar]
- 10.Judge TA, Lewis JD, Lichtenstein GR. Colonic dysplasia and cancer in inflammatory bowel disease. Gastrointest Endosc Clin N Am. 2002;12:495–523. doi: 10.1016/s1052-5157(02)00014-4. [DOI] [PubMed] [Google Scholar]
- 11.Labianca R, Milesi L, Mosconi S, Pessi MA, Beretta GD, Quadri A. The role of adjuvant chemotherapy in colon cancer. Surg Oncol. 2007;16(S1):S93–6. doi: 10.1016/j.suronc.2007.10.042. [DOI] [PubMed] [Google Scholar]
- 12.Monga DK, O'Connell MJ. Surgical adjuvant therapy for colorectal cancer: current approaches and future directions. Ann Surg Oncol. 2006;13:1021–34. doi: 10.1245/ASO.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 13.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–32. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 14.Chung DC. The genetic basis of colorectal cancer: insights into critical pathways of tumorigenesis. Gastroenterology. 2000;119:854–65. doi: 10.1053/gast.2000.16507. [DOI] [PubMed] [Google Scholar]
- 15.Cho KR, Vogelstein B. Genetic alterations in the adenoma--carcinoma sequence. Cancer. 1992;70(6S):1727–31. doi: 10.1002/1097-0142(19920915)70:4+<1727::aid-cncr2820701613>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe T, Kobunai T, Toda E, et al. Distal colorectal cancers with microsatellite instability (MSI) display distinct gene expression profiles that are different from proximal MSI cancers. Cancer Res. 2006;66:9804–8. doi: 10.1158/0008-5472.CAN-06-1163. [DOI] [PubMed] [Google Scholar]
- 17.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–7. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 18.Rhee I, Jair KW, Yen RW, Lengauer C, Herman JG, Kinzler KW, Vogelstein B, Baylin SB, Schuebel KE. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature. 2000;404:1003–7. doi: 10.1038/35010000. [DOI] [PubMed] [Google Scholar]
- 19.Fox EJ, Leahy DT, Geraghty R, Mulcahy HE, Fennelly D, Hyland JM, O'Donoghue DP, Sheahan K. Mutually exclusive promoter hypermethylation patterns of hMLH1 and O6-methylguanine DNA methyltransferase in colorectal cancer. J Mol Diagn. 2006;8:68–75. doi: 10.2353/jmoldx.2006.050084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Jatkoe T, Zhang Y, Mutch MG, Talantov D, Jiang J, McLeod HL, Atkins D. Gene expression profiles and molecular markers to predict recurrence of Dukes' B colon cancer. J Clin Oncol. 2004;22:1564–71. doi: 10.1200/JCO.2004.08.186. [DOI] [PubMed] [Google Scholar]
- 21.Giacomini CP, Leung SY, Chen X, Yuen ST, Kim YH, Bair E, Pollack JR. A gene expression signature of genetic instability in colon cancer. Cancer Res. 2005;65:9200–5. doi: 10.1158/0008-5472.CAN-04-4163. [DOI] [PubMed] [Google Scholar]
- 22.Barrier A, Lemoine A, Boelle PY, et al. Colon cancer prognosis prediction by gene expression profiling. Oncogene. 2005;24:6155–64. doi: 10.1038/sj.onc.1208984. [DOI] [PubMed] [Google Scholar]
- 23.Su LK, Kinzler KW, Vogelstein B, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–70. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 24.Edelmann W, Cohen PE, Kneitz B, et al. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat Genet. 1999;21:123–7. doi: 10.1038/5075. [DOI] [PubMed] [Google Scholar]
- 25.Van Dyke T, Jacks T. Cancer modeling in the modern era: progress and challenges. Cell. 2002;108:135–44. doi: 10.1016/s0092-8674(02)00621-9. [DOI] [PubMed] [Google Scholar]
- 26.Jonkers J, Berns A. Conditional mouse models of sporadic cancer. Nat Rev Cancer. 2002;2:251–65. doi: 10.1038/nrc777. [DOI] [PubMed] [Google Scholar]
- 27.Heyer J, Yang K, Lipkin M, Edelmann W, Kucherlapati R. Mouse models for colorectal cancer. Oncogene. 1999;18:5325–33. doi: 10.1038/sj.onc.1203036. [DOI] [PubMed] [Google Scholar]
- 28.Bhan AK, Mizoguchi E, Smith RN, Mizoguchi A. Colitis in transgenic and knockout animals as models of human inflammatory bowel disease. Immunol Rev. 1999;169:195–207. doi: 10.1111/j.1600-065x.1999.tb01316.x. [DOI] [PubMed] [Google Scholar]
- 29.Garcea G, Sharma RA, Dennison A, Steward WP, Gescher A, Berry DP. Molecular biomarkers of colorectal carcinogenesis and their role in surveillance and early intervention. Eur J Cancer. 2003;39:1041–52. doi: 10.1016/s0959-8049(03)00027-3. [DOI] [PubMed] [Google Scholar]
- 30.Syngal S, Clarke G, Bandipalliam P. Potential roles of genetic biomarkers in colorectal cancer chemoprevention. J Cell Biochem Suppl. 2000;34:28–34. doi: 10.1002/(sici)1097-4644(2000)77:34+<28::aid-jcb7>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 31.Barrier A, Boelle PY, Lemoine A, et al. Gene expression profiling of nonneoplastic mucosa may predict clinical outcome of colon cancer patients. Dis Colon Rectum. 2005;48:2238–48. doi: 10.1007/s10350-005-0175-9. [DOI] [PubMed] [Google Scholar]
- 32.Arango D, Laiho P, Kokko A, et al. Gene-expression profiling predicts recurrence in Dukes' C colorectal cancer. Gastroenterology. 2005;129:874–84. doi: 10.1053/j.gastro.2005.06.066. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi Y, Kitadai Y, Bucana CD, Cleary KR, Ellis LM. Expression of vascular endothelial growth factor and its receptor, KDR, correlates with vascularity, metastasis, and proliferation of human colon cancer. Cancer Res. 1995;55:3964–8. [PubMed] [Google Scholar]
- 34.Ishigami SI, Arii S, Furutani M, Niwano M, Harada T, Mizumoto M, Mori A, Onodera H, Imamura M. Predictive value of vascular endothelial growth factor (VEGF) in metastasis and prognosis of human colorectal cancer. Br J Cancer. 1998;78:1379–84. doi: 10.1038/bjc.1998.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ono T, Miki C. Factors influencing tissue concentration of vascular endothelial growth factor in colorectal carcinoma. Am J Gastroenterol. 2000;95:1062–7. doi: 10.1111/j.1572-0241.2000.01909.x. [DOI] [PubMed] [Google Scholar]
- 36.Cascinu S, Staccioli MP, Gasparini G, Giordani P, Catalano V, Ghiselli R, Rossi C, Baldelli AM, Graziano F, Saba V, Muretto P, Catalano G. Expression of vascular endothelial growth factor can predict event-free survival in stage II colon cancer. Clin Cancer Res. 2000;6:2803–7. [PubMed] [Google Scholar]
- 37.Los M, Roodhart JM, Voest EE. Target practice: lessons from phase III trials with bevacizumab and vatalanib in the treatment of advanced colorectal cancer. Oncologist. 2007;12:443–50. doi: 10.1634/theoncologist.12-4-443. [DOI] [PubMed] [Google Scholar]
- 38.Chen WS, Lazar CS, Poenie M, Tsien RY, Gill GN, Rosenfeld MG. Requirement for intrinsic protein tyrosine kinase in the immediate and late actions of the EGF receptor. Nature. 1987;328:820–3. doi: 10.1038/328820a0. [DOI] [PubMed] [Google Scholar]
- 39.Galizia G, Ferraraccio F, Lieto E, et al. Prognostic value of p27, p53, and vascular endothelial growth factor in Dukes A and B colon cancer patients undergoing potentially curative surgery. Dis Colon Rectum. 2004;47:1904–14. doi: 10.1007/s10350-004-0695-8. [DOI] [PubMed] [Google Scholar]
- 40.Resnick MB, Routhier J, Konkin T, Sabo E, Pricolo VE. Epidermal growth factor receptor, c-MET, beta-catenin, and p53 expression as prognostic indicators in stage II colon cancer: a tissue microarray study. Clin Cancer Res. 2004;10:3069–75. doi: 10.1158/1078-0432.ccr-03-0462. [DOI] [PubMed] [Google Scholar]
- 41.Galizia G, Lieto E, Ferraraccio F, De Vita F, Castellano P, Orditura M, Imperatore V, La Mura A, La Manna G, Pinto M, Catalano G, Pignatelli C, Ciardiello F. Prognostic significance of epidermal growth factor receptor expression in colon cancer patients undergoing curative surgery. Ann Surg Oncol. 2006;13:823–35. doi: 10.1245/ASO.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 42.Jonker DJ, O'Callaghan CJ, Karapetis CS, et al. Cetuximab for the treatment of colorectal cancer. N Engl J Med. 2007;357:2040–8. doi: 10.1056/NEJMoa071834. [DOI] [PubMed] [Google Scholar]
- 43.Cunningham D, Humblet Y, Siena S, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–45. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 44.Wu M, Rivkin A, Pham T. Panitumumab: Human monoclonal antibody against epidermal growth factor receptors for the treatment of metastatic colorectal cancer. Clin Ther. 2008;30:14–30. doi: 10.1016/j.clinthera.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 45.Frederiksen CM, Knudsen S, Laurberg S, Ørntoft TF. Classification of Dukes' B and C colorectal cancers using expression arrays. J Cancer Res Clin Oncol. 2003;129:263–71. doi: 10.1007/s00432-003-0434-x. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Jatkoe T, Zhang Y, Mutch MG, Talantov D, Jiang J, McLeod HL, Atkins D. Gene expression profiles and molecular markers to predict recurrence of Dukes' B colon cancer. J Clin Oncol. 2004;22:1564–71. doi: 10.1200/JCO.2004.08.186. [DOI] [PubMed] [Google Scholar]
- 47.Eschrich S, Yang I, Bloom G, et al. Molecular staging for survival prediction of colorectal cancer patients. J Clin Oncol. 2005;23:3526–35. doi: 10.1200/JCO.2005.00.695. [DOI] [PubMed] [Google Scholar]
- 48.Funovics MA, Alencar H, Montet X, Weissleder R, Mahmood U. Simultaneous fluorescence imaging of protease expression and vascularity during murine colonoscopy for colonic lesion characterization. Gastrointest Endosc. 2006;64:589–97. doi: 10.1016/j.gie.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 49.Alencar H, Funovics MA, Figueiredo J, Sawaya H, Weissleder R, Mahmood U. Colonic adenocarcinomas: near-infrared microcatheter imaging of smart probes for early detection--study in mice. Radiology. 2007;244:232–8. doi: 10.1148/radiol.2441052114. [DOI] [PubMed] [Google Scholar]
- 50.Funovics MA, Alencar H, Su HS, Khazaie K, Weissleder R, Mahmood U. Miniaturized multichannel near infrared endoscope for mouse imaging. Mol Imaging. 2003;2:350–7. doi: 10.1162/15353500200303166. [DOI] [PubMed] [Google Scholar]
- 51.Uedo N, Iishi H, Tatsuta M, et al. A novel videoendoscopy system by using autofluorescence and reflectance imaging for diagnosis of esophagogastric cancers. Gastrointest Endosc. 2005;62:521–8. doi: 10.1016/j.gie.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 52.Izuishi K, Tajiri H, Fujii T, Boku N, Ohtsu A, Ohnishi T, Ryu M, Kinoshita T, Yoshida S. The histological basis of detection of adenoma and cancer in the colon by autofluorescence endoscopic imaging. Endoscopy. 1999;31:511–6. doi: 10.1055/s-1999-57. [DOI] [PubMed] [Google Scholar]
- 53.Uedo N, Higashino K, Ishihara R, Takeuchi Y, Iishi H. Digestive Endoscopy. 2007;19(S1):S134–S138. doi: 10.1055/s-2007-966834. [DOI] [PubMed] [Google Scholar]
- 54.Pawley JB, editor. Handbook of Biological Confocal Microscopy. 3rd. Springer Verlag; New York: 2006. [Google Scholar]
- 55.Laemmel E, Genet M, Le Goualher G, Perchant A, Le Gargasson JF, Vicaut E. Fibered confocal fluorescence microscopy (Cell-viZio) facilitates extended imaging in the field of microcirculation. A comparison with intravital microscopy. J Vasc Res. 2004;41:400–11. doi: 10.1159/000081209. [DOI] [PubMed] [Google Scholar]
- 56.Wang TD, Friedland S, Sahbaie P, Soetikno R, Hsiung PL, Liu JT, Crawford JM, Contag CH. Functional imaging of colonic mucosa with a fibered confocal microscope for real-time in vivo pathology. Clin Gastroenterol Hepatol. 2007;5:1300–5. doi: 10.1016/j.cgh.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baum RP, Brümmendorf TH. Radioimmunolocalization of primary and metastatic breast cancer. Q J Nucl Med. 1998;42:33–42. [PubMed] [Google Scholar]
- 58.Dessureault S, Koven I, Reilly RM, et al. Pre-operative assessment of axillary lymph node status in patients with breast adenocarcinoma using intravenous 99mtechnetium mAb-170H.82 (Tru-Scint AD) Breast Cancer Res Treat. 1997;45:29–37. doi: 10.1023/a:1005878113826. [DOI] [PubMed] [Google Scholar]
- 59.Neri D, Carnemolla B, Nissim A, et al. Targeting by affinity-matured recombinant antibody fragments of an angiogenesis associated fibronectin isoform. Nat Biotechnol. 1997;15:1271–5. doi: 10.1038/nbt1197-1271. [DOI] [PubMed] [Google Scholar]
- 60.Weissleder R, Tung CH, Mahmood U, Bogdanov A. In vivo imaging of tumors with protease-activated near-infrared fluorescent probes. Nat Biotechnol. 1999;17:375–8. doi: 10.1038/7933. [DOI] [PubMed] [Google Scholar]
- 61.Tung CH, Mahmood U, Bredow S, Weissleder R. In vivo imaging of proteolytic enzyme activity using a novel molecular reporter. Cancer Res. 2000;60:4953–8. [PubMed] [Google Scholar]
- 62.Bremer C, Tung CH, Weissleder R. In vivo molecular target assessment of matrix metalloproteinase inhibition. Nat Med. 2001;7:743–8. doi: 10.1038/89126. [DOI] [PubMed] [Google Scholar]
- 63.Marten K, Bremer C, Khazaie K, Sameni M, Sloane B, Tung CH, Weissleder R. Detection of dysplastic intestinal adenomas using enzyme-sensing molecular beacons in mice. Gastroenterology. 2002;122:406–14. doi: 10.1053/gast.2002.30990. [DOI] [PubMed] [Google Scholar]
- 64.Hsiung PL, Hardy J, Friedland S, Soetikno R, Du CB, Wu AP, Sahbaie P, Crawford JM, Lowe AW, Contag CH, Wang TD. Detection of colonic dysplasia in vivo using a targeted heptapeptide and confocal microendoscopy. Nat Med. 2008;14:454–8. doi: 10.1038/nm1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kelly K, Alencar H, Funovics M, Mahmood U, Weissleder R. Detection of invasive colon cancer using a novel, targeted, library-derived fluorescent peptide. Cancer Res. 2004;64:6247–51. doi: 10.1158/0008-5472.CAN-04-0817. [DOI] [PubMed] [Google Scholar]
- 66.Newton JR, Kelly KA, Mahmood U, Weissleder R, Deutscher SL. In vivo selection of phage for the optical imaging of PC-3 human prostate carcinoma in mice. Neoplasia. 2006;8:772–80. doi: 10.1593/neo.06331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hinoi T, Akyol A, Theisen BK, Ferguson DO, Greenson JK, Williams BO, Cho KR, Fearon ER. Mouse Model of Colonic Adenoma-Carcinoma Progression Based on Somatic Apc Inactivation. Cancer Research. doi: 10.1158/0008-5472.CAN-07-2735. in press. [DOI] [PubMed] [Google Scholar]
- 68.Akyol A, Hinoi T, Feng Y, Bommer GT, Glaser TM, Fearon ER. Generating somatic mosaicism with a Cre recombinase-microsatellite sequence transgene. Nat Methods. 2008;5:231–3. doi: 10.1038/NMETH.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]