Abstract
Multidrug resistance protein 1 (ABCC1) is a member of the ‘C’ class of ATP-binding cassette transporters, which can give rise to resistance to chemotherapy via drug export from cells. It also acts as a leukotriene C4 transporter, and hence has a role in adaptive immune response. Most C-class members have an additional NH2-terminal transmembrane domain versus other ATP-binding cassette transporters, but little is known about the structure and role of this domain. Using electron cryomicroscopy of 2D crystals, data at 1/6 per Å−1 resolution was generated for the full-length ABCC1 protein in the absence of ATP. Analysis using homologous structures from bacteria and mammals allowed the core transmembrane domains to be localised in the map. These display an inward-facing conformation and there is a noteworthy separation of the cytoplasmic nucleotide-binding domains. Examination of non-core features in the map suggests that the additional NH2-terminal domain has extensive contacts on one side of both core domains, and mirrors their inward-facing configuration in the absence of nucleotide.
Keywords: Multidrug resistance, ATP-binding cassette, MRP1, ABCC1, Structure, Electron crystallography, Cryo-electron microscopy
1. Introduction
Overexpression of several ATP-binding cassette (ABC) transporters can give rise to the phenotype of multidrug resistance (MDR) in cancer cells (Szakacs et al., 2004, 2006). One of these is multidrug resistance protein 1 (MRP1/ABCC1) (Cole et al., 1992, 1994; Deeley and Cole, 2006; Haimeur et al., 2004). Clinical studies have shown that MRP1 is expressed in a range of solid and haematological malignancies and MRP1 expression has been correlated with negative response to treatment and disease outcome (Deeley and Cole, 2006). MRP1 is expressed in most tissues throughout the body; in polarized cells, MRP1 is found on the basolateral cellular surface. It is thought to provide protection from xenobiotics in these tissues.
Gene transfection studies have confirmed the role of MRP1 in conferring resistance to natural product chemotherapeutic agents and genotoxins by mediating cellular efflux of these substrates (Cole et al., 1994; Peklak-Scott et al., 2005). MRP1 also transports organic anions conjugated to glutathione (GSH) (e.g., cysteinyl leukotriene C4) (Loe et al., 1996) as well as glucuronate and sulfate conjugates (Leslie et al., 2005). Studies on mrp1−/− mice suggest that the transport of leukotriene C4 by MRP1 may play a significant role in the mediation of inflammatory responses (Mueller et al., 2009). MRP1 can also transport unconjugated organic anions such as methotrexate and GSH (Cole and Deeley, 2006). GSH has been shown to stimulate the transport of some conjugated compounds, and a GSH-induced change in the conformation of the transporter may be involved (Rothnie et al., 2006).
ABC transporters typically consist of two transmembrane domains (TMDs), each composed of six predicted transmembrane α-helices, and two nucleotide-binding domains (NBDs) (Hyde et al., 1990). The TMDs probably form the translocation pathway for substrates and appear to be major determinants of substrate specificity. Unlike most ABC transporters, MRP1, contains three rather than two TMDs (Bakos et al., 1996; Cole et al., 1992; Haimeur et al., 2004; Hipfner et al., 1999) as do several other ABCCC family members (Deeley and Cole, 2006). The additional TMD (TMD0) is predicted to contain five transmembrane α-helices and a glycosylated extracellular N-terminal region. TMD0 is connected to TMD1 by a large cytoplasmic loop of ~120 amino acids.
In the ABC transporters, which work against a concentration gradient, transport is coupled to the binding and hydrolysis of ATP by the NBDs, which appear to undergo a reversible dimerisation during the transport cycle. Accumulating structural data from X-ray crystallography of bacterial ABC transporters are mostly consistent with such a transport model (Dawson and Locher, 2007; Gerber et al., 2008; Hollenstein et al., 2007; Khare et al., 2009; Locher, 2004; Oldham et al., 2008).
So far, structural data for MRP1 has been limited and of relatively low resolution (22 Å), and consequently, there was no clear resolution of the location of TMD0 in these studies (Rosenberg et al., 2001). The TMD0 region, including the extended cytoplasmic loop, displays no homology with any other structure deposited in the protein data bank. Hence homology models may be constructed only for the core four domains of the protein (DeGorter et al., 2008). Here, we have determined the 3D structure of MRP1 by electron cryomicroscopy of 2D crystals with an in-plane resolution of 1/6 per Å−1 and with a missing cone of data of only 26° giving a resolution perpendicular to the crystal plane of about 1/10 per Å−1. At this combined resolution, long transmembrane α-helices may be resolved as cylindrical regions of high density in the map (Ford and Holzenburg, 2008).
2. Results
2.1. MRP1 purification, ATPase activity and substrate binding
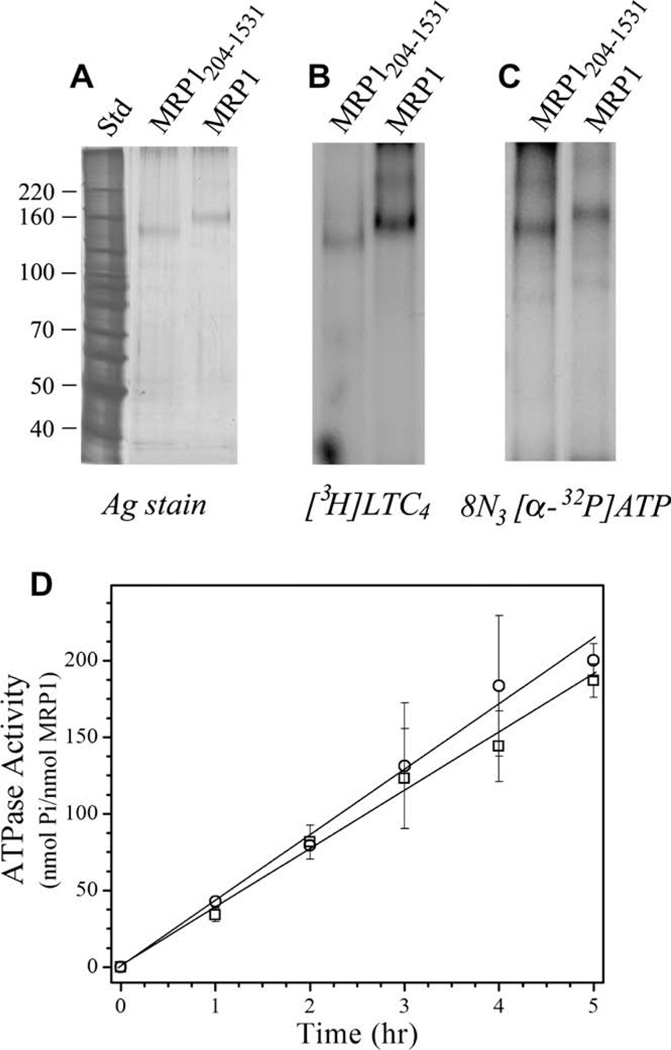
Wild-type MRP1 with hemagglutinin and hexa-histidine tags at the COOH-terminus was expressed in Pichia pastoris as described previously (Cai et al., 2001; Wu et al., 2005). A construct lacking TMD0 (MRP1204–1531) was generated and expressed in a similar manner. Membranes were solubilized in LPG, then diluted in DDM and subjected to a two-step purification procedure utilizing Co2+-IMAC resin and DE52 anion chromatography (Wu et al., 2005). Protein purity was judged to be greater than 90% by silver staining (Fig. 1A).
Figure 1.
Properties of purified recombinant MRP1 and MRP1204–1531. expressed in P. pastoris. (A) Purified MRP1 (35 ng) and ‘short’ MRP1204–1531 (28 ng) were resolved by SDS–PAGE and silver stained. Molecular weight markers are shown on the left. (B) Purified MRP1 (1.4 µg) and MRP1204–1531 (0.8 µg) were incubated with 8-azido[α-32P]ATP (2.3 µCi; 5 µM), irradiated at 302 nm, and then resolved by SDS–PAGE and processed for autoradiography. (C) Purified MRP1 (1.5 µg) and MRP1204–1531 (0.9 µg) were incubated with [3H]LTC4 (200 nM; 0.13 µCi), irradiated at 302 nm, and then resolved by SDS–PAGE and processed for autoradiography. (D) MRP1 (0.49 µg) (○) and MRP1204–1531 (0.42 µg) (□) were assayed for ATPase activity over a 5 h time period. Values obtained were corrected for ATP hydrolysis in the absence of protein. Each point represents the mean (±SD) of four determinations.
To determine whether the MRP1 was active after purification, its ability to bind the high affinity MRP1 substrate ([3H]LTC4) and nucleotide (8-azido-[α-32P]ATP), as well as its ATPase activity, were determined. As shown in Fig. 1B, DDM-solubilized, and purified MRP1 as well as MRP1204–1531 were readily photolabeled with [3H]LTC4. Purified MRP1 constructs were also photolabeled with 8-azido-[α-32P]ATP (Fig. 1C). No significant photolabeling of other protein bands was detected. Finally, the ATPase activity was determined for both constructs (Fig. 1D). These observations indicate that the full-length MRP1 protein used for the structural analysis described below had retained its function, and also that removal of the N-terminal TMD0 domain had no significant impact on the activity of the purified material.
2.2. Crystallisation of full-length MRP1
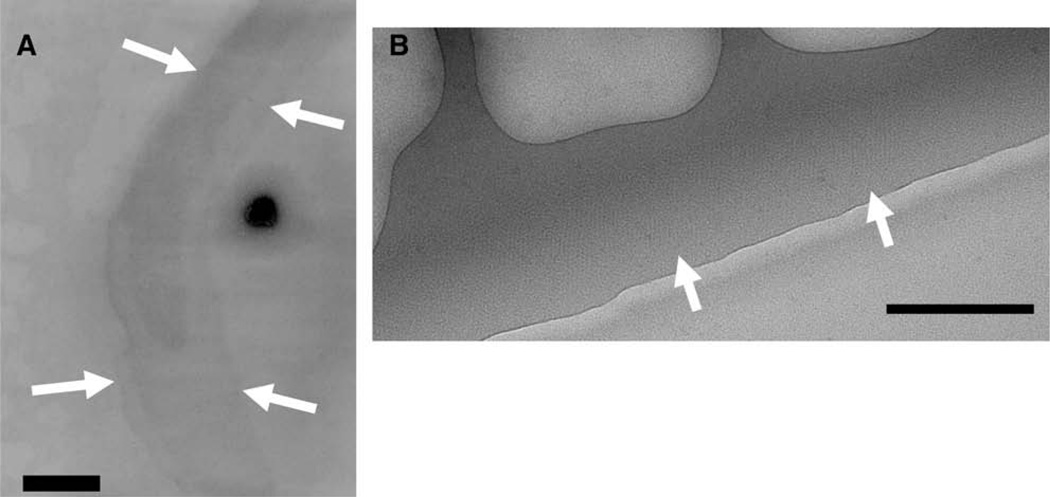
2D crystals were generated on the carbon surface of an electron microscope (EM) grid (Auer et al., 1999). 2-D crystals were usually observed in the square grid windows in a peripheral zone ~2 µm across which was visible at low magnification as shown in Fig. 2A (arrows). This zone presumably reflects optimal conditions for 2D crystal formation because of a local gradient of protein/precipitant within each window of the grid. A mosaic of 2D crystals is found in this zone, suggesting that individual crystals (typically 0.5–2 µm across) nucleate in this area and grow outwards until meeting another area. At strong defocus, the unstained 2-D crystals could be identified (Fig. 2B, arrows). A similar method of 2-D crystal formation was used by Auer et al. (1999) and successes have been achieved for other ABC transporters and other membrane proteins using this method (Auer et al., 1998; Awayn et al., 2005; Rosenberg et al., 2004, 2005; Scarborough, 1994). The MRP1 2D crystals show a p1 two-dimensional plane group. Other plane group symmetries were discounted by examining phase relationships for untilted 2D crystals using the program ALLSPACE (Valpuesta et al., 1994) (see Table 1), as well as by comparison of the unit cell dimensions of the 2D crystals versus the dimensions of other ABC proteins.
Figure 2.
Two-dimensional (2-D) crystals of MRP1. (A) Electron micrograph of an MRP1 crystal embedded in negative stain (2% w/v uranyl-acetate) and recorded at low-magnification (3500×). The edge of the crystal is indicated by the white arrows. The characteristic outline of these crystal areas was used to identify the crystals at low magnification (see Methods). Scale bar = 200 nm. (B) Electron micrograph of part of a MRP1 2D crystal embedded in ice. The edge of the crystal is shown by the white arrows. Scale bar = 100 nm.
Table 1.
Summary of electron crystallographic data for the MRP1 2D crystals.
| No. of crystal areas merged | 106 |
| Size of crystal areas (pixels) | 4096 × 4096 or 2048 × 2048 |
| Pixel size at the specimen level | 1.9 Å (CCD), 1.19 Å (film) |
| Unit cell parameters | a = 69.2 Å (SD = 0.4) (n = 7) |
| b = 78.3 Å (SD = 0.4)(n = 7) | |
| γ = 124.2° (SD = 0.2) (n = 7) | |
| Two-sided space group | p1 |
| Range of crystal tilts | 0–64° |
| Tilt range, (number of crystals) | 0–10° (10); 10–25° (11); 25–35° (10); |
| 35–40° (5); 40–45° (13); 45–50° (11); | |
| 50–60° (40); 60–64° (6) | |
| Range of underfocus | ~−2000–−30000 Å |
| Total no. of measurements | 11,730 |
| Resolution range | Phase residual (untilted crystals)b |
| 100–13.9 Å | 30.8° |
| 100–9.9 Å | 38.1° |
| 13.7–9.8 Å | 63.2° |
| 9.8–8.0 Å | 70.3° |
| Data completeness to 1/6 per Å−1 in-plane | 89% |
| Overall weighted interimage phase residuala | 38.8° |
| Overall weighted R-factor | 29.2% |
For fitted lattice lines, including data to IQ 7 and to a nominal in-plane resolution of 1/6 per Å−1.
Where random data would give rise to a mean phase residual of 90°.
2.3. Cryo EM data
Electron cryomicroscopy of individual crystals of MRP1 yielded well-sampled structure factors to about 1/6 per Å−1 resolution in the plane of the crystal and 1/10 per Å−1 resolution perpendicular to the plane (Supplementary data, Fig. 1A). After merging data from several untilted crystals, the interimage phase errors of multiply-sampled reflections with an arbitrary cut-off of 6 Å resolution were plotted (Supplementary data, Fig. 1B). The well-sampled data with low phase errors extend to approximately 10 Å resolution, whilst data to about 6 Å resolution are sampled less completely, and with larger phase errors. The overall interimage phase residual for the merging of the MRP1 data was 38° (Table 1), where a phase residual of 90° would correspond to random data. The distribution of amplitudes and phases along lattice lines showed the data extending to about 10 Å resolution perpendicular to the crystal plane (Supplementary data, Fig. 2A–D).
2.4. The MRP1 3D map
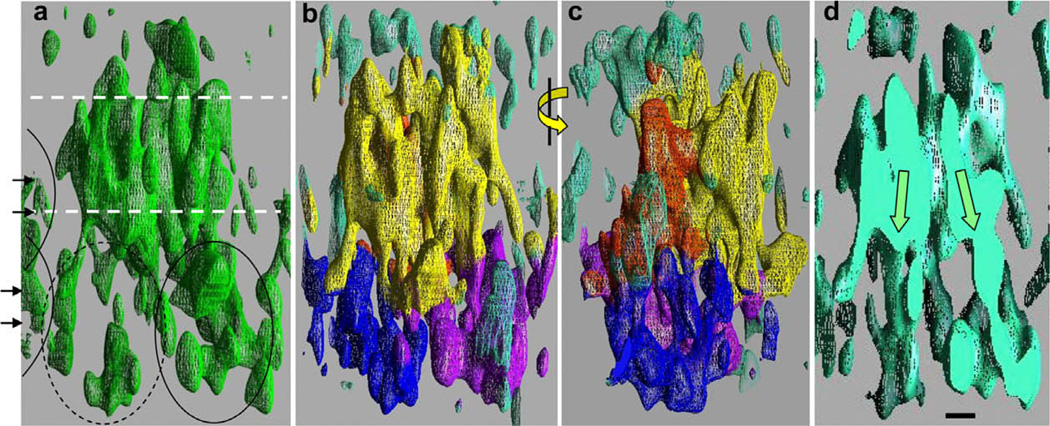
The 3D map generated from the MRP1 2D crystals is displayed in Fig. 3. At a high density threshold (2.0σ above the mean density level, left panel), a 50 Å – thick band of density is observed that is composed of closely packed cylindrical features, probably transmembrane α-helices. Below, and above this band of high density are the cytoplasmic and extracellular regions of the protein, which are ~60 Å and ~15 Å wide respectively. Two orthogonal views of the map, with a lower density threshold (1.25σ above the mean density level) are shown in Fig. 3, panels b and c. The map has been coloured according to the assignments discussed later: Yellow corresponds to regions assigned to TMDs 1 and 2; orange TMD 0; blue and purple NBDs and green to unassigned regions, mostly on the upper (extracellular) surface of the map. The NBDs of MRP1 are considerably further apart than in the ‘sandwich dimer’ configuration, consistent with the nucleotide-free conditions for crystal production (Aller et al., 2009; Hollenstein and Dawson, 2007; Oldham et al., 2008). A central slice through the map (Fig. 3d) provides evidence for an inward-facing configuration of the TMDs. The two halves of the transporter appear to lean towards each other, with an angle of approximately 20° (arrows). This angle is lower than that observed for the Escherichia coli MsbA structural model (Ward et al., 2007) but is similar to that displayed by the inward-facing murine P-glycoprotein structure (Aller et al., 2009).
Figure 3.
The MRP1 3D density map. (a) A view along the crystal plane, showing high density regions of the map (green mesh). Molecules in adjacent unit cells encroach at the extreme left (arrowheads, arcs). A region about 50 Å thick, consistent with the TMDs is indicated by the white dashed lines. Weaker density is displayed in the ~60 Å thick cytoplasmic region (bottom) and the ~15 Å thick extracellular region (top). Density for one of the presumed NBDs (right, ellipse) is stronger than for the other (left, dashed ellipse). (b) Similar view of the density map, but with a lower density threshold for the mesh. The map has been coloured according to the interpretation described in the main text, with yellow for TMDs 1 and 2, orange for TMD0, blue and purple for the two NBDs and turquoise for unassigned regions. (c) As for panel (b), but after a 90° rotation about the vertical axis as indicated. (d) Central slice through the map, viewed from the same orientation as in panels a and b. The two halves of the protein appear to lean inwards in an inverted ‘V’ shape, giving an inward-facing conformation to the TMDs of the transporter. The scale bar corresponds to 1 nm and applies to all panels. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.5. Interpretation of the MRP1 map
Structures of homologues of MRP1 (Sav1866 and P-glycoprotein) were firstly compared to, and docked within the MRP1 map using the Chimera ‘fit model to map’, and Situs ‘cores’ routines. Secondly, cylindrical regions of continuous density were traced by hand. Thirdly, NBDs were initially fitted by hand as a dimer, and then the local position of each NBD was individually refined as above.
The overall positions of the docked Sav1866 and P-glycoprotein structures within the MRP1 map were similar (data for Sav1866 shown in Supplementary data, Fig. 3), but the sandwich dimer of the Sav1866 NBDs was clearly different to the MRP1 map. The result of the fitting of the nucleotide-free P-glycoprotein structure is displayed in Fig. 4, with the MRP1 map viewed as in Fig. 3c. A good fit was apparent for the C-terminal half of P-glycoprotein: The rightwards slope of the transmembrane helices was reproduced in the MRP1 map, as was the positioning of the ‘L’ shaped NBD(2). At the centre of the map (Fig. 4b), the pseudo-twofold symmetry of the P-glycoprotein structure is matched as is the curved ‘V’ shape formed by TMD helices 6 and 12 of P-glycoprotein. On the cytoplasmic side, only one of the P-glycoprotein NBDs lies well within the MRP1 map. The N-terminal P-glycoprotein NBD (arrows) does not, as further shown by the rearmost slice through the map (Fig. 4c). A shift and rotation of the N-terminal NBD would be required to place it within the MRP1 map. The leftwards tilt of the TMD helices in P-glycoprotein is matched in the rearmost slice of the MRP1 map (Fig. 4c), and the extension of density into the extracellular region (red ellipse) probably explains why the N-terminal half of P-glycoprotein is favoured for this region of the map. The fitting exercise gave an indication of the regions containing density for TMD0 (Fig. 4, white dashed outlines).
Figure 4.
Comparison of the MRP1 map with the P-gp structural model (yellow ribbon trace). Three sequential slices through the MRP1 map (green netting) are displayed in the same orientation as in panel c of Figure 3. (a) Nearmost slice with the C-terminal half of P-gp. (b) Central slice. (c) Rearmost slice, with the N-terminal half of P-gp. Regions to the left in panels a–c, shown by the white dashed lines, were interpreted as corresponding to the additional TMD0 in MRP1. The fitting of the P-gp model to the MRP1 map (see main text for methodology) places its first NBD in a region of low density (arrows, panels b and c). The scale bar corresponds to 1 nm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
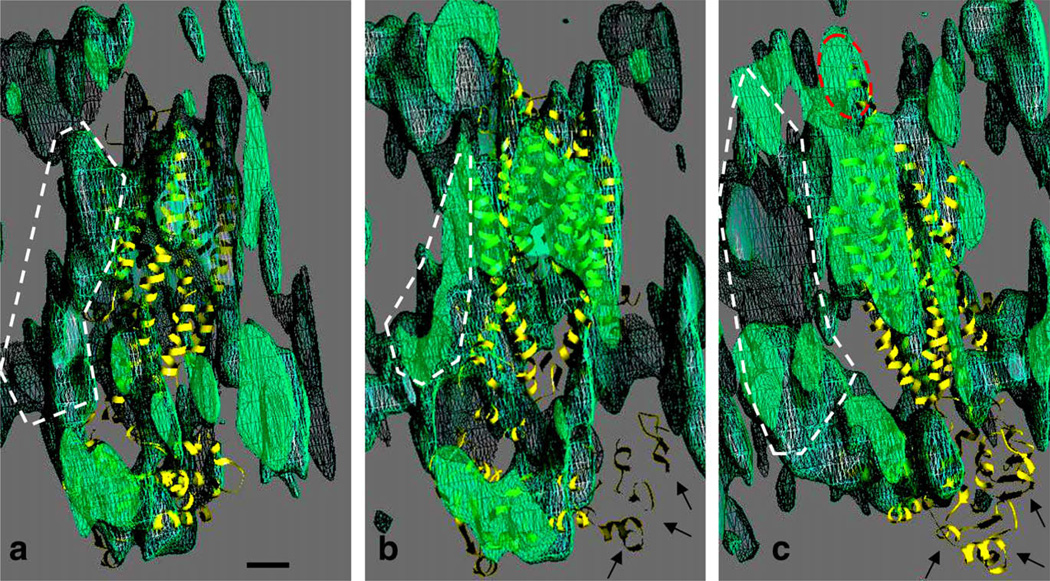
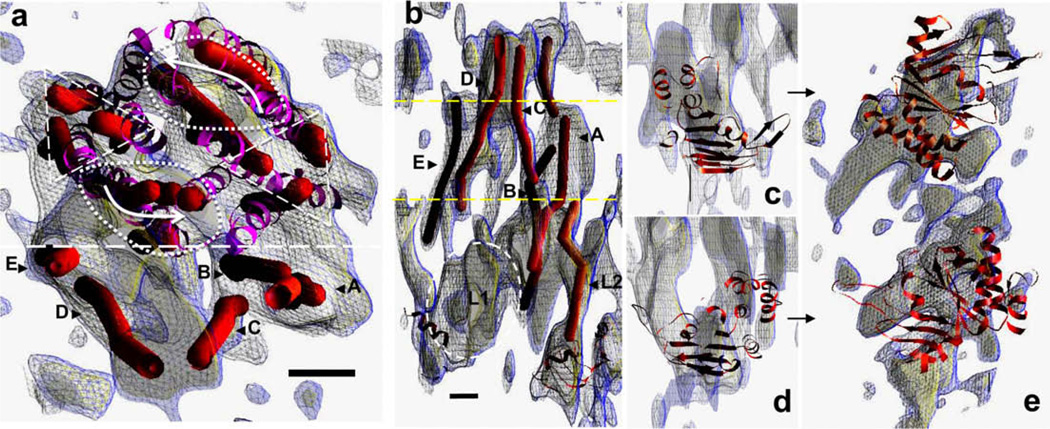
The manual tracing of cylindrical regions of density (red cylinders) in the MRP1 map (blue/yellow mesh) are displayed in Fig. 5. The position of the fitted P-glycoprotein structure is superimposed (purple ribbon trace). Panel a shows a view onto the TMDs from the extracellular surface. A cluster of cylinders (above the dashed line) shows some twofold symmetry as indicated by the triangular and oval dashed outlines, and where the curved arrows indicates the opposing directions of tilt of cylinders on each side of this region (see also Fig. 3a and c). This region of twofold symmetry roughly coincides with the fitted P-glycoprotein TMDs (ribbon trace). On the periphery of the diamond-shaped region (below the dashed line) lie several further cylindrical regions of density. Four of these (A–E) are candidates for TMD0 helices. A side-view of this peripheral region (Fig. 5b) reveals that two (C and D) extend into the extracellular region, whilst a fifth (B) may extend partially into the lipid bilayer (boundaries of which, as estimated by the fitted P-glycoprotein model, are indicated by the dashed lines, Fig. 5b). Hydropathy plots suggest that TMD0 will have a glycosylated N-terminal region of ~36 residues, and a third extracellular loop of ~18 residues. Interestingly, the fitted cylinders in the putative TMD0 region mirror the inverted ‘V’ shape of the rest of the TMD region, with the A,C pair leaning inwards to contact the D,E pair at the extracellular surface. A lozenge-shaped region of density (dashed ellipse) sits directly below the D,E cylinders in the cytoplasmic region of the map (L1) and likewise, a cylindrical region (L2) extends downwards from the vicinity of the A,B,C cylinders.
Figure 5.
Further interpretation of the MRP1 map. (a) Manual tracing of the paths (red cylinders) of cylindrical regions of continuous density in the MRP1 map (blue mesh at 0.9σ, yellow mesh at 1.25σ), using a slab of the high density region viewed from the expected extracellular side of the protein. The location of the fitted P-gp structure within the map is also displayed (purple ribbon trace), as well as local clusters of cylinders that are roughly related by a local twofold symmetry in terms of position (triangular and oval dashed outlines) as well as tilt (curved arrows). On one side of the region (separated by the dashed line) lie cylindrical densities (A–E) that are not matched by the P-gp structure, and may be part of TMD0. A side view of this region is displayed in panel b, with an estimate of the boundaries of the lipid bilayer (yellow dashed lines). Some of the cylindrical densities extend up into the extracellular region at the top (C,D), whilst some (A,B) extend downwards. A sixth cylindrical path (L2) extends down towards a region occupied by the C-terminal NBD of the fitted P-gp structure, whilst a lozenge-shaped region of density (L1, dashed ellipse) extends towards the opposite NBD. Individual fitting of the MetNI NBDs is shown in the relevant sections of the MRP1 map (panels c–e), as viewed from the same orientation as in Fig. 4 (panels c and d), or as viewed from the cytoplasmic side of the protein (panel e). Scale bars = 1 nm, panels b–e are at the same scale. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fitting of individual NBDs to the map (Fig. 5, panels c–e), yielded positions and orientations that were roughly consistent with previous ABC transporter structures (Aller et al., 2009; Hollenstein et al., 2007; Khare et al., 2009; Kadaba et al., 2008). One of the fitted NBDs was close to the C-terminal NBD P-glycoprotein model (panels d,e and Fig. 4), but the other was located differently, as anticipated (see Fig. 4c). The overall impression gained was of asymmetry, a 45° clockwise rotation of the latter NBD around the viewing direction in panel e.
3. Discussion
Studies of MRP1 have shown that residues in the TMDs 1 and 2 are important for binding and transport of a number of MRP1 substrates (Campbell et al., 2004; Conseil et al., 2006; Deeley and Cole, 2006; Haimeur et al., 2002, 2004; Zhang et al., 2004). Partial deletions and mutations of specific amino acids in TMD0 can interfere with protein processing and activity (Gao et al., 1998; Ito et al., 2003; Leslie et al., 2003). These latter mutations may interfere with folding of the protein, since complete removal of TMD0 does not prevent trafficking to its normal location nor transport action (Bakos et al., 1998; Westlake et al., 2003). However TMD0 and a small portion of the cytoplasmic loop connecting it to TMD1 are required for retention of MRP1 at the cell surface, and hence they contain at least partially redundant elements that contribute to the processing and trafficking of the protein (Westlake et al., 2005). MRP1 lacking TMD0 has similar ATPase and LTC4-binding activities to the full-length protein (Fig. 1). Previous structural studies of MRP1 in detergent showed monomers of 8–10 nm diameter and with a favoured projection of roughly pentagonal shape (Rosenberg et al., 2001). Interestingly, after reconstitution into lipid bilayers, the protein produced small and poorly ordered 2D crystals with a p2 plane group symmetry (Rosenberg et al., 2001). The (better ordered) 2D crystals in this study were generated with the monomeric detergent-solubilised protein, although the overall dimensions of the unit cell (12 × 8 × 7 nm) are roughly consistent with the previous studies (Rosenberg et al., 2001). Moreover, viewed along its long axis, the MRP1 map shows a roughly pentagonal outline, again consistent with the earlier low resolution data for the detergent-solubilised protein.
3.1. Interpretation of the 3D map of MRP1
Long transmembrane α-helices should give rise to discernible density in the present MRP1 map. MRP1 likely has 17 transmembrane α-helices, five in TMD0 and six each in TMDs 1 and 2. The region of high density in the MRP1 map (see Fig. 3a) contains tightly packed cylindrical densities, consistent with this number of transmembrane α-helices. The TMDs of MRP1 seem to be in an inward-facing conformation and there is a concomitant separation of the NBDs. This is in agreement with the nucleotide-free P-glycoprotein structure (Aller et al., 2009) as well as for MetNI, Mal- FGK and ModBC in the nucleotide-free states (Gerber et al., 2008; Kadaba et al., 2008; Khare et al., 2009; Ward et al., 2007). The unusual relative orientation of one of the NBDs in MRP1 may reflect the relative specialisation of the two NBDs in MRP1 (Gao et al., 2000). A more prosaic explanation could be that the latter NBD is partly disordered in the crystals. Neither explanation will be unprecedented (the E. coli MsbA structure has only partial density for the NBDs and the V. cholera MsbA has an unusual rotation of the NBDs away from the usually adopted configuration such as that found for S. typhimurium MsbA) (Gerber et al., 2008; Kadaba et al., 2008; Khare et al., 2009; Ward et al., 2007).
Additional density in the TMD region could be the location of TMD0, on one side of the core ABC transporter structure. Unequivocal assignments as to which segment of TMD0 interact with which segments in TMDs 1 and 2 are not possible. If the fit shown in Fig. 4 is correct, and the core TMDs of MRP1 are similar to other ABC transporters, then TMD0 would interact with TM helices 14,15 and 16 in TMD2 and 7 in TMD1 (see also Supplementary data, Fig. 4). TMD0 would come into contact with helices crossing over from one side of the protein to the other and domain-swapped intracytoplasmic loops. The putative TMD0 region emulates the inward-facing core TMDs. Hence, TMD0 may participate in large scale conformational changes. Whether TMD0 is an active participant, or passive passenger, is debatable. Evidence from TMD0-lacking MRP1 suggests a passive role in the mechanism of action is more likely.
TMD0 not only consists of transmembrane helical segments, but also contains cytoplasmic and extracellular loops. Extracellular loops in ABC proteins are generally short connecting turns, but longer loops (>5 residues) have been found to be simple extensions of membrane-spanning helices (e.g. in P-glycoprotein (Aller et al., 2009)) or in one example, a separate domain (MalF) (Oldham et al., 2007). The data presented here suggest that the TMD0 extracellular loops are more likely to be extensions of the membrane-spanning helices or short turns (Fig. 5b, top). In contrast, the cytoplasmic region close to the putative TMD0 helices displays a larger lozenge-shaped density, L1 and a smaller cylindrical density L2 (Fig. 5b, bottom). TMD0 is connected to TMD1 in MRP1 by a cytoplasmic loop of ~120 residues. This loop displays no significant homology to any known protein structures, and may be large enough to form a discrete domain. We would propose that the L1 density in the MRP1 map could be the site of such a domain.
4. Material and methods
4.1. Plasmid construction
The plasmid pHIL-MRP1cHA-His6 for expression of full-length MRP1 in Pichia pastoris has been described previously (Cai et al., 2001; Wu et al., 2005). To express an NH2-terminally truncated MRP1 lacking TMD0, a plasmid containing cDNA coding for MRP1 amino acids 204–1531 was created as follows. pHIL-MRP1cHA-His6 was digested with SacII and BamHI, and the resulting vector fragment containing MRP1 cDNA coding for amino acids 281–1531 was gel purified. A MRP1 cDNA fragment coding for amino acids 204–300 was then generated by PCR using the plasmid pcDNA3.1(−)MRP1K as template (Leslie et al., 2003). The upstream primer (5′-AGG CCG CGG AAA AAA ATG GAC CCT AAT CCC TGC CCA-3′) contained a SacII site immediately followed by a spacer of six adenosines, an ATG start codon, and a sequence coding for six amino acids starting from position 204. The downstream primer (5′-GAC GAT CAA AGC CTC CAC C-3′) contained a DNA sequence at a position 42 nucleotides beyond the BamHI site in the MRP1 cDNA. The PCR product (approximately 0.3 kb) was digested with SacII and BamHI, gel purified, and ligated to the pHIL vector fragment containing sequence encoding residues 281–1531. The fidelity of the PCR product was confirmed by sequencing. The resulting construct was designated pHIL-MRP1204–1531cHA-His6.
4.2. Protein purification, nucleotide-binding and hydrolysis, ATPase activity and substrate binding
Crude membranes from cells expressing either plasmid pHIL-MRP1cHA-His6 or pHIL-MRP1204–1531cHA-His6 were prepared using a French Pressure Cell (Thermo Electron Corporation, Waltham, MA). Recombinant MRP1 and MRP1204–1531 proteins were solubilized with LPG (4–6 mg ml−1) and then diluted with DDM (Sigma–Aldrich) to a final concentration of 0.02%. The solubilized proteins were purified by Co2+-IMAC (BD BioSciences Clontech) and DE52 anion chromatography. Protein purity and identification were confirmed by silver staining and by immunoblotting with MRP1-specific monoclonal antibody QCRL-1 (Cai et al., 2001) as before.
Purified MRP1 and MRP1204–1531 were photolabeled with 8-azido-[α-32P]ATP and [3H]LTC4. For 32P-azidoATP labelling, purified MRP1 and ‘short’ MRP1204–1531 (0.8–1.5 µg) in buffer containing 50 mM Tris–HCl, 250 mM sucrose, 0.187 mg ml−1 DDM, pH 7.4, were incubated with 5 µM (2.3 µCi) 8-azido-[α-32P]ATP (Affinity Labeling Technologies, Inc., Lexington, KY) and 5 mM MgCl2 on ice for 30 min and then irradiated at 302 nm for 8 min (CL-1000 Ultraviolet Crosslinker, DiaMed). Protein samples (Conseil and Deeley and Cole, 2006) were subjected to SDS–PAGE and the dried gels were exposed to film. For photolabeling with the high affinity MRP1 substrate LTC4, purified MRP1 and MRP1204–1531 (0.8–1.5 µg) were incubated with [3H]LTC4 (200 nM; 0.13–0.15 µCi) (Perkin-Elmer Life Sciences) and irradiated at 302 nm as before (25,41). [3H]LTC4-labeled proteins were then resolved by SDS–PAGE and processed for autoradiography. The ATPase activity of purified MRP1 and MRP1204–1531 was determined using a colorimetric assay essentially as described (Mao et al., 2000). Briefly, 0.25–0.75 µg of MRP1 and MRP1204–1531 were incubated at 37 °C in a reaction mixture containing 50 mM Tris–HCl (pH 7.4), 10% glycerol, 0.02% DDM, 5 mM ATP, and 5 mM MgCl2. Reactions were stopped at various times over a 5 h period by the addition of 18% SDS. The amount of inorganic phosphate released was determined immediately using a colorimetric method adapted from Chifflet et al. (1988) and Doige et al., 1992. Data were corrected for ATP hydrolysis in the absence of protein.
4.3. Two-dimensional crystals
2D crystals were grown on carbon films, supported on gold-plated copper 400 mesh electron microscope grids (Agar) from a solution containing 0.2 mg ml−1 MRP1, 2.8 mg ml−1 dodecymaltoside, 110 mM ammonium sulphate and 17% (w/v) PEG 4000 (Hampton Research) with the pH being adjusted to 8.0 with 50 mM Tris–HCl. Carbon-coated grids were heated to 160 °C for 1 h prior to the experiment. A 10 µl drop was placed in the well of a Hampton Research plate (Crystal Quick) equilibrated against 150 µl of 1M MgCl2. The grid was placed onto the drop with the grid bars facing the solution. The plate was sealed with Crystal Clear® film (Hampton Research) and left at 4 °C for 17 h in an incubator (Rumed, Jencons).
4.4. Specimen preparation and electron microscopy
Grids were picked up from the surface of the drops and either negatively stained with 2% (w/v) uranyl-acetate (Rosenberg et al., 2003) and viewed on a Tecnai 10 electron microscope, or blotted for five to ten seconds with filter paper (Whatman No. 4) and then frozen in liquid ethane using an FEI Vitrobot. The grids were transferred at liquid-nitrogen temperature into a Gatan cryotransfer specimen holder (Gatan, Pleasanton, CA) and observed at 95 K in a Philips CM200 FEG or a Tecnai 20 FEG operated at 200 kV and equipped with a Gatan 4096 × 4096 CCD camera with a pixel size of 15 µm. Images were collected at a magnification of 50,000 and 80,000×, and with a calibrated electron dose of 10–12 e/Å2. Some images were recorded on Kodak SO-163 film and developed for 12 min in a full-strength Kodak D19 developer. Selected films were subjected to densitometry using a Zeiss SCAI scanner set at 7 µm pixel size, corresponding to 1.14 Å at the specimen level. We did not observe any significant difference in the quality of the data recorded on film or on the CCD camera. Crystals were located in a fringe region around the edge of the grid windows (see Fig. 2A). Images were recorded with defocus varying between −0.2 and −3.0 µm, the mean defocus employed was −0.5 µm.
4.5. Image processing
Images were processed using the MRC-LMB image processing suite. After three cycles of lattice unbending to correct for lattice distortions (Crowther et al., 1996), correction of the contrast transfer function was carried out after determining defocus with CTF-FIND2 (Henderson et al., 1986). Structure factors were merged by refining to a common phase origin using ORIGTILTK. Amplitudes and phases were averaged as described previously (Henderson et al., 1986). The image amplitudes were scaled as a function of resolution, with bacteriorhodopsin as a reference. Density maps were calculated with CCP4 software (Collaborative Computational Project No. 4, 1994) as described previously (Rosenberg et al., 2005).
Homologues of MRP1 (Sav1866 and P-glycoprotein) were docked within the MRP1 map using the Chimera ‘fit model to map’, and Situs ‘cores’ routines (Pettersen et al., 2004; Wriggers et al., 1999), yielding similar positions for each algorithm. Docking of individual NBDs within the MRP1 map was carried out using an initial search with the widely-separated MetNI NBD dimer (with regulatory domains removed) (Kadaba et al., 2008). The position of each NBD was then individually refined over a smaller search space as above (Pettersen et al., 2004). A search using individual NBDs versus the entire MRP1 map did not yield a consistent docking position, probably because of the relatively small size and roughly globular shape of a single NBD relative to the map and the limited resolution of the latter.
Supplementary Material
Acknowledgments
We thank Drs. Neil Ranson and Peiyi Wang for facilitating access to electron microscopes at the Universities of Leeds and Sheffield, UK; and Dr. William Nicholson (University of Leeds) and Mr. Simon Oliver (University of Manchester) for assistance with software. We would also like to thank Drs. Daniel Levy and Jean-Louis Rigaud (Institut Curie, Paris) for microscopy assistance and the British Council for a grant to MFR to visit their laboratory). The work is supported by the Leukaemia Research Fund (London, UK) and the Canadian Institutes of Health Research (Grant MOP-10519). SPCC is a Canada Research Chair in Cancer Biology and Bracken Chair in Genetics and Molecular Medicine.
Abbreviations
- Co2+-IMAC
Co2+-immobilized metal affinity resin chromatography
- DDM
n-dodecyl β–d-maltoside
- HA
hemagglutinin
- LPG
lysophosphatidyl glycerol
- LTC4
leukotriene C4
- MRP1
multidrug resistance protein 1/ABCC1
- NBD
nucleotide-binding domain
- P-gp
P-glycoprotein/ABCB1
- SDS–PAGE
SDS–polyacrylamide gel electrophoresis
- TMD
transmembrane domain
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jsb.2010.01.011.
References
- Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer M, Scarborough GA, Kuhlbrandt W. Three-dimensional map of the plasma membrane H+-ATPase in the open conformation. Nature. 1998;392:840–843. doi: 10.1038/33967. [DOI] [PubMed] [Google Scholar]
- Auer M, Scarborough GA, Kuhlbrandt W. Surface crystallisation of the plasma membrane H+-ATPase on a carbon support film for electron crystallography. J. Mol. Biol. 1999;287:961–968. doi: 10.1006/jmbi.1999.2652. [DOI] [PubMed] [Google Scholar]
- Awayn NH, Rosenberg MF, Kamis AB, Aleksandrov LA, Riordan JR, Ford RC. Crystallographic and single-particle analyses of native and nucleotide-bound forms of the cystic fibrosis transmembrane conductance regulator (CFTR) protein. Biochem. Soc. Trans. 2005;33:996–999. doi: 10.1042/BST20050996. [DOI] [PubMed] [Google Scholar]
- Bakos E, Hegedus T, Hollo Z, Welker E, Tusnady GE, Zaman GJR, Flens MJ, Varadi A, Sarkadi B. Membrane topology and glycosylation of the human multidrug resistance-associated protein. J. Biol. Chem. 1996;271:12322–12326. doi: 10.1074/jbc.271.21.12322. [DOI] [PubMed] [Google Scholar]
- Bakos E, Evers R, Szakacs G, Tusnady GE, Welker E, Szabo K, De Haas M, Van Deemter L, Borst P, Varadi A, Sarkadi B. Functional multidrug resistance protein (MRP1) lacking the N-terminal transmembrane domain. J. Biol. Chem. 1998;273:32167–32175. doi: 10.1074/jbc.273.48.32167. [DOI] [PubMed] [Google Scholar]
- Cai J, Daoud R, Georges E, Gros P. Functional expression of multidrug resistance protein 1 in Pichia pastoris. Biochemistry. 2001;40:8307–8316. doi: 10.1021/bi010093c. [DOI] [PubMed] [Google Scholar]
- Campbell JD, Koike K, Moreau C, Sansom MSP, Deeley RG, Cole SPC. Molecular modeling correctly predicts the functional importance of Phe(594) in transmembrane Helix 11 of the multidrug resistance protein, MRP1(ABCC1) J. Biol. Chem. 2004;279:463–468. doi: 10.1074/jbc.M310711200. [DOI] [PubMed] [Google Scholar]
- Chifflet S, Torriglia A, Chiesa R, Tolosa S. A method for the determination of inorganic-phosphate in the presence of labile organic phosphate, high-concentrations of protein – application to lens ATPases. Anal. Biochem. 1988;168:1–4. doi: 10.1016/0003-2697(88)90002-4. [DOI] [PubMed] [Google Scholar]
- Cole SP, Deeley RG. Transport of glutathione and glutathione conjugates by MRP1. Trends Pharmacol. Sci. 2006;27:438–446. doi: 10.1016/j.tips.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Cole SPC, Bhardwaj G, Gerlach JH, Mackie JE, Grant CE, Almquist KC, Stewart AJ, Kurz EU, Duncan AMV, Deeley RG. Overexpression of a transporter gene in a multidrug-resistant human lung-cancer cell-line. Science. 1992;258:1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- Cole SPC, Sparks KE, Fraser K, Loe DW, Grant CE, Wilson GM, Deeley RG. Pharmacological characterization of multidrug-resistant Mrp-transfected human tumor-cells. Cancer Res. 1994;54:5902–5910. [PubMed] [Google Scholar]
- Conseil G, Deeley RG, Cole SPC. Functional importance of three basic residues clustered at the cytosolic interface of transmembrane helix 15 in the multidrug and organic anion transporter MRP1 (ABCC1) J. Biol. Chem. 2006;281:43–50. doi: 10.1074/jbc.M510143200. [DOI] [PubMed] [Google Scholar]
- Crowther RA, Henderson R, Smith JM. MRC image processing programs. J. Struct. Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- Dawson RJ, Locher KP. Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett. 2007;581:935–938. doi: 10.1016/j.febslet.2007.01.073. [DOI] [PubMed] [Google Scholar]
- Deeley RG, Cole SPC. Substrate recognition and transport by multidrug resistance protein 1 (ABCC1) FEBS Lett. 2006;580:1103–1111. doi: 10.1016/j.febslet.2005.12.036. [DOI] [PubMed] [Google Scholar]
- DeGorter MK, Conseil G, Deeley RG, Campbell RL, Cole SPC. Molecular modeling of the human multidrug resistance protein 1 (MRP1/ABCC1) Biochem. Biophys. Res. Commun. 2008;365:29–34. doi: 10.1016/j.bbrc.2007.10.141. [DOI] [PubMed] [Google Scholar]
- Doige CA, Yu X, Sharom FJ. ATPase activity of partially purified P-glycoprotein from multidrug- resistant Chinese hamster ovary cells. Biochim. Biophys. Acta. 1992;24:149–160. doi: 10.1016/0005-2736(92)90078-z. [DOI] [PubMed] [Google Scholar]
- Ford RC, Holzenburg A. Electron crystallography of biomolecules: mysterious membranes and missing cones. Trends Biochem. Sci. 2008;33:38–43. doi: 10.1016/j.tibs.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Gao M, Yamazaki M, Loe DW, Westlake CJ, Grant CE, Cole SPC, Deeley RG. Multidrug resistance protein – Identification of regions required for active transport of leukotriene C-4. J. Biol. Chem. 1998;273:10733–10740. doi: 10.1074/jbc.273.17.10733. [DOI] [PubMed] [Google Scholar]
- Gao M, Cui HR, Loe DW, Grant CE, Almquist KC, Cole SP, Deeley RG. Comparison of the functional characteristics of the nucleotide binding domains of multidrug resistance protein 1. J. Biol. Chem. 2000;275:13098–13108. doi: 10.1074/jbc.275.17.13098. [DOI] [PubMed] [Google Scholar]
- Gerber S, Comellas-Bigler M, Goetz BA, Locher KP. Structural basis of trans-inhibition in a molybdate/tungstate ABC transporter. Science. 2008;321:246–250. doi: 10.1126/science.1156213. [DOI] [PubMed] [Google Scholar]
- Haimeur A, Deeley RG, Cole SPC. Charged amino acids in the sixth transmembrane helix of multidrug resistance protein 1 (MRP1/ABCC1) are critical determinants of transport activity. J. Biol. Chem. 2002;277:41326–41333. doi: 10.1074/jbc.M206228200. [DOI] [PubMed] [Google Scholar]
- Haimeur A, Conseil G, Deeley RG, Cole SPC. The MRP-related and BCRP/ABCG2 multidrug resistance proteins: biology, substrate specificity and regulation. Curr. Drug Metab. 2004;5:21–53. doi: 10.2174/1389200043489199. [DOI] [PubMed] [Google Scholar]
- Henderson R, Baldwin JM, Downing KH, Lepault J, Zemlin F. Structure of purple membrane from Halobacterium–Halobium – recording, measurement and evaluation of electron-micrographs at 3.5 a resolution. Ultramicroscopy. 1986;19:147–178. [Google Scholar]
- Hipfner DR, Deeley RG, Cole SPC. Structural, mechanistic and clinical aspects of MRP1. Biochim. Biophys. Acta, Biomembr. 1999;1461:359–376. doi: 10.1016/s0005-2736(99)00168-6. [DOI] [PubMed] [Google Scholar]
- Hollenstein K, Dawson RJ, Locher KP. Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 2007;17:412–418. doi: 10.1016/j.sbi.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Hollenstein K, Frei DC, Locher KP. Structure of an ABC transporter in complex with its binding protein. Nature. 2007;446:213–216. doi: 10.1038/nature05626. [DOI] [PubMed] [Google Scholar]
- Hollenstein K, Dawson RJ, Locher KP. Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 2007;17:412–418. doi: 10.1016/j.sbi.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Hyde SC, Emsley P, Hartshorn MJ, Mimmack MM, Gileadi U, Pearce SR, Gallagher MP, Gill DR, Hubbard RE, Higgins CF. Structural model of ATP-binding proteins associated with cystic- fibrosis, multidrug resistance and bacterial transport. Nature. 1990;346:362–365. doi: 10.1038/346362a0. [DOI] [PubMed] [Google Scholar]
- Ito K, Weigl KE, Deeley RG, Cole SPC. Mutation of proline residues in the NH2-terminal region of the multidrug resistance protein, MRP1 (ABCC1): effects on protein expression, membrane localization, and transport function. Biochim. Biophys. Acta, Biomembr. 2003;1615:103–114. doi: 10.1016/s0005-2736(03)00228-1. [DOI] [PubMed] [Google Scholar]
- Kadaba NS, Kaiser JT, Johnson E, Lee A, Rees DC. The high-affinity E coli methionine ABC transporter: structure and allosteric regulation. Science. 2008;321:250–253. doi: 10.1126/science.1157987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare D, Oldham ML, Orelle C, Davidson AL, Chen J. Alternating access in maltose transporter mediated by rigid-body rotations. Mol. Cell. 2009;33:528–536. doi: 10.1016/j.molcel.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EM, Letourneau IJ, Deeley RG, Cole SPC. Functional and structural consequences of cysteine substitutions in the NH2 proximal region of the human multidrug resistance protein 1 (MRP1/ABCC1) Biochemistry. 2003;42:5214–5224. doi: 10.1021/bi027076n. [DOI] [PubMed] [Google Scholar]
- Leslie EM, Deeley RG, Cole SPC. Multidrug resistance proteins: role of P-glycoprotein, MRP1, MRP2, and BCRP (ABCG2) in tissue defense. Toxicol. Appl. Pharmacol. 2005;204:216–237. doi: 10.1016/j.taap.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Locher KP. Structure and mechanism of ABC transporters. Curr. Opin. Struct. Biol. 2004;14:426–431. doi: 10.1016/j.sbi.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Loe DW, Almquist KC, Deeley RG, Cole SPC. Multidrug resistance protein (MRP)-mediated transport of leukotriene C-4 and chemotherapeutic agents in membrane vesicles – demonstration of glutathione-dependent vincristine transport. J. Biol. Chem. 1996;271:9675–9682. doi: 10.1074/jbc.271.16.9675. [DOI] [PubMed] [Google Scholar]
- Mao Q, Deeley RG, Cole SP. Functional reconstitution of substrate transport by purified multidrug resistance protein MRP1 (ABCC1) in phospholipid vesicles. J. Biol. Chem. 2000;275:34166–34172. doi: 10.1074/jbc.M004584200. [DOI] [PubMed] [Google Scholar]
- Mueller CF, Becher MU, Zimmer S, Wassmann S, Keuler B, Nickenig G. Angiotensin II triggers release of leukotriene C(4) in vascular smooth muscle cells via the multidrug resistance-related protein 1. Mol. Cell Biochem. 2009;333:261–267. doi: 10.1007/s11010-009-0227-x. [DOI] [PubMed] [Google Scholar]
- Oldham ML, Khare D, Quiocho FA, Davidson AL, Chen J. Crystal structure of a catalytic intermediate of the maltose transporter. Nature. 2007;450:515–521. doi: 10.1038/nature06264. [DOI] [PubMed] [Google Scholar]
- Oldham ML, Davidson AL, Chen J. Structural insights into ABC transporter mechanism. Curr. Opin. Struct. Biol. 2008;18:726–733. doi: 10.1016/j.sbi.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peklak-Scott C, Townsend AJ, Morrow CS. Dynamics of glutathione conjugation and conjugate efflux in detoxification of the carcinogen, 4-nitroquinoline 1-oxide: contributions of glutathione, glutathione S-transferase, and MRP1. Biochemistry. 2005;44:4426–4433. doi: 10.1021/bi047810y. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera – a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rosenberg MF, Mao QC, Holzenburg A, Ford RC, Deeley RG, Cole SPC. The structure of the multidrug resistance protein 1 (MRP1/ABCC1) – crystallization and single-particle analysis. J. Biol. Chem. 2001;276:16076–16082. doi: 10.1074/jbc.M100176200. [DOI] [PubMed] [Google Scholar]
- Rosenberg MF, Kamis AB, Callaghan R, Higgins CF, Ford RC. Three-dimensional structures of the mammalian multidrug resistance P-glycoprotein demonstrate major conformational changes in the transmembrane domains upon nucleotide binding. J. Biol. Chem. 2003;278:8294–8299. doi: 10.1074/jbc.M211758200. [DOI] [PubMed] [Google Scholar]
- Rosenberg MF, Kamis AB, Aleksandrov LA, Ford RC, Riordan JR. Purification and crystallization of the cystic fibrosis transmembrane conductance regulator (CFTR) J. Biol. Chem. 2004;279:39051–39057. doi: 10.1074/jbc.M407434200. [DOI] [PubMed] [Google Scholar]
- Rosenberg MF, Callaghan R, Modok S, Higgins CF, Ford RC. Three-dimensional structure of P-glycoprotein: the transmembrane regions adopt an asymmetric configuration in the nucleotide-bound state. J. Biol. Chem. 2005;280:2857–2862. doi: 10.1074/jbc.M410296200. [DOI] [PubMed] [Google Scholar]
- Rothnie A, Callaghan R, Deeley RG, Cole SPC. Role of GSH in estrone sulfate binding and translocation by the multidrug resistance protein 1 (MRP1/ABCC1) J. Biol. Chem. 2006;281:13906–13914. doi: 10.1074/jbc.M600869200. [DOI] [PubMed] [Google Scholar]
- Scarborough GA. Large single-crystals of the neurospora-crassa plasma-membrane H(+)- ATPase – an approach to the crystallization of integral membrane-proteins. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1994;50:643–649. doi: 10.1107/S0907444993014283. [DOI] [PubMed] [Google Scholar]
- Szakacs G, Annereau JP, Lababidi S, Shankavaram U, Arciello A, Bussey KJ, Reinhold W, Guo Y, Kruh GD, Reimers M, Weinstein JN, Gottesman MM. Predicting drug sensitivity and resistance. profiling ABC transporter genes in cancer cells. Cancer cell. 2004;6:129–137. doi: 10.1016/j.ccr.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discovery. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- Valpuesta JM, Carrascosa JL, Henderson R. Analysis of electron-microscope images and electron-diffraction patterns of thin-crystals of 029-connectors in ice. J. Mol. Biol. 1994;240:281–287. doi: 10.1006/jmbi.1994.1445. [DOI] [PubMed] [Google Scholar]
- Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc. Natl. Acad. Sci. USA. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlake CJ, Qian YM, Gao M, Vasa M, Cole SPC, Deeley RG. Identification of the structural and functional boundaries of the multidrug resistance protein 1 cytoplasmic loop 3. Biochemistry. 2003;42:14099–14113. doi: 10.1021/bi035333y. [DOI] [PubMed] [Google Scholar]
- Westlake CJ, Cole SPC, Deeley RG. Role of the NH2-terminal membrane spanning domain of multidrug resistance protein 1/ABCC1 in protein processing and trafficking. Mol. Biol. Cell. 2005;16:2483–2492. doi: 10.1091/mbc.E04-12-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wriggers W, Milligan RA, Mccammon JA. Situs: a package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 1999;125:185–195. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- Wu P, Oleschuk CJ, Mao QC, Keller BO, Deeley RG, Cole SPC. Analysis of human multidrug resistance protein 1 (ABCC1) by matrix-assisted laser desorption ionization/time of flight mass spectrometry: toward identification of leukotriene C4 binding sites. Mol. Pharmacol. 2005;68:1455–1465. doi: 10.1124/mol.105.016576. [DOI] [PubMed] [Google Scholar]
- Zhang DW, Nunoya K, Vasa M, Gu HM, Theis A, Cole SPC, Deeley RG. Transmembrane helix 11 of multidrug resistance protein 1 (MRPI/ABCC1): identification of polar amino acids important for substrate specificity and binding of ATP at nucleotide binding domain 1. Biochemistry. 2004;43:9413–9425. doi: 10.1021/bi0495230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.