Summary
The PyrR protein regulates expression of pyrimidine biosynthetic (pyr) genes in many bacteria. PyrR binds to specific sites in the 5’ leader RNA of target operons and favors attenuation of transcription. Filter binding and gel mobility assays were used to characterize the binding of PyrR from Bacillus caldolyticus to RNA sequences (binding loops) from the three attenuation regions of the B. caldolyticus pyr operon. Binding of PyrR to the three binding loops and modulation of RNA binding by nucleotides was similar for all three RNAs. Apparent dissociation constants at 0° C ranged from 0.13 to 0.87 nM in the absence of effectors; dissociation constants were decreased by 3 to 12 fold by uridine nucleotides and increased by 40 to 200 fold by guanosine nucleotides. The binding data suggest that pyr operon expression is regulated by the ratio of intracellular uridine nucleotides to guanosine nucleotides; the effects of nucleoside addition to the growth medium on aspartate transcarbamylase (pyrB) levels in B. subtilis cells in vivo supported this conclusion. Analytical ultracentrifugation established that RNA binds to dimeric PyrR, even though the tetrameric form of unbound PyrR predominates in solution at the concentrations studied.
Keywords: RNA binding to proteins, regulation of attenuation, PyrR, Bacillus caldolyticus, ultracentrifugation, pyrimidine nucleotides
Introduction
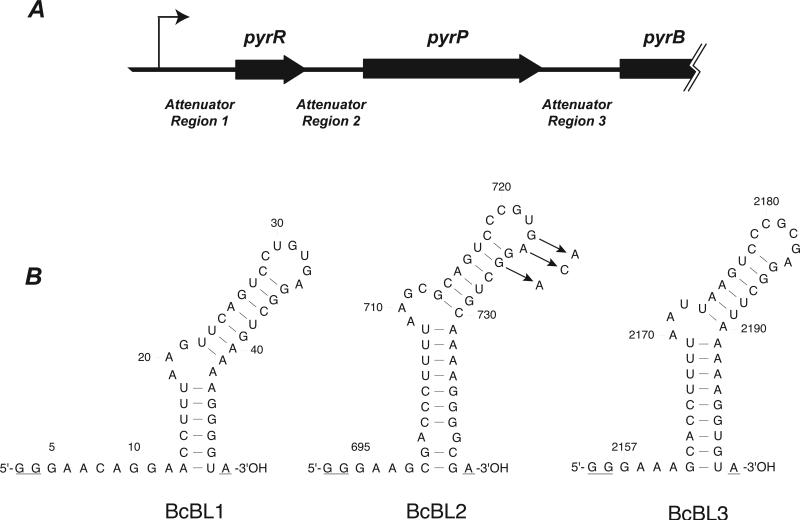
The PyrR protein regulates expression of the genes of de novo pyrimidine nucleotide biosynthesis (pyr genes) in nearly all Gram positive and many other bacteria by a transcription attenuation mechanism [1]. PyrR acts by binding to a segment of pyr mRNA with conserved sequence and secondary structure [1, 2]. When PyrR is bound, a downstream antiterminator stem-loop structure is prevented from forming, and formation of a transcription terminator is permitted. The affinity of PyrR for pyr mRNA is increased by uridine nucleotides [2, 3], so an elevated pyrimidine level in the cells results in greater termination of transcription at sites upstream of the open reading frames of the pyr genes. Three sites of PyrR binding and transcription attenuation have been identified in the pyr operons of Bacillus subtilis and related Bacillus species [1]. These are located in the 5’ untranslated leader of the operon (binding loop 1 or BL1), between the first cistron of the operon, pyrR, and the second cistron pyrP (BL2), and between pyrP and the third cistron pyrB (BL3) (Fig. 1A).
Figure 1.
(A) Map of the 5’-end of the B. caldolyticus pyr operon. The thin bent arrow represents the transcriptional start site; open reading frames are represented as thick arrows; the untranslated regions containing the three attenuator regions are shown as lines of medium thickness. (B) Sequence of the three pyr mRNA species (binding loops) bound by PyrR that were examined in this study. The BcBL1, BcBL2 and BcBL3 sequences were derived from portions of the DNA sequence of Attenuator Regions 1, 2 and 3, respectively, shown in A. Numbers refer to the nucleotide number in the B. caldolyticus pyr transcript with +1 as the transcriptional start site [13]. The secondary structures were predicted by MFOLD version 3.1 (http://www.bioinfo.rpi.edu/applications/mfold) [33]. Three nucleotides in each binding loop that are not part of the wild type pyr mRNA sequence are underlined: The two first G residues in each transcript are specified by the T7 promoter and the terminal A residue is added by Taq polymerase when used for preparation of templates for in vitro transcription by T7 polymerase. Arrows indicate three single-base substitution RNA variants in BcBL2 examined in this study.
All of the initial genetic [4-7] and biochemical [2, 3, 8, 9] studies of the regulation of pyr genes by PyrR in our laboratory were conducted with B. subtilis strains and PyrR purified from B. subtilis. However, PyrR from the closely related thermophilic organism, Bacillus caldolyticus offers advantages for biochemical studies. B. caldolyticus PyrR is more stable than the B. subtilis homologue. At the concentrations examined B. caldolyticus PyrR exists in solution as single aggregation state, the tetramer, and forms crystals that are highly suitable for X-ray crystallographic analysis [10, 11]. B. caldolyticus offers an excellent alternative system for studies of PyrR-dependent regulation of the pyr operon. The organization and regulation of the B. caldolyticus pyr operon is essentially the same as in B. subtilis [12, 13]. Plasmid-borne B. caldolyticus pyrR restores normal regulation by pyrimidines to a B. subtilis strain in which the pyrR gene was deleted [13]. The structures of PyrR proteins from both species have been determined at high resolution [8, 10] and the subunit and dimeric structures of the two homologues are essentially identical, although B. subtilis PyrR crystallizes as a hexamer or as a dimer, whereas B. caldolyticus PyrR is a tetramer [10]. The recent determination of the structure of B. caldolyticus PyrR with bound nucleotides led to the unexpected finding that both UMP and GMP bind to equivalent sites on the PyrR dimer [10]. The nucleotide binding sites do not overlap with the likely RNA binding site on PyrR. A preliminary RNA binding study demonstrated that guanosine nucleotides have effects on RNA binding by PyrR that are opposite to the effects of uridine nucleotides [10]. That is, GMP and GTP decrease the affinity of PyrR for pyr RNA, whereas UMP and UTP increase its affinity for RNA.
In the present work we have studied in detail the binding of B. caldolyticus PyrR to the three RNA sequences to which it binds in B. caldolyticus, which we called BcBL1, BcBL2 and BcBL3, and the effects of nucleotides on RNA binding. A rapid and convenient filter binding assay [14] was used for many of these experiments. Electrophoretic mobility shift assays and sedimentation velocity experiments were also used to characterize binding of PyrR to RNA. Use of the filter binding assay was frustrated in previous studies with B. subtilis PyrR because that protein tended to aggregate and failed to bind quantitatively to various hydrophobic filters. However, the filter binding method can be used to study RNA binding to PyrR from B. caldolyticus, perhaps because this protein has a lower overall negative electrostatic surface potential than the B. subtilis homologue [10] and does not aggregate. This study leads to a more refined characterization of the PyrR-RNA interaction, definition of binding stoichiometry as one RNA binding loop per PyrR dimer and a definition of the specificity of nucleotide effects on RNA binding. Implications of the current findings for the physiological regulation of pyrimidine biosynthesis are presented; the most important of these is that regulation of pyr operon expression by PyrR relies on shifts in the ratio of uridine nucleotides to guanosine nucleotides, not the intracellular concentration of uridine nucleotides alone.
Results
Uridine and guanosine nucleotides modulate PyrR binding to all three pyr mRNA binding loops
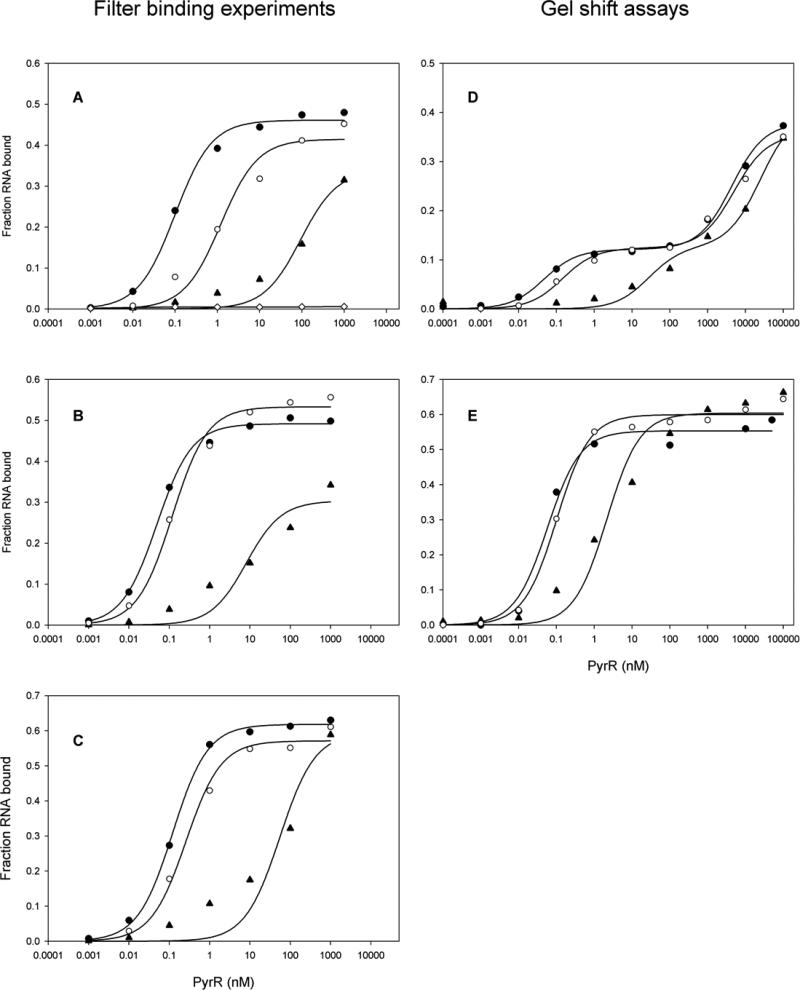
Predicted secondary structures of the three B. caldolyticus pyr mRNA binding loops (BcBL1, BcBL2 and BcBL3) examined in this study are shown in Fig. 1B. All three binding loops contain segments that are conserved in PyrR binding loops from homologous regulatory systems in other bacteria [2]. Conserved features include the predicted stem-loop structure with a purine-rich internal bulge, a terminal hexaloop containing the CNGNGA consensus sequence, and the UUUAA consensus sequence in the lower stem and internal bulge. Filter binding was used to estimate the affinity of the B. caldolyticus PyrR protein to each of the three binding loops (Fig. 2A-C). Binding was specific for pyr RNA, as shown by failure of a control RNA, the antisense strand to BcBL1 to bind to any concentration of PyrR tested (Fig. 2A).
Figure 2.
Representative filter binding experiment of the 32P-labeled PyrR binding loops BcBL1 (A), BcBL2 (B), and BcBL3 (C) to various concentrations of PyrR in the absence of effector (open circles), with 500 μM UMP (closed circles) or 500 μM GMP (closed triangles). Binding to a control RNA (the antisense strand of BcBL1) is shown as open diamonds (A). Representative eletrophoretic gel mobility shift assay with 32P-labeled BcBL1 (D) and BcBL2 (E) in the absence of effector (open circles), with 500 μM UMP (closed circles) or 500 μM GMP (closed triangles).
Binding of PyrR to BcBL2 and BcBL3 in standard binding buffer in the absence of effectors followed a binding curve (sigmoid on a semi-log plot of PyrR concentration versus % of total RNA bound) that was indicative of a simple PyrR-RNA binding isotherm (Fig. 2B and C). However, the binding curve for BcBL1 deviated consistently from the fitted curve (Fig. 2A). On the other hand, in the presence of 0.5 mM UMP, which stimulated binding for all three binding loops, PyrR binding to BcBL1 resembled the binding observed for the other two binding loops. The apparent dissociation constants (Kd values) for RNA binding are shown in Table 1. When no nucleotides were present, PyrR bound most tightly to BcBL2 and BcBL3 (Kd of 0.13 ± 0.02 nM and 0.2 ± 0.08 nM, respectively). The Kd value for PyrR binding to BcBL1, 0.9 ± 0.3 nM, corresponds to slightly looser binding. Addition of 0.5 mM UMP, UDP or UTP resulted in tighter binding, yielding to Kd values in the range of 0.04 nM to 0.09 nM for the three RNAs. PRPP and dUMP also stimulated binding, although not as effectively as UMP.
Table 1.
Apparent RNA dissociation constants (Kd-values) from filter binding determinations of PyrR binding to the three pyr operon binding loops
| Kd -values (nM) |
|||
|---|---|---|---|
| BcBL1 | BcBL2 | BcBL3 | |
| No effector | 0.87 ± 0.3 | 0.13 ± 0.02 | 0.21 ± 0.08 |
| UMP | 0.07 ± 0.02 | 0.04 ± 0.01 | 0.08 ± 0.05 |
| UDP | 0.07 ± 0.02 | 0.04 ± 0.01 | ND |
| UTP | 0.09 ± 0.02 | 0.04 ± 0.01 | ND |
| dUMP | 0.16 ± 0.08 | 0.05 ± 0.01 | ND |
| PRPP | 0.11 ± 0.03 | 0.06 ± 0.01 | ND |
| GMP | 79 ± 17 | 5.2 ± 2.9 | 49 ± 25 |
| GDP | 37 ± 12 | 2.7 ± 1.6 | ND |
| GTP | 12 ± 3 | 1.1 ± 0.2 | ND |
| dGMP | 9 ± 5 | 0.73 ± 0.01 | ND |
The effectors were present at 0.5 mM. The data are averages of at least three independent determinations and include standard deviations of the mean value. ND, not determined.
The apparent Kd values for binding of B. caldolyticus pyr binding loops BcBL1, BcBL2 and BcBL3 to PyrR were increased in the presence of GMP by 90-fold, 40-fold and 200-fold, respectively, relative to their values in the absence of effector, indicative of reduced affinity for RNA (Table 1). However, these constants were difficult to determine precisely because the binding data were not adequately fit by a simple binding equation, as illustrated in Fig. 2A, B and C. GDP, GTP and dGMP also inhibited binding, although they were less effective at saturating concentrations than GMP (Table 1).
Because all three binding loops bound with similar affinity to PyrR and the effects of nucleotides on RNA binding were similar for all three RNAs, we conducted most of the subsequent studies with a single RNA, BcBL2, because the binding of the homologous B. subtilis RNA, BsBL2, was most thoroughly investigated in a previous study [2].
Concentrations of nucleotides required for activation or inhibition of PyrR binding to pyr binding loops
The concentrations of nucleotides that modulate PyrR binding to RNA in vitro were determined, so these values could be compared to likely intracellular concentrations of the nucleotides. Measurements of the binding of RNA to PyrR over a wide range of nucleotide concentrations in the filter binding assay yielded the concentration at which the effect of the nucleotide was half-maximal (Table 2). As a function of concentration, UMP was ten-fold more effective than UTP at stimulating binding of PyrR to BcBL1 and 100-fold more effective than UTP at stimulating binding to BcBL2. Additionally, the UTP concentration necessary for activation of PyrR was almost ten-fold lower for BcBL1 than for BcBL2. As a function of concentration, GTP was a much more effective inhibitor of RNA binding than GMP. Even though addition of a saturating GMP concentration resulted in a higher apparent dissociation constant for RNA than did GTP (Table 1), the concentration required to achieve this inhibition was much higher for GMP (Table 2). The high concentration of GMP needed to affect RNA binding, as compared to GTP, suggests that GTP is the more likely physiological regulator, especially given that nucleoside triphosphate levels are usually several fold higher than levels of the corresponding nucleoside monophosphate in vivo.
Table 2.
Half-maximum concentrations of nucleotides or PRPP required for either activation (UMP, UTP and PRPP) or inhibition (GMP and GTP) of binding of PyrR to BcBL1 and BcBL2
| Half-maximum concentration (μM) |
||
|---|---|---|
| BcBL1 | BcBL2 | |
| UMP | 0.04 ± 0.02 | 0.02 ± 0.01 |
| UTP | 0.3 ± 0.2 | 2.4 ± 0.7 |
| PRPP | 0.7 ± 0.3 | 2.0 ± 1.4 |
| GMP | 269 ± 143 | 232 ± 162 |
| GTP | 18 ± 6 | 8 ± 6 |
The data are averages of at least two independent determinations.
The guanosine to uridine nucleotide ratio governs PyrR binding to pyr RNA
Table 3 shows the effects of varying the ratio between effectors that increase and effectors that decrease binding of PyrR to BcBL2 RNA with the total concentration of the two effectors held constant. When the GMP to UMP ratio was increased from 0.11 to 19, the apparent dissociation constant for RNA increased 18-fold, documenting the antagonism of the two effectors. When the ratio of GTP to UTP was varied over the same range, the effects were similar to the effects of GMP and UMP; the values for the apparent Kd for BcBL2 varied over a 10-fold range. The effects of PRPP on RNA binding to PyrR were similar to those of uridine nucleotides (Table 1); GMP and GTP also antagonized the effects of PRPP (data not shown), as expected if PRPP and the nucleotides bind at the same site. From these observations we predict that the most important factor regulating the affinity of PyrR for target pyr RNA sites in vivo is the intracellular ratio of guanosine nucleotides to uridine nucleotides, rather than the concentration of the individual nucleotides.
Table 3. Effects of the ratio of guanosine to uridine nucleotide concentrations on binding of PyrR to BcBL2.
The total concentration of nucleotides was held constant at 1 mM.
| Concentration of nucleotide (μM) |
Kd value for RNA (nM) | Concentration of nucleotide (μM) |
Kd value for RNA (nM) | ||
|---|---|---|---|---|---|
| GMP | UMP | GTP | UTP | ||
| 0 | 0 | 0.13 | 0 | 0 | 0.13 |
| 0 | 1000 | 0.06 | 0 | 1000 | 0.05 |
| 100 | 900 | 0.06 | 50 | 950 | 0.06 |
| 250 | 750 | 0.05 | 150 | 850 | 0.09 |
| 500 | 500 | 0.11 | 250 | 750 | 0.11 |
| 750 | 250 | 0.29 | 500 | 500 | 0.17 |
| 900 | 100 | 0.84 | 750 | 250 | 0.40 |
| 950 | 50 | 1.1 | 900 | 100 | 0.61 |
| 1000 | 0 | 9.8 | 1000 | 0 | 0.75 |
Structural requirements of effectors for affecting PyrR binding to BcBL2
To learn more about how PyrR distinguishes purine and pyrimidine nucleotides, we tested the ability of purine and pyrimidine nucleotide structural variants to activate or inhibit binding of BcBL2 to PyrR (see Supplementary Material and Table S1). In general, RNA binding to PyrR was activated by pyrimidine nucleotides regardless of structure, whereas the specificity of purine nucleotide effects on RNA binding indicated that both the exocyclic oxo and amino groups of the purine ring and the 2’-hydroxyl group of ribose in GMP contribute significantly to its action. These observations suggest specific interactions between PyrR and the purine ring of purine nucleotides that do not occur with pyrimidine nucleotides, even though such interactions have not been observed in the presently available x-ray structures of PyrR-nucleotide complexes [10, 11].
Effects of Mg2+, pH and temperature on binding of PyrR to BcBL2
Experiments characterizing the effects of Mg2+ ion concentration, pH and temperature on the binding of BcBL2 RNA to PyrR in the filter binding assay are shown in detail in the Supplemental Data. Three important conclusions were derived from these studies. First, Mg2+ ions at a concentration of 10 mM or higher were essential for tight binding of RNA. Inclusion of Mg2+ ions in the electrophoresis gel was subsequently found to be crucial for obtaining tight binding of RNA in the gel shift assay. Second, the affinity of PyrR for BcBL2 RNA was 50-fold higher at pH 7.5 than at pH 5.5, and the effect of GMP on RNA binding was strongly pH dependent, whereas the effect of UMP was much less so (Fig. S1A, Supplementary Material). Ionization of one of four histidine residues in B. caldolyticus PyrR may mediate the pH dependence of the GMP effect on RNA binding. Finally, the binding studies in this report were conducted at 0°C to ensure stability of the components, for convenience in maintaining a constant temperature, and for comparison to results of previous gel shift studies. However, an increase in temperature promotes dissociation of a protein-RNA complex; an increase in temperature from 0°C to 50°C, which is close to the growth temperature for B. caldolyticus, increased the apparent Kd for BcBL2 binding to PyrR by about 40 fold to 4.5 ± 0.2 nM (Fig. S1B, Supplementary Material).
Direct comparison of the filter binding and electrophoretic mobility shift methods with BcBL1 and BcBL2
It was desirable to confirm the fundamental conclusions of the preceding RNA filter binding studies using an alternative method. Previous studies [2] of pyr RNA binding by B. subtilis PyrR used an electrophoretic gel mobility shift method. Some of these prior findings were different from those just described for binding of pyr RNA by B. caldolyticus PyrR (see Discussion). Therefore, it was important to compare directly the two methods for measuring RNA binding. B. caldolyticus PyrR and radiolabeled B. caldolyticus BcBL1 and BcBL2 were used for this comparison, because B. subtilis PyrR cannot be used for the filter binding method, because this protein is not quantitatively retained by hydrophobic filters.
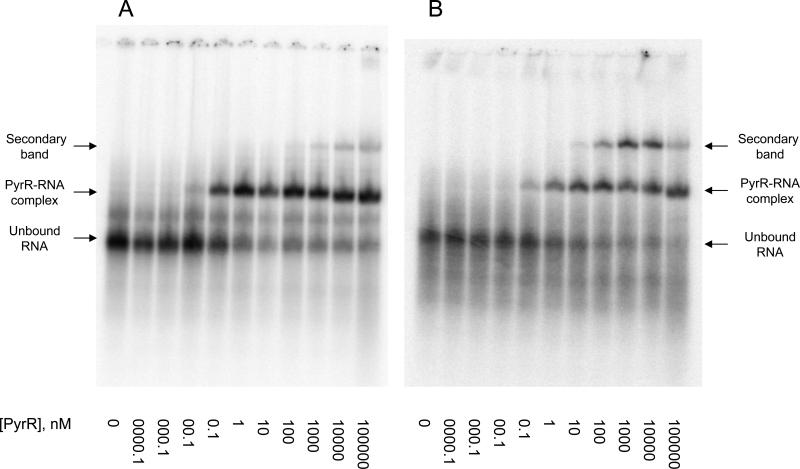
Inclusion of 1 mM Mg2+-acetate in the electrophoresis gel was necessary to observe binding of either BcBL1 or BcBL2 to B. caldolyticus PyrR at concentrations up to 100 μM protein, even though 10 mM Mg2+ was included in the binding mixture prior to electrophoresis, the electrophoresis buffer contained 1 mM Mg2+, and the gel was subjected to prior electrophoresis for 90 min before loading the samples. With this modification of the previously used method [2], tight binding of B. caldolyticus PyrR to BcBL1 and BcBL2 was observed by the gel shift method (Fig. 2 D and E, Fig. 3, Table 4). The binding of BcBL1 to PyrR was clearly resolved into two phases (Fig. 2D), one corresponding to tight binding (Kd1 of Table 4) and another that was detected only at high concentrations of PyrR, well above those that could be studied in the filter binding studies. The significance of the species observed at PyrR concentrations greatly in excess of those needed to saturate the RNA is questionable, as non-specific binding to RNA cannot be excluded. The binding of BcBL2 was described by a single tight binding curve, although in the presence of 0.5 mM GMP the binding curve was broad and fit less well to a simple binding equation (Fig. 2E), as was observed on filter binding of BcBL2 under the same conditions (Fig. 2B). In addition, a second, more slowly migrating PyrR-BcBL2 complex was detected at high concentrations of PyrR when 0.5 mM GMP was present (Fig. 3B). In the absence of nucleotide (Fig. 3A) or when 0.5 mM UMP was present (not shown) this species was barely detectable. Again, the significance of this loosely binding complex is open to question. Importantly, the values for Kd (Kd1 for BcBL1) and the effects of UMP and GMP (Table 4) agreed reasonably with the corresponding values obtained with the filter binding method (Table 1). We also found that addition of Mg2+ to the gel was necessary to obtain tight binding of BcBL2 (Kd = 4 nM) to B. subtilis PyrR (data not shown). Thus, if care was taken to include 1 mM Mg2+ in the electrophoresis gel, similar results for the tight binding RNA curves were obtained by both methods, a finding that provides confidence in their validity.
Figure 3.
Analysis of the binding of 32P-labeled BcBL2 to PyrR by the electrophoretic gel mobility shift method in the absence of effector (A) and in the presence of 500 μM GMP (B). The concentration of PyrR (nM subunit) present in each lane is indicated below the figure. The apparent dissociation constants derived from these experiments are shown in Table 5. The presence of the unbound BcBL2 RNA, the PyrR-BcBL2 complex as well as a more slowly migrating secondary band are indicated on the side of each gel.
Table 4.
Apparent RNA dissociation constants (Kd-values) in electrophoretic gel shift assays of binding of BcBL1 and BcBL2 to PyrR
| BcBL1 |
BcBL2 |
||
|---|---|---|---|
| Kd1 (nM) | Kd2 (nM) | Kd (nM) | |
| No effector | 0.18 ± 0.04 | 7650 ± 2500 | 0.11 ± 0.04 |
| UMP | 0.06 ± 0.02 | 6300 ± 4900 | 0.07 ± 0.01 |
| GMP | 19 ± 8 | 16800 ± 7400 | 3.3 ± 1.9 |
UMP and GMP were present at 0.5 mM. The data are averages of three to four independent determinations.
Binding of BcBL2 structural variants to PyrR
The binding of RNA to B. caldolyticus PyrR exhibits high RNA sequence specificity, as expected from previous genetic and biochemical studies with B. subtilis PyrR [2, 6]. This was established by filter binding assay of B. caldolyticus PyrR to three variants of B. caldolyticus BL2 containing single base substitutions (Fig. 1). Analogous variants of B. subtilis BL2 were observed in previous gel shift studies with B. subtilis PyrR to have very different apparent Kd values relative to native BL2 [2]. With two of the three structural variants tested, the data (Supplemental Data, Table S2) indicated that a single base substitution in a highly conserved portion of the binding loop RNA (G723A) caused reduced binding to PyrR, whereas a substitution in a non-conserved nucleotide (G726A) did not. However, with a third structural variant, A724C, the binding observed by filter binding was much tighter than that detected by the gel mobility shift method (Supplementary Material). Binding of this structural variant was clearly altered from the wild type RNA, however, and additional experiments indicated that the A724C variant RNA differs from the wild type BcBL2 in its interaction with Mg2+ (Supplementary Material).
Effects of uridine and guanosine supplementation on pyr gene expression in vivo
If PyrR-mediated regulation of the pyr operon in Bacillus species is largely responsive to the ratio of uridine to guanosine nucleotides, as suggested by the effects of these nucleotides on binding of PyrR to binding loop RNA in vitro, then addition of guanosine or uridine to the bacterial growth medium would be expected to stimulate or repress, respectively, expression of pyr genes. Assays of aspartate transcarbamylase (ATCase), the enzyme encoded by pyrB, the third cistron of the operon, provided a convenient measure of operon expression in such experiments. Inclusion of guanosine in the growth medium increased the level of ATCase in B. subtilis cells by about 45%, as compared to a control culture without supplementation; inclusion of uridine decreased ATCase levels by almost two fold (Table 5). When both uridine and guanosine were included in the medium in equal amounts, the ATCase level was largely repressed, but expression increased substantially as the ratio of guanosine to uridine was increased. The results demonstrate competition between the effects of guanosine and uridine in the medium. As expected, the effects of nucleoside addition were not seen in a mutant strain of B. subtilis [4] in which the pyrR gene was deleted. These observations demonstrate that the effects of nucleotides on RNA binding to PyrR in vitro correlate with their predicted effects on pyr gene expression in vivo.
Table 5.
Effects of nucleoside supplementation in the growth medium on the expression of B. subtilis aspartate transcarbamylase (ATCase)
| Strain | Addition to the medium (μg/ml) | ATCase specific activity (nmol/min/mg) | |
|---|---|---|---|
| Guanosine | Uridine | ||
| DB104 | None | None | 86 ± 5 |
| DB104 | 50 | None | 120 ± 11 |
| DB104 | None | 50 | 56 ± 4 |
| DB104 | 50 | 50 | 47 ± 6 |
| DB104 | 50 | 10 | 110 ± 14 |
| DB104 | 50 | 2 | 130 ± 9 |
| DB104 ΔpyrR | None | None | 1300 ± 390 |
| DB104 ΔpyrR | 50 | None | 1300 ± 390 |
| DB104 ΔpyrR | None | 50 | 1400 ± 420 |
It should be noted that the effects of guanosine on ATCase expression shown in Table 5 were obtained with cells grown with succinate as the carbon source. Similar, but even larger, effects could be observed with glucose-grown cells only in cultures harvested at the end of exponential growth on limiting glucose; if the cells were harvested during growth on excess glucose, the stimulation of ATCase levels by guanosine was not observed, although strong repression by uridine was observed. We suggest that these results indicate that guanosine uptake and/or conversion to nucleotides is repressed by growth on glucose [15], which masks the effect of guanosine on pyr operon expression under such conditions.
Studies of RNA binding to PyrR by analytical ultracentrifugation
The quaternary structure of B. caldolyticus PyrR in solution was determined from both sedimentation velocity and equilibrium sedimentation experiments at high and low protein concentrations and in the presence and absence of 0.1M NaCl. Results of the sedimentation velocity studies are summarized in Table S3 (Supplementary Material). The calculated weight average mass for native PyrR ranged from 83 to 101 kDa and from 94 to 99 kDa for the His-tagged PyrR used in sedimentation velocity studies of RNA binding described below. The masses calculated from sequence of the native and His-tagged PyrR in the tetrameric form are 79.8 and 91.2 kDa, respectively. Since these weight average masses are calculated from the change in shape of moving boundary during the run, and the data are susceptible to various systematic errors, the variation observed in the mass in Table S3 is within experimental error.
Data from a sedimentation equilibrium study and an approach to equilibrium analysis of native PyrR over the concentration range of 0.25 to 25 μM subunit (Fig. S3 and S4, Supplementary Material) fit adequately to sedimentation of a single tetrameric species of calculated weight average mass of 78.3 kDa, although an alternative fit of the data to a model for sedimentation of a dimeric and tetrameric species in equilibrium could not be excluded (Supplementary Material). A similar sedimentation equilibrium study with His-tagged PyrR (0.25 to 25 μM) gives results similar to native PyrR except that the fitted weight average mass was 91.6 kDa. Altogether, the sedimentation velocity and equilibrium studies show that both native and His-tagged PyrR exist largely as tetramers in solution at concentrations greater than 1 μM, which is in accord with previous results obtained with size exclusion chromatography and X-ray crystallography [10]. These data and conclusions are discussed in greater detail in the Supplementary Material section.
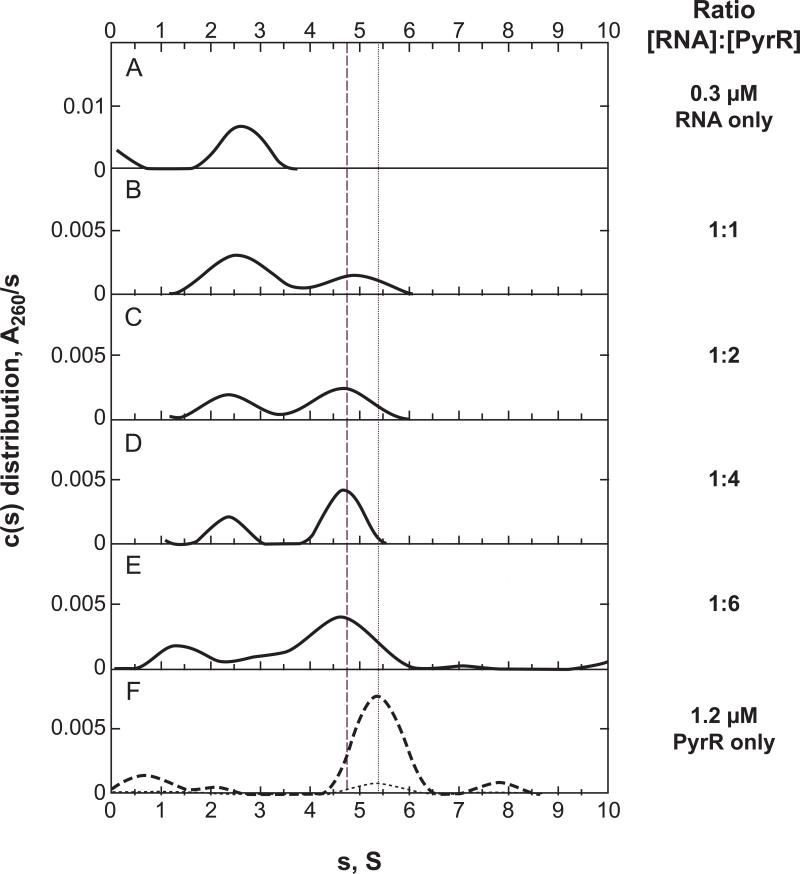
Sedimentation velocity was also used to analyze the binding of RNA to PyrR. Purified His-tagged PyrR was used for these studies because the native PyrR contained traces of ribonuclease, which might have degraded the RNA during the three-day duration of the titration experiment. As shown in Fig 4A, a 36 nt pyr binding loop RNA derived from BcBL2 sedimented as a single RNA species (s20,w = 2.63 S , MW = 12,900 Da) (MW calculated from sequence = 11,600 Da). This BcBL2 sample was titrated by adding aliquots of concentrated PyrR (Fig. 4B-E), so that up to 6 equivalents of monomer were added without significant dilution (<7%) of the RNA. Species analysis using either the basic non-interacting model of SEDPHAT [16, 17] or the more powerful hybrid local continuous distribution/global discrete species model [16, 17] showed that only two sedimenting species were present at significant concentrations in the range of 0.1 S to 5 S for each aliquot added (Table S4 in the Supplemental Data section). The first of these (s20,w = 2.6 S) corresponds to the free RNA. A second species appeared (s20,w = 4.9 S) that must correspond to an RNA-PyrR complex, since added PyrR will not contribute more than 1-2% to the total 260 nm absorbance at the concentrations added. On titrating the RNA with increasing amounts of PyrR, the loading concentration of the peak corresponding to free RNA declined, and that for the second peak increased, as shown by the area under the peaks in the c(s) distribution shown in Fig. 4. An additional shoulder at ~3 S, whose shape and position are somewhat variable, is evident in Panel E of Fig. 4 (and Fig, S5, Supplementary Material), where the protein concentration is ~ 3 times that necessary to saturate the RNA with the PyrR dimer. Based on the species analysis above, we strongly suspect that this shoulder is an artifact that results from the sensitivity of the c(s) distribution to boundary effects. As with the filter binding assays, some of the RNA (~30%) remained unbound at greater than saturating concentrations of PyrR. In Panel F of Fig 4, the PyrR stock solution was diluted to 1.2 μM subunits into the same buffer and centrifuged under the same conditions as used for the other panels in Fig. 4. Most of the protein sedimented as a tetramer with an s20,w = 5.5S with a minor species at about 10% of the tetramer concentration with an s20,w = 2.3 S and an estimated MW = 18,000 Da, which is likely a nonparticipating PyrR monomer (sequence MW = 22,800 Da). The sedimentation coefficients and buoyant mass variation observed with increasing PyrR concentrations are summarized in Table S4 (Supplementary Material). The s values in Table S4 for the free RNA peak decreased significantly with increasing PyrR concentration. We demonstrated that the pyr RNA appeared to be electrophoretically intact following the 3-day experiment at 20°C (data not shown), so the decrease in s value for the RNA is not the result of RNA degradation. The sedimentation coefficient of the new species (4.6 to 4.9 S) is significantly lower than that of free PyrR (5.4 S); a complex of RNA with the PyrR tetramer would be expected to have a larger s value than free PyrR, barring a large, unexpected increase in the hydrodynamic radius. Thus, the complex of RNA with PyrR must involve association with the protein in a form smaller than the tetramer. If one subtracts the buoyant mass of the RNA from that of the complex and calculates the molecular weight of the remaining protein, using a partial specific volume of 0.74 (calculated from the amino acid composition of PyrR) and a solvent density of 1.0 g/mL, the value obtained is 37,100 for the protein component. This is in reasonable agreement with the mass of a His-tagged PyrR dimer, 44,000. Finally, Fig. 5 shows a plot of the free RNA remaining against the ratio of PyrR subunit concentration to the initial RNA concentration (Table S4, Supplementary Material). The trend in the data is that of a typical stoichiometry plot where the RNA concentration is in large excess of its dissociation constant for PyrR. The data are consistent with a stoichiometry of one RNA molecule per PyrR dimer in the complex with about 30% of the RNA that does not bind under these conditions. Thus, we conclude that the complex has the composition of (PyrR)2-RNA.
Figure 4.
A size-distribution analysis of sedimenting species observed during a titration of pyr binding loop RNA with increasing amounts of PyrR. A plot of c(s) distributions against the uncorrected sedimentation coefficient, s, is shown for RNA only (A), for molar ratios RNA to PyrR subunit (B-E), and for PyrR (F) from absorbance data collected at 260 nm. The c(s) values in the PyrR panel (F) were multiplied by factors of 10 (dotted line) and 100 (dashed line) to make them visible on the same scale as used for the other panels. The initial concentration of RNA was 0.3 μM for Panel A and 4 separate aliquots of PyrR were added giving the final ratios shown in the figure. A concentration of PyrR of1.2 μM was used for Panel F. The sedimentation distributions, c(s), were calculated using SEDPHAT; 72 scans were collected at 3-min intervals. Further experimental details are given in Materials and Methods. The vertical dotted line in the figure relates the protein peak to the other panels and the vertical dashed line does the same for the PyrR-RNA complex.
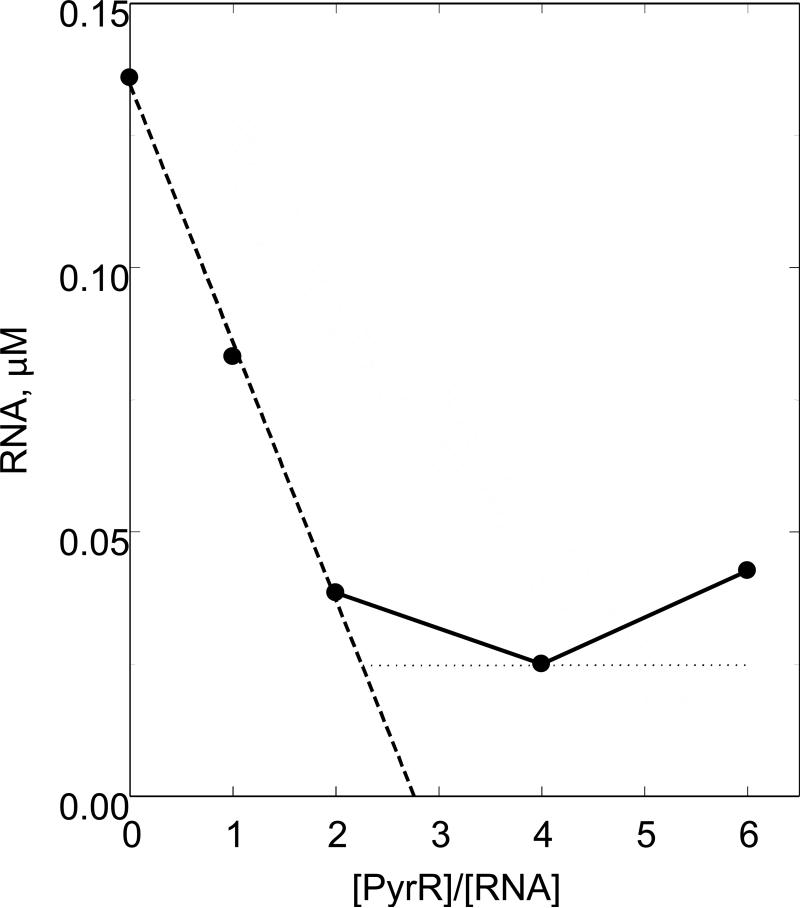
Figure 5.
A plot of A260 for the free RNA peak, which is obtained by integrating the area under the peak in the s range of 2 to 2.6 S, against the molar ratio of PyrR to RNA in the sample. The data were obtained from the analytical ultracentrifugation experiment described in Fig. 4. The free RNA values were corrected for a slight loss of total A260 ranging from 5% to 20%, of which a maximum of 5% was due to dilution. The dotted line shows a least -squares fit through the first 3 points. The horizontal dotted line shows the concentration of non-binding RNA from Fig. 4, Panel D in which the RNA peak is clearly defined. The intersection between the dashed and dotted lines indicates that 2 subunits of PyrR bind to 1 RNA stem-loop.
Discussion
Complexity of RNA binding to PyrR
The complex binding curves for BcBL1 and for all three binding loop RNAs in the presence of guanosine nucleotides (Fig. 2) indicate that the binding of RNA to PyrR cannot be fully described by a simple binding equilibrium. While the biophysical basis for the complexity of the RNA binding curves is not established, we suggest that it arises from multiple PyrR conformational and/or aggregation states that differ in their affinity for RNA and possibly also for nucleotides. PyrR conformation is implicated because the heterogeneity in RNA binding is strongly affected by uridine and guanosine nucleotides, which are known to bind to the UPRTase active site of PyrR [10]. The simplest model that fits our observations posits the existence of two PyrR conformations, one with higher affinity for RNA than the other. The high affinity state is favored by binding of either uridine nucleotides or binding of RNA itself in the case of BcBL1. The low affinity state is favored by the binding of guanosine nucleotides. Thus, RNA binding involves at least two coupled reactions, RNA binding to PyrR and nucleotide binding to PyrR.
The demonstration by analytical ultracentrifugation that the PyrR tetramer dissociates into dimers when RNA binds adds yet another reaction that is likely coupled to the RNA and nucleotide binding reactions discussed above. It is likely that, at high dilution, tetrameric PyrR dissociates to dimers in the absence of RNA, but this could not be conclusively demonstrated at the lowest concentration (1 μM) that could be analyzed by analytical ultracentrifugation. We note, however, that all of the filter binding experiments were conducted at PyrR concentrations well below this value, where some or all of the PyrR may be present in dimeric form. We propose that the tetrameric form of PyrR has low affinity for RNA, because the likely RNA binding site is known from the crystal structures to be occluded in the center of the tetramer [10, 11]. The dimeric form of PyrR, in which the RNA binding site would be exposed to the solvent, is likely to have higher affinity for RNA. Coupling of the dimer-tetramer equilibrium to the equilibria for PyrR-RNA binding and PyrR-nucleotide binding could explain the complex binding curves seen in our experiments, especially when RNA binding in the presence of guanosine nucleotides was examined.
The involvement of multiple coupled equilibria—PyrR tetramer-dimer association together with binding of RNA and nucleotides to dimer and tetramer with different affinities for each state of aggregation—in the experimentally observed RNA binding in this work dictates that one should not regard the apparent Kd values for RNA or the half-maximal values for nucleotide effects on RNA binding as simple equilibrium constants. Hence, we have consistently used the term “apparent Kd” to describe the concentrations of PyrR that yielded half-maximal RNA binding in our experiments.
Correlations between results of filter binding studies and electrophoretic mobility shift studies of RNA binding
Direct comparison of the binding of BcBL1 and BcBL2 to B. caldolyticus PyrR by the filter binding and gel shift methods demonstrated that, as long as Mg2+ was included in the electrophoresis gel, there was good agreement between the two methods. However, agreement was much poorer with the A724C structural variant of BcBL2 RNA, even with a high Mg2+ concentration in the gel. The sensitivity to Mg2+ and to the structure of the RNA studied suggests that the gel shift method can give highly misleading results in some cases. An RNA that dissociates rapidly from PyrR may appear to bind poorly, or not to bind at all, in the gel shift assay. We conclude that for protein-RNA binding studies in general it would be prudent to confirm electrophoretic mobility shift conclusions when possible by an alternative method, such as a filter binding assay.
In light of our current observations on the importance of Mg2+ in gel shift assays with PyrR and pyr binding loop RNAs, the studies of the specificity of RNA binding of B. subtilis PyrR should be re-examined. Our findings with the native and the G723A and G726A sequence variants of BcBL2 indicate that the effects on affinity observed previously for native BsBL2 and its structural variants [2] are valid, at least qualitatively. However, the previous observations that B. subtilis PyrR binding to BsBL1 and BsBL3 was weak and barely affected by uridine nucleotides were misleading and probably resulted from the dissociation of the required cation Mg2+ from these two RNAs, but not BsBL2, during electrophoresis. In fact, we now have evidence that PyrR from both Bacillus species binds tightly to all three binding loop RNAs from both species and that binding of all three RNAs is significantly modulated by nucleotides (data not shown).
Physiological implications of these studies
The data in Tables 1-3 indicate that uridine nucleotides and guanosine nucleotides are the primary metabolite modulators of PyrR binding to pyr attenuator region RNA, and hence the primary regulators of pyrimidine biosynthesis. Uridine-nucleotide stimulation of RNA binding is easily understood in terms of feedback regulation, i.e. end-product repression of the pyr operon, because binding of PyrR to RNA leads to increased termination of transcription prior to transcription of pyr genes coding for biosynthetic enzymes. UMP and UTP exert their effects on RNA binding in vitro in the micromolar concentration range, well below their estimated concentrations in exponentially growing cells, which are 1 mM for UTP and 0.1 to 0.3 mM for UMP [19]. GTP is probably the primary physiological guanosine nucleotide modulator, because it is effective as at a much lower concentration than GMP (Table 2) and is present in growing cells at about 0.6 mM [20], well above the concentrations at which it exerts its effects on RNA binding to PyrR. The effects of GMP are unlikely to be significant in vivo, because intracellular GMP levels [20, 21] do not reach the concentration needed to antagonize RNA binding.
Of the other metabolites studied, none seems likely to serve as a physiologically important modulator of pyr operon expression. In particular, we suggested in previous publications [4, 9] that PRPP might may be a feed-forward regulator because it would be expected to compete at uridine nucleotide binding sites. However, the present data indicate that PRPP actually behaves like UMP and UTP to stimulate RNA binding to PyrR. PRPP also fails to antagonize the effects of GTP on RNA binding nearly as effectively as UMP or UTP (data not shown). Thus, the PRPP effects seem not to be physiologically important in regulation of pyr expression.
The intracellular concentrations of uridine nucleotides exceed the half-maximal concentrations required for activation of PyrR binding to RNA by two or three orders of magnitude (Table 2), even allowing for an increase of up to 40 fold in the Kd values for PyrR binding to RNA measured at 0° C when shifted to the physiological growth temperature for B. caldolyticus (Fig. S1B). The values of the apparent dissociation constants suggest that PyrR functions as a regulator of pyr gene expression largely under conditions where the affinity of PyrR for pyr attenuator sites is substantially reduced by the antagonistic effect of guanosine nucleotides. Since guanosine and uridine nucleotides can compete for binding to the same sites on PyrR [10], the affinity of the protein for pyr RNA will be determined by the ratio of their intracellular concentrations, not by the concentration of the individual nucleotides. Table 3 illustrates how the affinity of PyrR for BcBL2 RNA varied over a 10 to 20 fold range as a function of GMP/UMP or GTP/UTP ratio. The experiments in Table 3 were conducted at a total nucleotide concentration of 1 mM, which is near their physiological concentration and well above the concentrations at which the individual nucleotides exert their effects on RNA binding. Most importantly, our studies of the effects of nucleoside addition to the growth medium on B. subtilis pyrB (ATCase) expression in vivo (Table 5) support the conclusion that the ratio of guanosine to uridine nucleotides dominates PyrR action. The crucial role of guanosine nucleotides in the regulation of pyr genes in bacilli was not fully appreciated before these studies were conducted.
The cross-regulation of pyrimidine biosynthesis by guanosine nucleotides is part of a more general phenomenon. The accumulation of high levels of intracellular guanosine nucleotides indicates that cells have adequate carbon, nitrogen and energy sources for RNA and DNA synthesis. Thus, accumulated purine nucleotides constitute appropriate feed-forward metabolites to stimulate pyrimidine nucleotide biosynthesis. Other examples of such feed-forward activation of pyrimidine biosynthesis in Bacillus species include the activation by GTP of the pyrimidine-repressible carbamyl phosphate synthetase [22], of UMP kinase [23] and of CTP synthetase (for example, see [24]). Furthermore, GTP levels are used as regulatory signals governing a variety of more global regulatory circuits in Bacillus cells. Examples of this regulatory function of the GTP pool in B. subtilis include the regulation of ribosomal RNA synthesis [25], initiation of sporulation [26], activation of the CodY regulon in nutrient-starved cells [27], and the conversion of GTP to ppGpp and pppGpp during the stringent response to amino acid starvation [26]. A reduction in intracellular GTP without a corresponding decline in uridine nucleotide pools would result in much tighter binding of PyrR to attenuation region RNA and reduced expression of the pyr operon, which is an appropriate response to the original nutrient limitation.
Materials and methods
In vitro transcription and purification of 32P-labeled RNA
For synthesis of radiolabeled RNA by in vitro transcription, plasmids containing templates that contained the T7 RNA polymerase promoter followed by DNA specifying one of the three PyrR binding loops (BcBL1, BcBL2 and BcBL3) from the B. caldolyticus pyr operon were constructed. The isolated plasmids were used as templates in PCR reactions to generate products of approximately 150 base pairs in length that were used as templates for in vitro transcription with T7 RNA polymerase. The PCR templates were purified using the GFX purification kit prior to in vitro transcription using the MaxiScript kit from Ambion (Austin, TX, USA). The procedure recommended by the manufacturer was used, except that the reactions were incubated for two h at 37°C, and the RNA was labeled by including 10 μM [α-32P]GTP (800 Ci/mmol; MP Biomedicals Inc., Irvine, CA, USA) in the reaction mix together with 500 μM of each of ATP, UTP and CTP. Unincorporated nucleotides were separated by running the sample through a G-50 Micro Column from Amersham Biosciences/GE Healthcare (Piscataway, NJ, USA. The labeled RNA was purified using denaturing (8 M urea) 15% polyacrylamide gel electrophoresis as described previously [10]. Following phenol extraction of RNA eluted from a cut-out piece of gel, the RNA was precipitated over night at -20°C with 1 μg of yeast tRNA as carrier RNA and resuspended in 200 μl buffer (25 mM Tris-acetate pH 7.5, 50 mM K-acetate and 1 mM EDTA). The concentration of the RNA was determined by duplicate liquid scintillation counting of a 10-fold dilution of the RNA sample. Prior to use, the labeled RNA was denatured at 85°C for 15 min and allowed to refold for 15 min at 37°C, after which Mg-acetate was added to a final concentration of 10 mM.
Filter binding assays of RNA binding to PyrR
A double-filter method for filter binding [14] was used as described previously [10]. Unless noted otherwise, the reaction mixture contained 25 mM Tris-acetate, pH 7.5, 50 mM K-acetate, 10 mM Mg-acetate, 25 pM 32P-labeled RNA, 0.04 U/μl of RNase inhibitor (Superase-In; Ambion), 100 μg/ml (~4 μM) of yeast tRNA, 50 μg/ml of acetylated BSA, nucleotides at the indicated concentrations and various concentrations of native B. caldolyticus PyrR, which was purified as described previously [10]. The concentration of the PyrR protein was determined using the Bradford assay purchased from Bio-Rad. The 50 μl reaction mixtures were allowed to incubate for 40 min on ice prior to filtering through the two membranes followed by a single wash with 50 μl binding buffer. Filtration and washing were complete within 60 sec. For some experiments, the pH was varied by use of a 25 mM Tris-25 mM 2-(N-morpholino)ethanesulfonate-acetate buffer. For experiments where the temperature was varied, the pH of binding buffer was adjusted at each temperature examined to the pH of binding buffer at room temperature. The filters and the washing buffer were all equilibrated at the relevant temperature for at least 30 min prior to the experiments and the filtration apparatus was used at the temperature indicated. After overnight exposure of the membranes to a PhosphorImager screen, the radioactivity was determined using a PhosphorImager and quantified using ImageQuant software (Molecular Dynamics/GE Healthcare, Sunnyvale, CA, USA). Dissociation constants were determined by fitting the data to a simple binding equation using SigmaPlot 9.0 (Systat Software, Inc.. San Jose, CA, USA) as described previously [10] or, when two phases were clearly identified, to a two-state binding curve. Since the RNA concentration of 25 pM was close in some cases to the apparent Kd, these data were also fitted to a quadratic binding equation [28], but the calculated apparent Kd values obtained were not significantly different from those obtained with the simple binding equation. To determine the concentration of nucleotides required for half-maximum effect on PyrR binding to the RNA, the obtained Kd values from at least two independent experiments were plotted against the nucleotide concentration, and the half-maximum concentration and standard deviation were calculated by SigmaPlot 9.0. Only about 50 to 60% of the total radioactive RNA was bound by saturating levels of PyrR; this was consistently observed for all three binding loop RNAs (Fig. 2); a similar result was noted in sedimentation velocity experiments. We suggest that the unbound RNA consists of species that do not fold into the native secondary structure needed for binding.
Electrophoretic gel mobility shift assays of RNA binding to PyrR
Gel shift analysis was performed essentially as described previously [2] with the following modifications. The gels were run in a Bio-Rad (Hercules, CA. USA) Protean IIxi apparatus cooled to 4°C and contained 6% acrylamide (37.5:1 acrylamide/bis solution), 12.5mM Tris-acetate pH 7.5, 2.5% glycerol and 1mM Mg-acetate. The running buffer contained 12.5 mM Tris-acetate pH 7.5 and 1mM Mg-acetate. The gels were pre-run for 1.5 h at 150V followed immediately by loading of 15 μl sample, electrophoresis for 15 min at 50V and for 3 h at 300V. After electrophoresis, the gels were dried on filter paper and exposed overnight to a PhosphorImager screen. Data were analyzed as described above. The RNA binding mixture (50 μl) used for gel shifts was the same as for the filter binding experiments, except that BSA was omitted and 0.0125% loading dye (xylene cyanol) and 5% glycerol were added. Binding of RNA to appreciably higher concentrations of PyrR could be characterized by the gel shift method than with the filter binding method described above, because concentrations of PyrR in excess of 1 μM were not quantitatively retained by the nitrocellulose filter.
Preparation of ribonuclease-free PyrR for analytical ultracentrifugation
His-tagged B. caldolyticus PyrR was used in the ultracentrifugation experiments. Direct comparison of the binding of BcBL2 RNA to His-tagged PyrR and to native PyrR under identical conditions by the filter binding methods showed that binding of RNA to His-tagged PyrR was modulated by nucleotides in the same manner as native PyrR, but the apparent Kd for RNA was about 100 fold larger. His-tagged PyrR was prepared as follows. The PyrR coding sequence was amplified from pSHCO2, a plasmid containing B. caldolyticus PyrR [10], and ligated into plasmid pET412b, which is a pET41 derivative encoding a C-terminal octa-histidine tag preceded by cleavage site for tobacco etch virus protease (Etti Harms, Purdue University, personal communication); the resultant plasmid is pCalCHIS. E.coli strain BL21-DE3 was transformed with pCalCHIS for pyrR expression. Cultures of E. coli BL21-DE3/pCalCHIS were grown in LB medium at 37°C until A600 reached 0.8, induced with 0.4 mM IPTG and grown 4 h at 37°C. Cells were harvested and lysed by sonication in buffer containing 20 mM Tris, pH 7.5, 500 mM NaCl. The lysate was centrifuged at 15,000 rpm, 4°C for 30 min. Because PyrR at high concentration is insoluble in the high-salt buffers used for Ni-affinity purification, the purification protocol employed high ionic strength prior to elution, and low ionic strength during elution. Following filtration through a 0.4 micron membrane, the supernatant was batch-bound to Ni-Sepharose resin (Amersham) in buffer containing 20 mM Tris, pH 7.5, 500 mM NaCl supplemented with 100 mM imidazole to minimize binding of E. coli proteins. The resin was washed in 20 mM Tris, pH 7.5 and 100 mM imidazole, packed into a Biorad Econo-Column and eluted in 20 mM Tris, pH 7.5 with a 0.1 to 1.0 M imidazole gradient. The protein eluted at approximately 600 mM imidazole. Fractions were pooled and dialyzed against 20 mM Tris, pH 7.5, 5% glycerol. Remaining traces of RNase contaminants were by chromatography of purified PyrR on an Amersham Superdex 200 HiLoad16/60 column, eluting with 0.1 M Tris-HCl, pH 7.5 and 0.15 M NaCl. The pooled fractions were dialyzed against buffer containing 20 mM Tris, pH 7.5. The RNase-free status of purified PyrR was confirmed by detection of no degraded RNA species on gel electrophoresis of samples that had been incubated with radiolabeled RNA for 2 weeks at room temperature.
Analytical ultracentrifugation
Sedimentation equilibrium and velocity experiments using either native PyrR or His-tagged PyrR were run on a Beckman XL-I Analytical Ultracentrifuge (Beckman-Coulter, Fullerton, CA, USA) at 20°C. Carbon filled epon two sector centerpieces and sapphire windows were used for all experiments. In all experiments, a buffer of 20 mM Tris-Cl, pH7.5 and some cases 10 mM MgCl2 and 0.1 M NaCl were included. The cell and rotor were equilibrated under vacuum and at 0 RPM for at least 1 h prior to the start of the run so that the system could reach thermal equilibrium. For the velocity experiments the centrifuge was started from 0 RPM and data collection was started immediately with scans collected every 3 min. Both absorbance and interference data were usually collected. For the equilibrium experiments, both approach to equilibrium data and data at equilibrium were collected and the samples were pre-equilibrated as above. The condition of equilibrium was tested using the program Winmatch 0.99 from the RASMB software archive, www.rasmb.bbri.org/rasmb. The sample took 40 h to initially reach equilibrium and 24 h between speed changes.
Multiwavelength sedimentation velocity experiments were undertaken to define the stoichiometry of the RNA-PyrR complex in solution. However, only data obtained at 260 nm had sufficient signal to noise ratio to be used for the entire range of the titration. His-tagged PyrR was prepared by dialysis against buffer containing 20 mM Tris-Cl, pH 7.5, 10mM MgCl2. BcBL2 RNA (nucleotide nos. 702 through 737, see Fig. 1B), which was chemically synthesized by Integrated DNA Technologies (Coralville, IA, USA) was prepared in the same buffer. These experiments were run in a Beckman Optima XL-I ultracentrifuge. A 12 mm path length cell containing a double-sector carbon-filled epon centerpiece with sapphire windows was used for all experiments, and this cell was treated with RNaseZAP (Ambion) for 24 h to remove nucleases. The reference and sample sectors were filled with 400 μL of dialysis buffer and sample, containing 0.3 μM RNA, respectively. After thermal equilibration of the sample (at 20°C for at least 1 h at 0 rpm), all sedimentation velocity experiments were run at 50,000 rpm at 20°C and scans were recorded at three-minute intervals using both absorbance (at 278 nm and 260 nm) and interference optics.
The initial sedimentation velocity analysis contained only 0.3 μM RNA, which has a calculated molecular mass of 11,564 Da. Immediately after completion of this first experiment, a 5 μL aliquot of dialysis buffer was added to the reference sector, and a 5 μL aliquot of concentrated PyrR protein added to the RNA in the sample chamber. This equimolar mix of PyrR subunit to RNA was sedimented as well. Subsequent sedimentation velocity analyses were performed by titrating the RNA with increasing amounts of PyrR in varying molar ratios. The total time of the binding experiment was 3 days. A final experiment was run in the absence of added RNA at a PyrR subunit concentration of 1.2 μM.
Data were considered fit when plots of the fitting residuals were observed to be evenly distributed around zero and the r.m.s.d. was lower than the signal to noise determined from the plateau region of the first scan. Data were usually analyzed using the hybrid local continuous/global discrete model [16] with 0 to 3 discrete species included. For all discrete species, the buoyant mass was calculated, so that masses from molecules with dissimilar partial specific volumes (protein and RNA) would be additive. PyrR has no tryptophan residues, and at the concentrations used in these experiments, A280 and A260 would be no more than 0.0075 AU at the highest protein concentration (1.8 μM).
Solvent densities and viscosities were calculated to be 1.00 g/mL and 0.01 Poise, respectively, for the Tris buffer and 1.0039 g/ml and 0.01 Poise for the NaCl-containing buffer using SEDNTERP [29]. The partial specific volume of 0.7398 cm3/g was also calculated with SEDNTERP from the sequence of His-tagged PyrR and 0.7450 was calculated from the sequence of the native protein; an approximate value for RNA, 0.51 cm3/g, was obtained from Table 2 of Durchschlag and Zipper [30]. The data were fit to various models using the appropriate algorithms in SEDFIT9.3b [31] and SEDPHAT4.1b [16], which were obtained from www.analyticalultracentrifugation.com.
Growth of B. subtilis cells and ATCase assay
For studies of the effects on supplementation of the growth medium with nucleosides on ATCase levels in B. subtilis, cultures strains DB104 and DB104ΔpyrR [4] were grown on buffered minimal medium [5] that included 0.1% (w/v) casamino acids (Becton Dickinson/Difco, Franklin Lakes, NJ, USA), 50 mg per ml histidine and 6 mg per ml disodium succinate, supplemented as shown in Table 5 with nucleosides. Cells were grown at 37°C with aeration, harvested in the exponential growth phase by centrifugation, and ruptured by sonication. Cell debris was removed by a 10 min centrifugation (14,000 × g) and the supernatant was used in assays.
ATCase activity was determined by a radiometric procedure [32]. Assays were performed at 30°C in 100 μl final volume containing the following: 50 mM Tris-acetate (pH 8.3), 50 mM [14C]-aspartate (final specific activity of 10 μCi/mmol, Amersham), and 10 mM carbamoyl phosphate (Sigma, St. Louis, MO, USA). Reactions were initiated by addition of enzyme and stopped with 900 μl 0.2 N acetic acid after 30 min. A 950 μl portion of each reaction was added to a column containing Dowex AG-50W-X8 (200-400 mesh hydrogen form, Bio-Rad) to retain unreacted aspartate. Columns were washed 4 times with 400 μl 0.2 N acetic acid with the eluate collected in scintillation vials. Aquasol-2 (Packard Research/Perkin-Elmer, Meriden, CT, USA) was added to the collected eluate, total counts were determined using liquid scintillation counting and used to calculate ATCase activity. Protein concentrations were determined using the BCA protein assay kit (Pierce, Rockford, IL, USA), using bovine serum albumin as the protein standard.
Supplementary Material
Supplementary Material
Descriptive text
Table S1. Structural variants of purine and pyrimidine nucleotides that affect binding of PyrR to BcBL2 as determined by filter binding
Table S2. Apparent RNA dissociation constants (Kd-values) for binding of structural variants of BcBL2 to PyrR
Table S3. Sedimentation velocity analysis of native and His-tagged PyrR
Table S4. Sedimentation velocity constants from the titration of BcBL2 with PyrR
Figure S1. Effects of the pH of the binding buffer on reactions incubated at 0°C and of temperature on reactions at pH 7.5 on binding of BcBL2 to PyrR as determined by filter binding
Figure S2. Sedimentation velocity studies on native and His-tagged PyrR at moderate ionic strength
Figure S3. An approach to equilibrium study with the native PyrR at low ionic strength
Figure S4. Equilibrium sedimentation studies of native and His-tagged PyrR at low ionic strength and concentrations shown
Supplementary Material
to
pyr RNA binding to the Bacillus caldolyticus PyrR attenuation protein. Characterization and regulation by uridine and guanosine nucleotides
by
Casper Møller Jørgensen, Christopher J. Fields, Preethi Chander, Desmond Watt, John W. Burgner, II, Janet L. Smith and Robert L. Switzer
Doc. S1. Supplementary Results and Discussion
Structural requirements of effectors for affecting PyrR binding to BcBL2
To learn more about how PyrR differentiates between purine and pyrimidine nucleotides, we tested the ability of purine and pyrimidine nucleotide structural variants to activate or inhibit binding of BcBL2 to PyrR (Table S1). All of the pyrimidine nucleotides tested were capable of activating RNA binding to PyrR, which suggests that pyrimidine nucleotides with bulky additions to the pyrimidine ring (OMP and TMP) can bind to PyrR. This is in agreement with the crystal structure [1], in which this part of the pyrimidine ring does not make direct contacts with PyrR.
The structural requirements for the effects of purine nucleotides are more restrictive than for pyrimidine nucleotides. Substitution of the 2-exocyclic amino group in GMP (R5 in Table S1) with an oxo-group in XMP essentially abolished the effect of the nucleotide on RNA binding. Removal of this amino group (IMP) permitted inhibition of binding to BcBL2, but the apparent dissociation constant for binding of RNA was 10-fold lower than with GMP. AMP did not affect binding of BcBL2 to PyrR. The data suggest that both the exocyclic oxo and amino groups of GMP make functionally important contacts to PyrR.
Both 2’-GMP and 3’-GMP increased the apparent dissociation constant for BcBL2 binding by two to three-fold over the no effector level, in contrast to the 40-fold effect of 5’-GMP. These observations, together with their low intracellular concentrations, suggest that these nucleotides do not play a significant role in regulation of pyr gene expression in vivo. 2’, 3’-cGMP did not affect binding of PyrR to BcBL2, but 3’, 5’-cGMP increased the apparent dissociation constant by almost eight-fold. The physiological relevance of these findings is doubtful, but the data indicate that the 2’-hydroxyl group of ribose in GMP is important for binding of the nucleotide to PyrR or modulating its effect on RNA binding.
Binding of BcBL2 structural variants to PyrR
We wanted to determine whether the binding of RNA to B. caldolyticus PyrR exhibits the high sequence specificity expected from previous genetic and biochemical studies with B. subtilis PyrR. To examine this question, we used the filter binding assay to examine binding of B. caldolyticus PyrR to three B. caldolyticus PyrR binding loops containing single base substitutions (Fig. 1B) that were observed in previous gel shift studies with B. subtilis PyrR to have very different apparent Kd values [2].
The G726A sequence variant of BcBL2 has a changed base pair in the upper stem predicted not to affect the structure of the binding loop and does not alter a conserved nucleotide. B. caldolyticus PyrR bound to the G726A structural variant with the same apparent Kd as the wild type binding loop, both in the presence and absence of nucleotides (Table S2). Gel shift assay of the corresponding structural variant in B. subtilis (G728A) also showed tight binding to PyrR in the presence of UMP [2].
The G at position 723 in BcBL2 is part of a sequence of nucleotides in the terminal loop that is conserved in PyrR binding loops from many bacteria; replacement of this nucleotide is predicted to disrupt binding to PyrR. Filter binding studies of the B. caldolyticus G723A BcBL2 replacement determined that the apparent Kd in the presence of UMP was increased by more than 1300-fold relative to the Kd of wild type BcBL2 (Table 6). Impaired binding of PyrR to the G723A structural variant was even more pronounced without nucleotide effectors or in the presence of 0.5 mM GMP. Gel mobility shift assays with B. subtilis PyrR also indicated that replacement of this G with an A (G725A in BsBL2) disrupted PyrR binding in the presence of UMP; the variant RNA bound with an apparent Kd that was more than 800-fold larger than the Kd for the wild type BsBL2 binding loop [2].
The A724C replacement in BcBL2 (BcBL2-A724C) is expected to reduce binding by PyrR, since it, like BcBL2-G723A, changes a conserved residue in the terminal loop. In previous gel mobility shift assays with this structural variant (A726C in B. subtilis) and B. subtilis PyrR, a very high apparent Kd was observed (20 μM in the presence of UMP [2]). However, we did not observe the same large effect on binding of the BcBL2-A724C RNA variant in the filter binding studies with B. caldolyticus PyrR (Table 6). In the presence of UMP, the apparent Kd was close to that of wild type BcBL2. With no effector and with GMP, on the other hand, decreased affinity of PyrR for the A724C structural variant was observed, indicating that the A724 nucleotide is indeed involved in PyrR binding to BcBL2.
The binding of the A724C structural variant of BcBL2 was investigated further using the gel shift method and B. caldolyticus PyrR. Strikingly, no binding of the BcBL2 A724C RNA to PyrR (up to 10 μM PyrR) was detected under the conditions used for the studies in Table 4 and Fig. 3. This was in sharp contrast to the relatively tight binding of the same RNA observed by filter binding. However, when the Mg2+ concentration in the electrophoresis gel was increased to 10 mM, binding of this RNA variant was detected (Kd of 3500 nM in the absence of effector, bimodal binding with Kd1 =1.2 nM and Kd2 >20,000 nM in the presence of 0.5 mM UMP). These values are much larger than those obtained by filter binding (in which the concentration of Mg2+ is also 10 mM). These observations suggest that the A724C variant RNA differs from the wild type BcBL2 in its interaction with Mg2+. They also point out an unusual instance in which the filter binding and gel shift assays yielded highly divergent results even though the same RNA and PyrR were used under as close to identical conditions as possible.
Effects of Mg2+, pH and temperature on binding of PyrR to BcBL2
The standard buffer used in filter binding assays of RNA binding contained 10 mM Mg2+ and 50 mM K+; varying the concentration of Mg2+ in the range from 1 mM to 50 mM or the K+ concentration between 10 mM and 300 mM did not affect binding of PyrR to BcBL2 or BcBL3 (data not shown). However, when Mg2+ was omitted from the binding buffer, BcBL2 RNA bound to PyrR with an affinity about 15-fold lower than when Mg2+ was present. Inclusion of 10 mM EDTA in the binding reaction without added Mg2+ further reduced the apparent affinity of PyrR for BcBL2 by about 100 fold. This documents a requirement of the system for divalent cations for RNA binding and suggests that traces of such cations may contaminate the reaction components.
The effects of changing the pH of the binding buffer in the range of pH 5.5 to 8.5 on the binding of BcBL2 to PyrR are shown in Fig. S1A. With no effector present, PyrR bound 50-fold less tightly to BcBL2 at pH 5.5 than at the standard pH of 7.5 used in this study. When UMP was present in the reaction mix, the same change in pH resulted in less than a three-fold change in the apparent Kd. The addition of GMP, on the other hand, had an opposite and very large effect on binding of PyrR to BcBL2, i.e., a more than 300-fold higher apparent Kd was observed at pH 8.5 than at pH 6.5. These results indicate that when the pH is changed, a residue in PyrR that is important for binding of RNA or nucleotides undergoes altered ionization. This residue could be a histidine, since the pKa value for the imidazole side chain of N-acetyl-histidine ethyl ester is 7. No ionizable group in the GMP binding site in the crystal structure accounts for the observed pH dependence. Although none of the histidine residues in PyrR lie closer to a purine nitrogen of GMP than 8 Å [1], His82 and His21 are candidates for mediating the observed pH effect. His82 resides on a flexible loop adjacent to the nucleotide binding site, and may be proximal to the guanine base in unseen conformations of the enzyme. His21 lies in the putative RNA binding face of PyrR [1, 3], and the equivalent residue in B. subtilis PyrR, His22, was strongly implicated in RNA binding by the mutagenesis studies of Savacool and Switzer [4]. In B. caldolyticus PyrR, His21 is in the interface between dimers that forms the closed tetramer, a position in which its ionization state may influence tetramer-dimer equilibration. The intracellular pH in B. subtilis cells has been determined to lie in the range from 7.6 to 8.3 in growth media with pH buffered at values from 6 to 8 [5] and is likely similar in B. caldolyticus. Thus, the marked differences in GMP and UMP effects on RNA binding occur at a physiologically relevant pH.
Analytical ultracentrifugation of PyrR and PyrR-RNA complexes
Several sedimentation velocity studies were performed on both native and His-tagged PyrR as a function of protein and salt concentrations. Since Mg2+ is required for RNA binding, 10 mM Mg2+ was also included in all cases. Figure S2 shows the results of sedimentation velocity studies on both the native (Panels A, B, and C) and His-tagged (Panels D, E, and F) PyrR. The data in both cases were analyzed with the program SEDFIT [6]. Panels A and D show boundary movement as a function of time with dotted lines, the solid lines show fits to the data based on the c(s) distributions as a function of s20,w shown in Panels C and F. Panels B and E show the residuals between observed and fit data. The distributions of PyrR as a function of s20,w in Panels C and F indicate that nearly all of the PyrR, regardless of the source, sediments with s values near 5 S (see Figure S2 legend for further details). The calculated masses of the 5 S particles are 83 and 100 kDa for the native and His-tagged PyrR, respectively. A sedimentation velocity coefficient of 5.2 S was calculated from the Protein Data Base file for the native PyrR x-ray structure (1XZN) using the bead modeling program HYDROPRO [7], which is in good agreement with the observed s20,w values. The calculated mass values are somewhat greater than the sequence mass for a tetramer (Table S3), which is likely the result of rapid, weak formation of higher order oligomers (octomers, etc.). Further characterization of these larger oligomers was not attempted. The values for s20,w given in legend of Figure S2 were further corrected for protein concentration using SEDINTERP; the frictional ratio (f/f0) was estimated from these values and the corresponding sequence masses of the tetramer (Table S3). The value of this ratio was calculated using the Teller algorithm from the program SEDNTERP v1.09 (which was obtained from http://www.rasmb.bbri.org/rasmb/ms_dos/) and assuming 0.4 g of water per 1.0 g of protein [8]; it is about 1.28 for the oligomeric form of both native and His-tagged PyrR, which suggests that the addition of the His-tag does not significantly alter the shape of the tetramer. Thus, we suggest that using the His-tagged PyrR for the long-term titration studies shown in Figures 5 and S5 to avoid possible ribonuclease contamination is reasonable.
As noted in the Discussion, crystallographic studies on this PyrR suggest that the potential binding site for RNA is sterically hindered in the tetramer. Titration studies in the analytical ultracentrifuge (Figures 4 and 5) indicate that the PyrR-RNA complex likely consists of a dimer of PyrR and a single molecule of RNA. To investigate the tetramer-dimer equilibrium further, we undertook sedimentation equilibrium experiments with both the native and His-tagged PyrR at monomer concentrations ranging from 0.25 μM to 25 μM, which represents the lower end of the useful range for detecting an equilibrium distribution of PyrR in the ultracentrifuge. As part of this experiment, we also collected “approach to equilibrium” data [9] at the slowest speed used (8000 and 10,000 rpm for the native and His-tagged PyrR, respectively). Figure S3 shows the approach to equilibrium data for three different concentrations of native PyrR; each pair of plots, reading horizontally, shows the interference and absorbance data as well as (below) the calculated residuals at the PyrR concentrations shown in the legend. The first 10 absorbance and interference scans were used such that the sample was close to but not at equilibrium. (Including an excess of scans at equilibrium or very near equilibrium conditions seems to prevent an accurate estimation of particle mass, according to studies with other proteins; Burgner, J. W. unpublished observations). The data at each concentration were fitted to a single species model using SEDPHAT 4.1. Masses of 61, 71, and 82 kDa were estimated from low to high PyrR concentrations. In our hands, this method gives estimates of the mass for simple systems within 1 to 5% of the sequence mass, but the estimate of the sedimentation coefficient is much more variable, as should be expected from the lack of sharpness in the boundaries. The decrease in the weight average mass, which is calculated here, with decreasing protein concentration is consistent with a dissociation constant for the tetramer that is less than 10-7 M.
Figure S4 shows the absorbance distribution as a function of rotational speed for native (Panel A) and His-tagged (Panel B) at equilibrium. The equilibrium data were collected at ~60 h for the slowest speed (see Figure S4 legend) and after an additional 40 h at each of the remaining speeds. These absorbance and interference (not shown) data were collected (see Figure S3, additional details) and fitted to a single species model using mass conservation. The calculated mass the native PyrR is 78.3 kDa for both the absorbance data alone and a combined set of both. The calculated mass for the His-tagged PyrR is 90.7 and 91.6 kDa for the absorbance and combined data, respectively. These values compare well with the sequence masses of the native (79.7 kDa) and His-tagged (89.2 kDa) PyrR. As indicated in the Results section, these data were also fit using a dimer-tetramer formation model using SEDPHAT and mass conservation between speeds. The estimated value for the Kd for a tetramer to dimer disassociation model is about 3 × 10-8 M for both PyrR forms. The interference data were not included in these calculations because blank data were unfortunately not collected; thus, corrections for changes in the baseline as a function of speed could not be made with sufficient accuracy. This is not the case for either the equilibrium absorbance data or all of the approach to equilibrium data, where time and radial independent noise can be removed algebraically [9]. Values for the global reduced χ2 were calculated for each fit and appear in the legend of Figure S4. It would appear from these values that the dimer-tetramer model does not provide a significantly better fit to the data than a tetramer-only model. However, the decrease in observed masses with decreased PyrR concentration observed in Figure S3 is suggestive of dissociation of the tetramer. Thus, assuming the weight average molecular mass describes only a dimer-tetramer equilibrium, the observed 60 kDa at the lowest native PyrR concentration would predict a Kd of about 2 × 10-8 M, which is in reasonable agreement with that calculated from the equilibrium data.
Figure S5 shows the sedimentation velocity data from the titration of His-tagged PyrR with RNA as well as the c(s) distribution plots for interference, A260 and A278 data and the residual plots from these fits. The distribution plots obtained for each titration point as well as for the reactants are qualitatively the same. While there are sufficient data to determine, by solving the appropriate simultaneous equations, the ratio of RNA to PyrR in the new ~4.7S peak found, the signal to noise prevented an accurate assessment of this ratio by multiwavelength analysis. However, the plot of unreacted RNA against the ratio of PyrR added to total RNA predicts a 2 to 1 ratio of PyrR to RNA (see Figure 5, main text).
References
1. Chander P, Halbig, KM, Miller JK, Fields CJ, Bonner HKS, Grabner GK, Switzer RL & Smith, JL (2005) Structure of the nucleotide complex of PyrR, the pyr attenuation protein from Bacillus caldolyticus, suggests dual regulation by pyrimidine and purine nucleotides. J Bacteriol 187, 1773-1782.
2. Bonner ER, D'Elia JN, Billips BK & Switzer RL (2001) Molecular recognition of pyr mRNA by the Bacillus subtilis attenuation regulatory protein PyrR, Nucleic Acids Res 29, 4851-4865.
3. Tomchick DR, Turner RJ, Switzer RL & Smith JL (1998) Adaptation of an enzyme to regulatory function: structure of Bacillus subtilis PyrR, a bifunctional pyr RNA-binding attenuation protein and uracil phosphoribosyltransferase. Structure 6, 337-350.
4. Savacool HK & Switzer RL (2002) Characterization of the interaction of Bacillus subtilis PyrR with pyr mRNA by site-directed mutagenesis of the protein. J Bacteriol 184, 2521-2528.
5. Breeuwer P, Drocourt J-L, Rombouts FM & Abee T (1996) A novel method for continuous determination of the intracellular pH in bacteria with the internally conjugated fluoresecent probe 5 (and 6-)-carboxyfluorescein succinimidyl ester. Appl Environ Microbiol 62, 178-183.
6. Schuck P (2000) Size distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J 78, 1606-1619.
7. Garcia de la Torre J, Navarro S, Lopez Martinez MC, Diaz FG & Lopez Cascales JJ (1994) HYDRO: a computer software for the prediction of hydrodynamic properties of macromolecules. Biophys J 67, 530-531.
8. Laue TM, Shah BD, Ridgeway TM & Pelletier SL (1992) Computer-aided interpretation of analytical sedimentation data for proteins. in Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding SE, Rowe AJ & Horton JC, eds) pp. 90-125, The Royal Society of Chemistry, Cambridge, UK.
9. Schuck P & Demeler B (1999) Direct sedimentation analysis of interference-optical data in analytical ultracentrifugation. Biophys J 76, 2288-2296.
Table S1. Structural variants of purine and pyrimidine nucleotides that affect binding of PyrR to BcBL2 as determined by filter binding
Table S2. Apparent RNA dissociation constants (Kd-values) for binding of structural variants of BcBL2 to PyrR
Table S3. Sedimentation velocity analysis of native and His-tagged PyrR
Table S4. Sedimentation velocity constants from the titration of BcBL2 with PyrR
Supplementary Material: Figure Legends
Figure S1. Effects of the pH of the binding buffer on reactions incubated at 0°C (A) and of temperature on reactions at pH 7.5 (B) on binding of BcBL2 to PyrR as determined by filter binding. Open circles: no effector; closed circles: 500 μM UMP; closed triangles: 500 μM GMP.
Figure S2. Sedimentation velocity studies on native and His-tagged PyrR at moderate ionic strength. Panels A, B, and C show the results of a sedimentation velocity run with native PyrR in 0.1 M NaCl at 50,000 rpm and 20°C. Panel A shows changes in absorbance as a function of radial position (dots) at six min intervals and the results (solid lines) from a c(s) distribution analysis using the program SEDPHAT 4.1. Panel B shows the calculated residuals against radial position for each scan in Panel A, and Panel C shows the c(s) distribution against s corrected to s20,w. The weight average sedimentation coefficient from the integration of the principal peak is 4.83 S. Panels D, E, and F show the results from an identical sedimentation velocity experiment and analysis using His-tagged PyrR. The weight average s20,w calculated from these results is 5.34 S. Additional details for both experiments are found in the Data Supplement text and Table S3.
Figure S3. An approach to equilibrium study with the native PyrR at low ionic strength. The initial concentrations of PyrR studied were: Panels A and B, 0.25 μM; Panels C and D, 1.25 μM; Panels E and F, 25 μM. The data were collected at 8000 rpm. Rayleigh interference optics were used for Panels A, C, and E; absorbance optics at 232 nm were used for Panels B and D; and absorbance at 278 nm was used for Panel F. The calculated values for s20,w and observed mass based on a global single species fit to both data sets with the program SEDPHAT 4.1 are 5.7 S and 61 kDa, respectively, for a native PyrR concentration of 0.25 μM . The calculated values for s20,w and mass for PyrR at an initial concentration of 1.25 μM are 3.7 S and 71 kDa , respectively, and a loading concentration of 25 μM PyrR, the calculated values for s20,w and mass are 5.1 S and 82 kDa. The solid lines in each panel are the fitted results from the single species model (see Data Supplement text); the calculated residuals against radial position for each scan are shown in the boxes below each panel.
Figure S4. Equilibrium sedimentation studies of native and His-tagged PyrR at low ionic strength and concentrations shown. The absorbance data in the upper panel (native PyrR) and the lower panel (His-tagged PyrR) were obtained from a continuation of the experiments shown in Figure S2 in that the samples were allowed to come to equilibrium at 8 k, 12 k, 1k 17 k, and 20 k rpm and absorbance scans across the cell were collected at A232, A232, and A278, respectively, from left to right. The solid lines were calculated based on a single discrete species model, with mass conservation, using SEDPHAT 4.1 and a mass of 78.3 kDa. The data in the lower panel were obtained using the same procedures as in the upper panel, except that the samples were allowed to come to equilibrium at 12 k, 15 k, 17 k, and 20 k rpm. Absorbance scans were collected at the same wavelengths indicated in the upper panel. The solid lines were calculated based on a single discrete species model, with mass conservation, using SEDPHAT4.1 and a mass of 91.6 kDa. Calculated residuals against radial position for each scan are shown in the boxes below each panel.
Figure S5. Sedimentation velocity studies of the interaction between RNA and His-tagged PyrR. The concentrations of PyrR and RNA in each sample are shown at the left of each row of panels; additional details appear in the legend of Figure 4. The three sets of scans shown in each row of panels were collected using Rayleigh interference, A 260 and A278, from left to right, respectively. The data were fitted using the c(s) distribution algorithm of SEDPHAT as described in the Methods section; c(s) distributions were calculated form interference data (red lines), absorbance at 260 nm (green lines), absorbance at 278 nm (blue lines).
Acknowledgements
This research was supported by United States Public Health Service Grants GM47112 to RLS, DK42303 to JLS and from the Purdue University Bindley Bioscience Center to JWB.
References
- 1.Switzer RL, Turner RJ, Lu Y. Regulation of the Bacillus subtilis pyrimidine biosynthetic operon by transcriptional attenuation: control of gene expression by an mRNA-binding protein. Prog Nucl Acids Res Mol Biol. 1999;62:329–367. doi: 10.1016/s0079-6603(08)60512-7. [DOI] [PubMed] [Google Scholar]
- 2.Bonner ER, D'Elia JN, Billips BK, Switzer RL. Molecular recognition of pyr mRNA by the Bacillus subtilis attenuation regulatory protein PyrR. Nucleic Acids Res. 2001;29:4851–4865. doi: 10.1093/nar/29.23.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner RJ, Bonner ER, Grabner GK, Switzer RL. Purification and characterization of Bacillus subtilis PyrR, a bifunctional pyr mRNA-binding attenuation protein/uracil phosphoribosyltransferase. J Biol Chem. 1998;273:5932–5938. doi: 10.1074/jbc.273.10.5932. [DOI] [PubMed] [Google Scholar]
- 4.Turner RJ, Lu Y, Switzer RL. Regulation of the Bacillus subtilis pyrimidine biosynthetic (pyr) gene cluster by an autogenous transcriptional attenuation mechanism. J Bacteriol. 1994;176:3708–3722. doi: 10.1128/jb.176.12.3708-3722.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu Y, Turner RJ, Switzer RL. Roles of the three transcriptional attenuators of the Bacillus subtilis pyrimidine biosynthetic operon in the regulation of its expression. J Bacteriol. 1995;177:1315–1325. doi: 10.1128/jb.177.5.1315-1325.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghim S-Y, Switzer RL. Characterization of cis-acting mutations in the first attenuator region of the B. subtilis pyr operon that are defective in regulation of expression by pyrimidines. J Bacteriol. 1996;178:2351–2355. doi: 10.1128/jb.178.8.2351-2355.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghim S-Y, Switzer RL. Mutations in B. subtilis PyrR, the pyr regulatory protein with defects in regulation by pyrimidines. FEMS Microbiol Lett. 1996;137:13–18. doi: 10.1111/j.1574-6968.1996.tb08075.x. [DOI] [PubMed] [Google Scholar]
- 8.Tomchick DR, Turner RJ, Switzer RL, Smith JL. Adaptation of an enzyme to regulatory function: structure of Bacillus subtilis PyrR, a bifunctional pyr RNA-binding attenuation protein and uracil phosphoribosyltransferase. Structure. 1998;6:337–350. doi: 10.1016/s0969-2126(98)00036-7. [DOI] [PubMed] [Google Scholar]
- 9.Lu Y, Switzer RL. Transcriptional attenuation of the Bacillus subtilis pyr operon by the PyrR regulatory protein and uridine nucleotides in vitro. J Bacteriol. 1996;178:7206–7211. doi: 10.1128/jb.178.24.7206-7211.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chander P, Halbig KM, Miller JK, Fields CJ, Bonner HKS, Grabner GK, Switzer RL, Smith JL. Structure of the nucleotide complex of PyrR, the pyr attenuation protein from Bacillus caldolyticus, suggests dual regulation by pyrimidine and purine nucleotides. J Bacteriol. 2005;187:1773–1782. doi: 10.1128/JB.187.5.1773-1782.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chander P. PhD thesis. Purdue University; West Lafayette, IN: 2006. Structural analysis of the regulatory protein of pyrimidine biosynthesis, PyrR. [Google Scholar]
- 12.Ghim S-Y, Nielsen P, Neuhard J. Molecular characterization of pyrimidine biosynthesis genes from the thermophile Bacillus caldolyticus. J Gen Microbiol. 1994;140:479–491. doi: 10.1099/00221287-140-3-479. [DOI] [PubMed] [Google Scholar]
- 13.Ghim S-Y, Neuhard J. The pyrimidine biosynthesis operon of the thermophile Bacillus caldolyticus includes genes for uracil phosphoribosyltransferase and uracil permease. J Bacteriol. 1994;176:3698–3707. doi: 10.1128/jb.176.12.3698-3707.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong L, Lohmann TM. A double-filter method for nitrocellulose-filter binding: Application to protein-nucleic acid interactions. Proc Nat Acad Sci USA. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuch R, Garibian A, Saxild HH, Piggot PJ, Nygaard P. Nucleosides as a carbon source in Bacillus subtilis: characterization of the drm-pupG operon. Microbiology (Reading) 1999;145:2957–2966. doi: 10.1099/00221287-145-10-2957. [DOI] [PubMed] [Google Scholar]
- 16.Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- 17.Balbo A, Minor KH, Velikovsky CA, Mariuzza RA, Peterson CB, Schuck P. Studying multiprotein complexes by multisignal sedimentation velocity analytical ultracentrifugation. Proc Nat Acad Sci USA. 2005;102:81–86. doi: 10.1073/pnas.0408399102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savacool HK, Switzer RL. Characterization of the interaction of Bacillus subtilis PyrR with pyr mRNA by site-directed mutagenesis of the protein. J Bacteriol. 2002;184:2521–2528. doi: 10.1128/JB.184.9.2521-2528.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Møllgaard H, Neuhard J. Deoxycytidylate deaminase from Bacillus subtilis. J Biol Chem. 1978;253:3536–3542. [PubMed] [Google Scholar]
- 20.Saxild HH, Nygaard P. Regulation of levels of purine biosynthetic enzymes in Bacillus subtilis: effects of changing purine nucleotide pools. J Gen Microbiol. 1991;137:2387–2394. doi: 10.1099/00221287-137-10-2387. [DOI] [PubMed] [Google Scholar]
- 21.Bernlohr DA, Switzer RL. Regulation of Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase inactivation in vivo. J Bacteriol. 1983;153:937–949. doi: 10.1128/jb.153.2.937-949.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulus TJ, Switzer RL. Synthesis and inactivation of the carbamyl phosphate synthetase isozymes of Bacillus subtilis during growth and sporulation. J Bacteriol. 1979;140:769–773. doi: 10.1128/jb.140.3.769-773.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gagyi C, Burcurenci N, Sirbu O, Labesse G, Ionescu M, Ofiteru A, Assairi L, Landais S, Danchin A, Barzu O, Gilles A-M. UMP kinase from the Gram-positive bacterium Bacillus subtilis is strongly dependent on GTP for optimal activity. Eur J Biochem. 2003;270:3196–3204. doi: 10.1046/j.1432-1033.2003.03702.x. [DOI] [PubMed] [Google Scholar]
- 24.Willemöes M, Sigurskjold BW. Steady-state kinetics of the glutaminase reaction of CTP synthase from Lactococcus lactis. the role of the allosteric activator GTP in coupling betyween glutamine hydrolysis and CTP synthesis. Eur J Biochem. 2002;19:4772–4779. doi: 10.1046/j.1432-1033.2002.03175.x. [DOI] [PubMed] [Google Scholar]
- 25.Krasny L, Gourse RL. An alternative strategy for bacteria ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO Journal. 2004;23:4473–4483. doi: 10.1038/sj.emboj.7600423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez JM, Dromerick A, Freese E. Response of guanosine 5'-triphosphate concentration to nutritional changes and its significance for Bacillus subtilis sporulation. J Bacteriol. 1981;146:605–613. doi: 10.1128/jb.146.2.605-613.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ratnayake-Lecamwasam M, Serror P, Wong KW, Sonenshein AL. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dav. 2001;15:1093–1103. doi: 10.1101/gad.874201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaak JE, Yakhnin H, Bevilqcqua PC, Babitzke P. A Mg2+-dependent RNA tertiary structure forms in the Bacillus subtilis trp operon leader transcript and appears to interfere with trpE translation control by inhibiting TRAP binding. J Mol Biol. 2003;332:555–574. doi: 10.1016/s0022-2836(03)00969-0. [DOI] [PubMed] [Google Scholar]
- 29.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, Rowe AJ, Horton JC, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. The Royal Society of Chemistry; Cambridge, UK: 1992. pp. 90–125. [Google Scholar]
- 30.Durschlag H, Zipper P. Calculation of partial specific volumes and other volumetric properties of small molecules and polymers. J Appl Cryst. 1997;30:803–807. [Google Scholar]
- 31.Schuck P. Size distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perbal B, Herve G. Biosynthesis of Escherichia coli aspartate transcarbamylase I. parameters of gene expression and sequential biosynthesis of the subunits. J Mol Biol. 1972;70:511–529. doi: 10.1016/0022-2836(72)90556-6. [DOI] [PubMed] [Google Scholar]
- 33.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breeuwer P, Drocourt J-L, Rombouts FM, Abee T. A novel method for continuous determination of the intracellular pH in bacteria with the internally conjugated fluoresecent probe 5 (and 6-)-carboxyfluorescein succinimidyl ester. Appl Environ Microbiol. 1996;62:178–183. doi: 10.1128/aem.62.1.178-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia de la Torre J, Navarro S, Lopez Martinez MC, Diaz FG, Lopez Cascales JJ. HYDRO: a computer software for the prediction of hydrodynamic properties of macromolecules. Biophys J. 1994;67:530–531. doi: 10.1016/S0006-3495(94)80512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuck P, Demeler B. Direct sedimentation analysis of interference-optical data in analytical ultracentrifugation. Biophys J. 1999;76:2288–2296. doi: 10.1016/S0006-3495(99)77384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Descriptive text
Table S1. Structural variants of purine and pyrimidine nucleotides that affect binding of PyrR to BcBL2 as determined by filter binding
Table S2. Apparent RNA dissociation constants (Kd-values) for binding of structural variants of BcBL2 to PyrR
Table S3. Sedimentation velocity analysis of native and His-tagged PyrR
Table S4. Sedimentation velocity constants from the titration of BcBL2 with PyrR
Figure S1. Effects of the pH of the binding buffer on reactions incubated at 0°C and of temperature on reactions at pH 7.5 on binding of BcBL2 to PyrR as determined by filter binding
Figure S2. Sedimentation velocity studies on native and His-tagged PyrR at moderate ionic strength
Figure S3. An approach to equilibrium study with the native PyrR at low ionic strength
Figure S4. Equilibrium sedimentation studies of native and His-tagged PyrR at low ionic strength and concentrations shown
Supplementary Material
to
pyr RNA binding to the Bacillus caldolyticus PyrR attenuation protein. Characterization and regulation by uridine and guanosine nucleotides
by
Casper Møller Jørgensen, Christopher J. Fields, Preethi Chander, Desmond Watt, John W. Burgner, II, Janet L. Smith and Robert L. Switzer
Doc. S1. Supplementary Results and Discussion
Structural requirements of effectors for affecting PyrR binding to BcBL2
To learn more about how PyrR differentiates between purine and pyrimidine nucleotides, we tested the ability of purine and pyrimidine nucleotide structural variants to activate or inhibit binding of BcBL2 to PyrR (Table S1). All of the pyrimidine nucleotides tested were capable of activating RNA binding to PyrR, which suggests that pyrimidine nucleotides with bulky additions to the pyrimidine ring (OMP and TMP) can bind to PyrR. This is in agreement with the crystal structure [1], in which this part of the pyrimidine ring does not make direct contacts with PyrR.
The structural requirements for the effects of purine nucleotides are more restrictive than for pyrimidine nucleotides. Substitution of the 2-exocyclic amino group in GMP (R5 in Table S1) with an oxo-group in XMP essentially abolished the effect of the nucleotide on RNA binding. Removal of this amino group (IMP) permitted inhibition of binding to BcBL2, but the apparent dissociation constant for binding of RNA was 10-fold lower than with GMP. AMP did not affect binding of BcBL2 to PyrR. The data suggest that both the exocyclic oxo and amino groups of GMP make functionally important contacts to PyrR.
Both 2’-GMP and 3’-GMP increased the apparent dissociation constant for BcBL2 binding by two to three-fold over the no effector level, in contrast to the 40-fold effect of 5’-GMP. These observations, together with their low intracellular concentrations, suggest that these nucleotides do not play a significant role in regulation of pyr gene expression in vivo. 2’, 3’-cGMP did not affect binding of PyrR to BcBL2, but 3’, 5’-cGMP increased the apparent dissociation constant by almost eight-fold. The physiological relevance of these findings is doubtful, but the data indicate that the 2’-hydroxyl group of ribose in GMP is important for binding of the nucleotide to PyrR or modulating its effect on RNA binding.
Binding of BcBL2 structural variants to PyrR
We wanted to determine whether the binding of RNA to B. caldolyticus PyrR exhibits the high sequence specificity expected from previous genetic and biochemical studies with B. subtilis PyrR. To examine this question, we used the filter binding assay to examine binding of B. caldolyticus PyrR to three B. caldolyticus PyrR binding loops containing single base substitutions (Fig. 1B) that were observed in previous gel shift studies with B. subtilis PyrR to have very different apparent Kd values [2].
The G726A sequence variant of BcBL2 has a changed base pair in the upper stem predicted not to affect the structure of the binding loop and does not alter a conserved nucleotide. B. caldolyticus PyrR bound to the G726A structural variant with the same apparent Kd as the wild type binding loop, both in the presence and absence of nucleotides (Table S2). Gel shift assay of the corresponding structural variant in B. subtilis (G728A) also showed tight binding to PyrR in the presence of UMP [2].
The G at position 723 in BcBL2 is part of a sequence of nucleotides in the terminal loop that is conserved in PyrR binding loops from many bacteria; replacement of this nucleotide is predicted to disrupt binding to PyrR. Filter binding studies of the B. caldolyticus G723A BcBL2 replacement determined that the apparent Kd in the presence of UMP was increased by more than 1300-fold relative to the Kd of wild type BcBL2 (Table 6). Impaired binding of PyrR to the G723A structural variant was even more pronounced without nucleotide effectors or in the presence of 0.5 mM GMP. Gel mobility shift assays with B. subtilis PyrR also indicated that replacement of this G with an A (G725A in BsBL2) disrupted PyrR binding in the presence of UMP; the variant RNA bound with an apparent Kd that was more than 800-fold larger than the Kd for the wild type BsBL2 binding loop [2].
The A724C replacement in BcBL2 (BcBL2-A724C) is expected to reduce binding by PyrR, since it, like BcBL2-G723A, changes a conserved residue in the terminal loop. In previous gel mobility shift assays with this structural variant (A726C in B. subtilis) and B. subtilis PyrR, a very high apparent Kd was observed (20 μM in the presence of UMP [2]). However, we did not observe the same large effect on binding of the BcBL2-A724C RNA variant in the filter binding studies with B. caldolyticus PyrR (Table 6). In the presence of UMP, the apparent Kd was close to that of wild type BcBL2. With no effector and with GMP, on the other hand, decreased affinity of PyrR for the A724C structural variant was observed, indicating that the A724 nucleotide is indeed involved in PyrR binding to BcBL2.
The binding of the A724C structural variant of BcBL2 was investigated further using the gel shift method and B. caldolyticus PyrR. Strikingly, no binding of the BcBL2 A724C RNA to PyrR (up to 10 μM PyrR) was detected under the conditions used for the studies in Table 4 and Fig. 3. This was in sharp contrast to the relatively tight binding of the same RNA observed by filter binding. However, when the Mg2+ concentration in the electrophoresis gel was increased to 10 mM, binding of this RNA variant was detected (Kd of 3500 nM in the absence of effector, bimodal binding with Kd1 =1.2 nM and Kd2 >20,000 nM in the presence of 0.5 mM UMP). These values are much larger than those obtained by filter binding (in which the concentration of Mg2+ is also 10 mM). These observations suggest that the A724C variant RNA differs from the wild type BcBL2 in its interaction with Mg2+. They also point out an unusual instance in which the filter binding and gel shift assays yielded highly divergent results even though the same RNA and PyrR were used under as close to identical conditions as possible.
Effects of Mg2+, pH and temperature on binding of PyrR to BcBL2
The standard buffer used in filter binding assays of RNA binding contained 10 mM Mg2+ and 50 mM K+; varying the concentration of Mg2+ in the range from 1 mM to 50 mM or the K+ concentration between 10 mM and 300 mM did not affect binding of PyrR to BcBL2 or BcBL3 (data not shown). However, when Mg2+ was omitted from the binding buffer, BcBL2 RNA bound to PyrR with an affinity about 15-fold lower than when Mg2+ was present. Inclusion of 10 mM EDTA in the binding reaction without added Mg2+ further reduced the apparent affinity of PyrR for BcBL2 by about 100 fold. This documents a requirement of the system for divalent cations for RNA binding and suggests that traces of such cations may contaminate the reaction components.
The effects of changing the pH of the binding buffer in the range of pH 5.5 to 8.5 on the binding of BcBL2 to PyrR are shown in Fig. S1A. With no effector present, PyrR bound 50-fold less tightly to BcBL2 at pH 5.5 than at the standard pH of 7.5 used in this study. When UMP was present in the reaction mix, the same change in pH resulted in less than a three-fold change in the apparent Kd. The addition of GMP, on the other hand, had an opposite and very large effect on binding of PyrR to BcBL2, i.e., a more than 300-fold higher apparent Kd was observed at pH 8.5 than at pH 6.5. These results indicate that when the pH is changed, a residue in PyrR that is important for binding of RNA or nucleotides undergoes altered ionization. This residue could be a histidine, since the pKa value for the imidazole side chain of N-acetyl-histidine ethyl ester is 7. No ionizable group in the GMP binding site in the crystal structure accounts for the observed pH dependence. Although none of the histidine residues in PyrR lie closer to a purine nitrogen of GMP than 8 Å [1], His82 and His21 are candidates for mediating the observed pH effect. His82 resides on a flexible loop adjacent to the nucleotide binding site, and may be proximal to the guanine base in unseen conformations of the enzyme. His21 lies in the putative RNA binding face of PyrR [1, 3], and the equivalent residue in B. subtilis PyrR, His22, was strongly implicated in RNA binding by the mutagenesis studies of Savacool and Switzer [4]. In B. caldolyticus PyrR, His21 is in the interface between dimers that forms the closed tetramer, a position in which its ionization state may influence tetramer-dimer equilibration. The intracellular pH in B. subtilis cells has been determined to lie in the range from 7.6 to 8.3 in growth media with pH buffered at values from 6 to 8 [5] and is likely similar in B. caldolyticus. Thus, the marked differences in GMP and UMP effects on RNA binding occur at a physiologically relevant pH.
Analytical ultracentrifugation of PyrR and PyrR-RNA complexes
Several sedimentation velocity studies were performed on both native and His-tagged PyrR as a function of protein and salt concentrations. Since Mg2+ is required for RNA binding, 10 mM Mg2+ was also included in all cases. Figure S2 shows the results of sedimentation velocity studies on both the native (Panels A, B, and C) and His-tagged (Panels D, E, and F) PyrR. The data in both cases were analyzed with the program SEDFIT [6]. Panels A and D show boundary movement as a function of time with dotted lines, the solid lines show fits to the data based on the c(s) distributions as a function of s20,w shown in Panels C and F. Panels B and E show the residuals between observed and fit data. The distributions of PyrR as a function of s20,w in Panels C and F indicate that nearly all of the PyrR, regardless of the source, sediments with s values near 5 S (see Figure S2 legend for further details). The calculated masses of the 5 S particles are 83 and 100 kDa for the native and His-tagged PyrR, respectively. A sedimentation velocity coefficient of 5.2 S was calculated from the Protein Data Base file for the native PyrR x-ray structure (1XZN) using the bead modeling program HYDROPRO [7], which is in good agreement with the observed s20,w values. The calculated mass values are somewhat greater than the sequence mass for a tetramer (Table S3), which is likely the result of rapid, weak formation of higher order oligomers (octomers, etc.). Further characterization of these larger oligomers was not attempted. The values for s20,w given in legend of Figure S2 were further corrected for protein concentration using SEDINTERP; the frictional ratio (f/f0) was estimated from these values and the corresponding sequence masses of the tetramer (Table S3). The value of this ratio was calculated using the Teller algorithm from the program SEDNTERP v1.09 (which was obtained from http://www.rasmb.bbri.org/rasmb/ms_dos/) and assuming 0.4 g of water per 1.0 g of protein [8]; it is about 1.28 for the oligomeric form of both native and His-tagged PyrR, which suggests that the addition of the His-tag does not significantly alter the shape of the tetramer. Thus, we suggest that using the His-tagged PyrR for the long-term titration studies shown in Figures 5 and S5 to avoid possible ribonuclease contamination is reasonable.
As noted in the Discussion, crystallographic studies on this PyrR suggest that the potential binding site for RNA is sterically hindered in the tetramer. Titration studies in the analytical ultracentrifuge (Figures 4 and 5) indicate that the PyrR-RNA complex likely consists of a dimer of PyrR and a single molecule of RNA. To investigate the tetramer-dimer equilibrium further, we undertook sedimentation equilibrium experiments with both the native and His-tagged PyrR at monomer concentrations ranging from 0.25 μM to 25 μM, which represents the lower end of the useful range for detecting an equilibrium distribution of PyrR in the ultracentrifuge. As part of this experiment, we also collected “approach to equilibrium” data [9] at the slowest speed used (8000 and 10,000 rpm for the native and His-tagged PyrR, respectively). Figure S3 shows the approach to equilibrium data for three different concentrations of native PyrR; each pair of plots, reading horizontally, shows the interference and absorbance data as well as (below) the calculated residuals at the PyrR concentrations shown in the legend. The first 10 absorbance and interference scans were used such that the sample was close to but not at equilibrium. (Including an excess of scans at equilibrium or very near equilibrium conditions seems to prevent an accurate estimation of particle mass, according to studies with other proteins; Burgner, J. W. unpublished observations). The data at each concentration were fitted to a single species model using SEDPHAT 4.1. Masses of 61, 71, and 82 kDa were estimated from low to high PyrR concentrations. In our hands, this method gives estimates of the mass for simple systems within 1 to 5% of the sequence mass, but the estimate of the sedimentation coefficient is much more variable, as should be expected from the lack of sharpness in the boundaries. The decrease in the weight average mass, which is calculated here, with decreasing protein concentration is consistent with a dissociation constant for the tetramer that is less than 10-7 M.
Figure S4 shows the absorbance distribution as a function of rotational speed for native (Panel A) and His-tagged (Panel B) at equilibrium. The equilibrium data were collected at ~60 h for the slowest speed (see Figure S4 legend) and after an additional 40 h at each of the remaining speeds. These absorbance and interference (not shown) data were collected (see Figure S3, additional details) and fitted to a single species model using mass conservation. The calculated mass the native PyrR is 78.3 kDa for both the absorbance data alone and a combined set of both. The calculated mass for the His-tagged PyrR is 90.7 and 91.6 kDa for the absorbance and combined data, respectively. These values compare well with the sequence masses of the native (79.7 kDa) and His-tagged (89.2 kDa) PyrR. As indicated in the Results section, these data were also fit using a dimer-tetramer formation model using SEDPHAT and mass conservation between speeds. The estimated value for the Kd for a tetramer to dimer disassociation model is about 3 × 10-8 M for both PyrR forms. The interference data were not included in these calculations because blank data were unfortunately not collected; thus, corrections for changes in the baseline as a function of speed could not be made with sufficient accuracy. This is not the case for either the equilibrium absorbance data or all of the approach to equilibrium data, where time and radial independent noise can be removed algebraically [9]. Values for the global reduced χ2 were calculated for each fit and appear in the legend of Figure S4. It would appear from these values that the dimer-tetramer model does not provide a significantly better fit to the data than a tetramer-only model. However, the decrease in observed masses with decreased PyrR concentration observed in Figure S3 is suggestive of dissociation of the tetramer. Thus, assuming the weight average molecular mass describes only a dimer-tetramer equilibrium, the observed 60 kDa at the lowest native PyrR concentration would predict a Kd of about 2 × 10-8 M, which is in reasonable agreement with that calculated from the equilibrium data.
Figure S5 shows the sedimentation velocity data from the titration of His-tagged PyrR with RNA as well as the c(s) distribution plots for interference, A260 and A278 data and the residual plots from these fits. The distribution plots obtained for each titration point as well as for the reactants are qualitatively the same. While there are sufficient data to determine, by solving the appropriate simultaneous equations, the ratio of RNA to PyrR in the new ~4.7S peak found, the signal to noise prevented an accurate assessment of this ratio by multiwavelength analysis. However, the plot of unreacted RNA against the ratio of PyrR added to total RNA predicts a 2 to 1 ratio of PyrR to RNA (see Figure 5, main text).
References
1. Chander P, Halbig, KM, Miller JK, Fields CJ, Bonner HKS, Grabner GK, Switzer RL & Smith, JL (2005) Structure of the nucleotide complex of PyrR, the pyr attenuation protein from Bacillus caldolyticus, suggests dual regulation by pyrimidine and purine nucleotides. J Bacteriol 187, 1773-1782.
2. Bonner ER, D'Elia JN, Billips BK & Switzer RL (2001) Molecular recognition of pyr mRNA by the Bacillus subtilis attenuation regulatory protein PyrR, Nucleic Acids Res 29, 4851-4865.
3. Tomchick DR, Turner RJ, Switzer RL & Smith JL (1998) Adaptation of an enzyme to regulatory function: structure of Bacillus subtilis PyrR, a bifunctional pyr RNA-binding attenuation protein and uracil phosphoribosyltransferase. Structure 6, 337-350.
4. Savacool HK & Switzer RL (2002) Characterization of the interaction of Bacillus subtilis PyrR with pyr mRNA by site-directed mutagenesis of the protein. J Bacteriol 184, 2521-2528.
5. Breeuwer P, Drocourt J-L, Rombouts FM & Abee T (1996) A novel method for continuous determination of the intracellular pH in bacteria with the internally conjugated fluoresecent probe 5 (and 6-)-carboxyfluorescein succinimidyl ester. Appl Environ Microbiol 62, 178-183.
6. Schuck P (2000) Size distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J 78, 1606-1619.
7. Garcia de la Torre J, Navarro S, Lopez Martinez MC, Diaz FG & Lopez Cascales JJ (1994) HYDRO: a computer software for the prediction of hydrodynamic properties of macromolecules. Biophys J 67, 530-531.
8. Laue TM, Shah BD, Ridgeway TM & Pelletier SL (1992) Computer-aided interpretation of analytical sedimentation data for proteins. in Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding SE, Rowe AJ & Horton JC, eds) pp. 90-125, The Royal Society of Chemistry, Cambridge, UK.
9. Schuck P & Demeler B (1999) Direct sedimentation analysis of interference-optical data in analytical ultracentrifugation. Biophys J 76, 2288-2296.
Table S1. Structural variants of purine and pyrimidine nucleotides that affect binding of PyrR to BcBL2 as determined by filter binding
Table S2. Apparent RNA dissociation constants (Kd-values) for binding of structural variants of BcBL2 to PyrR
Table S3. Sedimentation velocity analysis of native and His-tagged PyrR
Table S4. Sedimentation velocity constants from the titration of BcBL2 with PyrR
Supplementary Material: Figure Legends
Figure S1. Effects of the pH of the binding buffer on reactions incubated at 0°C (A) and of temperature on reactions at pH 7.5 (B) on binding of BcBL2 to PyrR as determined by filter binding. Open circles: no effector; closed circles: 500 μM UMP; closed triangles: 500 μM GMP.
Figure S2. Sedimentation velocity studies on native and His-tagged PyrR at moderate ionic strength. Panels A, B, and C show the results of a sedimentation velocity run with native PyrR in 0.1 M NaCl at 50,000 rpm and 20°C. Panel A shows changes in absorbance as a function of radial position (dots) at six min intervals and the results (solid lines) from a c(s) distribution analysis using the program SEDPHAT 4.1. Panel B shows the calculated residuals against radial position for each scan in Panel A, and Panel C shows the c(s) distribution against s corrected to s20,w. The weight average sedimentation coefficient from the integration of the principal peak is 4.83 S. Panels D, E, and F show the results from an identical sedimentation velocity experiment and analysis using His-tagged PyrR. The weight average s20,w calculated from these results is 5.34 S. Additional details for both experiments are found in the Data Supplement text and Table S3.
Figure S3. An approach to equilibrium study with the native PyrR at low ionic strength. The initial concentrations of PyrR studied were: Panels A and B, 0.25 μM; Panels C and D, 1.25 μM; Panels E and F, 25 μM. The data were collected at 8000 rpm. Rayleigh interference optics were used for Panels A, C, and E; absorbance optics at 232 nm were used for Panels B and D; and absorbance at 278 nm was used for Panel F. The calculated values for s20,w and observed mass based on a global single species fit to both data sets with the program SEDPHAT 4.1 are 5.7 S and 61 kDa, respectively, for a native PyrR concentration of 0.25 μM . The calculated values for s20,w and mass for PyrR at an initial concentration of 1.25 μM are 3.7 S and 71 kDa , respectively, and a loading concentration of 25 μM PyrR, the calculated values for s20,w and mass are 5.1 S and 82 kDa. The solid lines in each panel are the fitted results from the single species model (see Data Supplement text); the calculated residuals against radial position for each scan are shown in the boxes below each panel.
Figure S4. Equilibrium sedimentation studies of native and His-tagged PyrR at low ionic strength and concentrations shown. The absorbance data in the upper panel (native PyrR) and the lower panel (His-tagged PyrR) were obtained from a continuation of the experiments shown in Figure S2 in that the samples were allowed to come to equilibrium at 8 k, 12 k, 1k 17 k, and 20 k rpm and absorbance scans across the cell were collected at A232, A232, and A278, respectively, from left to right. The solid lines were calculated based on a single discrete species model, with mass conservation, using SEDPHAT 4.1 and a mass of 78.3 kDa. The data in the lower panel were obtained using the same procedures as in the upper panel, except that the samples were allowed to come to equilibrium at 12 k, 15 k, 17 k, and 20 k rpm. Absorbance scans were collected at the same wavelengths indicated in the upper panel. The solid lines were calculated based on a single discrete species model, with mass conservation, using SEDPHAT4.1 and a mass of 91.6 kDa. Calculated residuals against radial position for each scan are shown in the boxes below each panel.
Figure S5. Sedimentation velocity studies of the interaction between RNA and His-tagged PyrR. The concentrations of PyrR and RNA in each sample are shown at the left of each row of panels; additional details appear in the legend of Figure 4. The three sets of scans shown in each row of panels were collected using Rayleigh interference, A 260 and A278, from left to right, respectively. The data were fitted using the c(s) distribution algorithm of SEDPHAT as described in the Methods section; c(s) distributions were calculated form interference data (red lines), absorbance at 260 nm (green lines), absorbance at 278 nm (blue lines).