Abstract
For several Class I aminoacyl-tRNA synthetases (aaRSs), the rate determining step in aminoacylation is the dissociation of the charged tRNA from the enzyme. In this study, the following factors affecting release of the charged tRNA from the aaRS are computationally explored: the protonation states of amino acids and substrates present in the active site and the presence and absence of AMP and the elongation factor Tu (EF-Tu).
Through molecular modeling, internal pKa calculations, and molecular dynamics simulations, distinct, mechanistically relevant post-transfer states with the charged tRNA (Glu-tRNAGlu) bound to glutamyl-tRNA synthetase from Thermus thermophilus are considered. The behavior of these non-equilibrium states is characterized as a function of time using dynamical network analysis, local energetics, and changes in free energies to estimate transitions that occur during the release of the tRNA. The hundreds of nanoseconds of simulation time reveal system characteristics that are consistent with recent experimental studies.
The energetic and network results support the previously proposed mechanism in which the transfer of the amino acid to the tRNA is accompanied by the protonation of AMP to H-AMP. Subsequent migration of the proton to water reduces the stability of the complex and loosens the interface both in the presence and absence of AMP. The subsequent undocking of AMP or tRNA then proceeds along thermodynamically competitive pathways. Release of the tRNA acceptor stem is further accelerated by the deprotonation of the α–ammonium group on the charging amino acid. The proposed general base is Glu41, a residue binding the α–ammonium group that is conserved in both structure and sequence across nearly all Class I aaRSs. This universal handle is predicted through pKa calculations to be part of a proton relay system for destabilizing the bound charging amino acid following aminoacylation. Addition of EF-Tu to the aaRS·tRNA complex stimulates the dissociation of the tRNA core and acceptor stem.
Keywords: glutamyl-tRNA synthetase, dissociation, free energy of binding, molecular dynamics simulation, network analysis
Introduction
The aminoacyl-tRNA synthetases (aaRSs) help maintain the genetic code by recognizing their cognate tRNAs and amino acids from the pool of competing reactants within the cell [1, 2]. In the majority of cases, the formation of the aminoacylated tRNAs (charged tRNA) within the active site occurs via a two step process (see Figure 1). In the first step, the amino acid is activated by ATP, forming an aminoacyl-adenylate and pyrophosphate. In the second step, the amino acid moiety on the adenylate is transferred to the 2′ hydroxyl group at the 3′ end of the tRNA with the simultaneous formation of an AMP product. We refer to the aaRS·tRNA complexes before and after the amino acid transfer as the pre- and post-transfer states, respectively. Previous biochemical studies and crystal structures have provided valuable information about the first step of aminoacylation, the binding site of the adenylate, and the mode of interactions between the identity elements on the tRNA and the aaRS (see [3] and references therein). More elusive are the details of how the charged tRNA dissociates from the aaRS prior to binding the elongation factor Tu (EF-Tu) for delivery to the ribosome. In the Class I aaRSs, tRNA dissociation is the rate determining step for tRNA aminoacylation, which has been shown to be stimulated in the presence of EF-Tu [4, 5]. Dissociation has been hypothesized to begin with the charged 3′ end of the tRNA exiting the active site while the anticodon remains strongly bound to the aaRS [6],
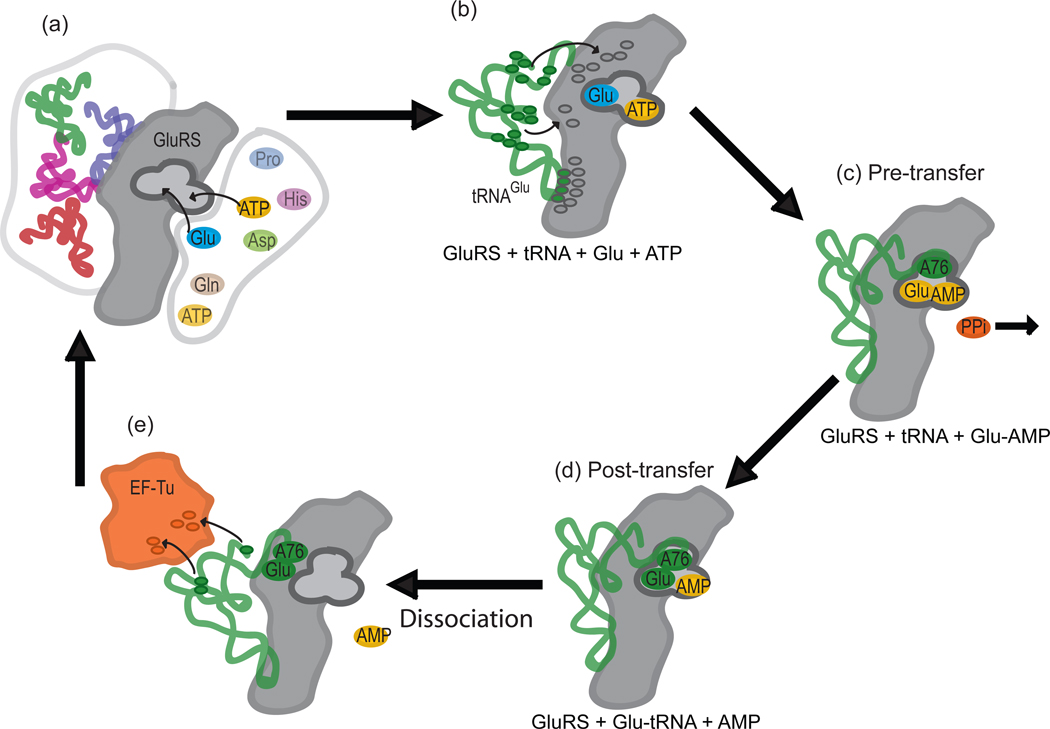
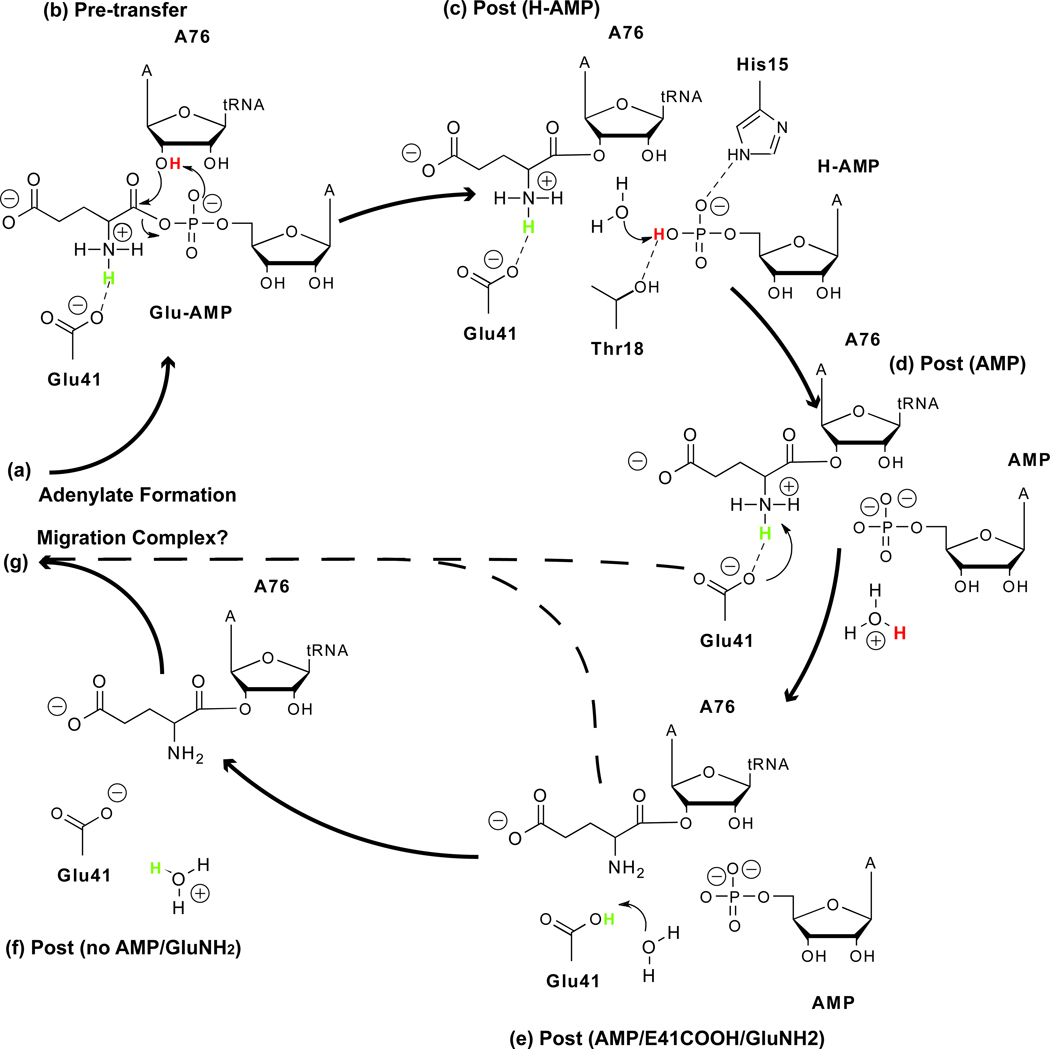
Figure 1.
Summary of aminoacylation reaction for GluRS·tRNAGlu. Panel (a) shows the apo-GluRS attracting ATP and selecting the cognate amino acid glutamate and tRNAGlu from the cellular pool. Panel (b) indicates the recognition of tRNAGlu identity elements (green ovals) by interactions with highly conserved residues (gray ovals) in GluRS. Panel (c) shows the Pre-transfer state(s) with the formation of the adenylate. Panel (d) contains the Post-transfer state(s) with the newly charged tRNA. Panel (e) shows the charged tRNA dissociating from the synthetase before association with the EF-Tu through the tuning elements (green ovals to orange ovals) and subsequent transportation to the ribosome. The system can sample a large ensemble of states at each stage of the reaction.
In this study, we investigate the series of events occurring in the active site that control tRNA dissociation. We use the structure of the glutamyl-tRNA synthetase (GluRS) complexed with tRNAGlu from Thermus thermophilus [7] as a representative of monomeric Class I aaRSs. Although GluRS is atypical of Class I aaRSs in that it requires tRNA to be bound before the aminoacyl adenylate can be formed, the final process of AMP and aa-tRNA dissociation involves an analogous set of molecules in all Class I aaRSs. The modeled post-transfer states are differentiated by protonation of AMP and its neighboring amino acid residues and by the presence or absence of AMP in the active site. These states have been selected based on suggested reaction mechanisms [8] and internal pKa calculations. Through comparative analyses of each system state’s behavior with the pre-transfer state and experimental results, the undocking of AMP and changes in protonation states are evaluated as possible exit strategies for tRNA dissociation. Our results indicate that both factors assist in the release of the charged tRNA from the enzyme.
Structure and Phylogenetic Background of the GluRS·tRNAGlu Complex
The aaRSs are divided into two classes based on the structurally distinct, conserved core or catalytic domain (CD) containing the active site [9, 10]. The CD of the Class I aaRSs forms a Rossman fold with a three layered αβα topology containing a parallel β-sheet architecture. The active site is located at the C-terminal loops of the β-strands (see Figure 2a). Within the active site are the evolutionarily conserved HIGH and KMSK sequence motifs, which bind ATP during adenylate formation. The histidines in the HIGH motif form contacts with the phosphates while the KMSK loop is located near the adenine base. Located between the two halves of the Rossman fold (RF-N and RF-C), the connective polypeptide (CP1) insertion binds the 3′ end of the tRNA during aminoacylation. Class I aaRSs are further differentiated by the fold of the anticodon binding (ACB) domain [11, 12]. GluRS is part of the Class Ib subgroup with a set of α-helices (four-helix junction or 4HJ) connecting the CD to the C-terminal, α-helical ACB domain. GluRS in T. thermophilus has been crystallized in a variety of states prior to the second step of aminoacylation [13, 14, 15, 7]. The crystal structure used for the current study was obtained by the Yokoyama group and contains GluRS (468 residues) with a Glu-AMP analog and transcribed tRNAGlu in the active site [PDB code: 1N78] [7] (see Figure 2a for the structure and b for the standard tRNA cloverleaf schematic). Use of the analog creates an unreactive substrate complex mimicking the pre-transfer system state, which serves as a starting point for this study.
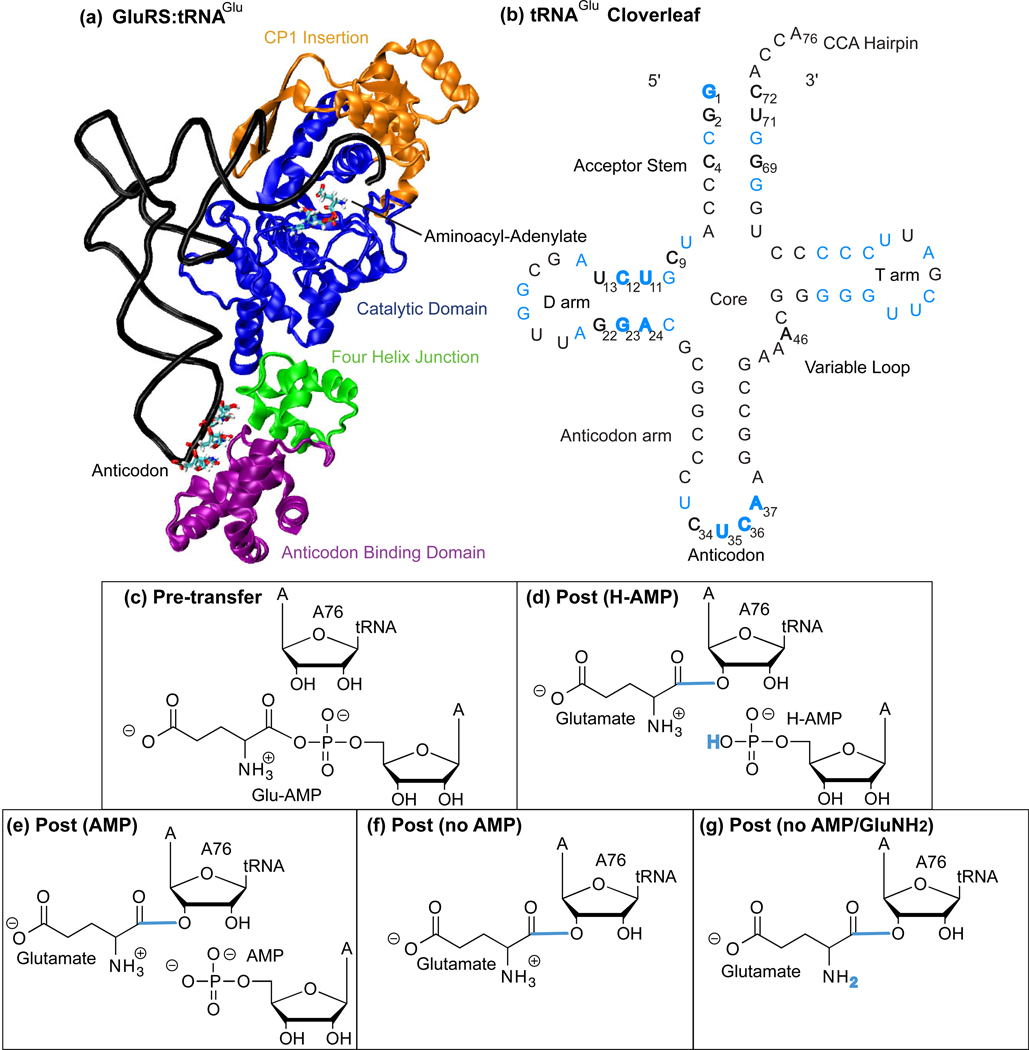
Figure 2.
GluRS·tRNAGlu system used in simulations: (a) GluRS with functional domains labeled, (b) tRNAGlu cloverleaf schematic where nucleotides with greater than 75% sequence identity across the evolutionary profile are colored blue. Identity elements are in bold. Panels (c–g) show the small molecule substrate/products in the active site for the different system states. Using the Pre-transfer state as the control, the bonds and atoms colored blue in each post-transfer state indicate the changes made to the AMP or charging amino acid moiety. Specific system state names are capitalized in the text.
GluRS has a divergent evolutionary history which has led to several classes of GluRS as well as GlnRS [16, 2, 17]. GluRS enzymes are divided into α (bacterial) and β (archaeal/eukaryal) types based primarily on the fold of their nonhomologous ACB domain structures. Both classes have discriminating (D-GluRS) and nondiscriminating versions based upon the ability of GluRS to charge tRNAGlu and/or tRNAGln with glutamate. The α-type GluRS in some bacterial organisms has evolved specific residues to recognize the third anticodon base, discriminating between tRNAGlu and tRNAGln [18]. Those bacteria with the discriminating GluRS have either acquired the eukaryal-type GlnRS through horizontal gene transfer [19] or evolved a GluRS2, which specifically recognizes and misacylates tRNAGln with glutamate for subsequent reduction [20, 21, 22]. Due to the lack of crystal structures containing tRNA docked to GluRS in these various subgroups, the scope of the current study is limited to the discriminating α-type represented by the GluRS in T. thermophilus.
MD Simulations and Analysis Methods for aaRSs
Molecular dynamics (MD) is a powerful method that has been used to study correlations [23], signaling pathways [24, 25], editing [26], and binding free energies [27, 28, 29, 30, 31] in aaRSs. Here, we perform long-timescale simulations of the entire GluRS·tRNAGlu complex with explicit solvent and both mono- and divalent ions to determine the dynamical and energetic behavior for the various pre- and post-transfer states. We characterize the behavior of these states as a function of time using dynamical networks, local energetics, and changes in free energy to estimate the transitions that occur during tRNA dissociation.
Results
Modeling Pre- and Post-transfer states
Pre-transfer state
The adenylate analog in the GluRS·tRNAGlu crystal structure (PDB code 1N78) is chemically inert because it lacks the α-carbonyl group on the glutamate backbone. Upon replacing the analog with Glu-AMP and equilibrating the system, small rearrangements occur around the α-carbonyl of the glutamate moiety and phosphate of the AMP moiety. In our simulations, the 2′-hydroxyl of A76 reorients towards the α-carbonyl group (see Figure 3). This reorientation positions the reactants such that the aminoacylation can take place. To accommodate these changes, the distance between the 3′-hydroxyl of A76 and the α-ammonium group on the charging glutamate lengthens. The contact distances between the highly conserved residues Ala7, Ser9, and Glu41 and the α-ammonium group shorten. In addition, contacts between the charging glutamate sidechain and other active site residues lengthen slightly as the system relaxes. The final configuration of the active site was within 2 Å of the crystal structure, demonstrating that the initial protonation states and modification of the analog produced minor perturbations (see Supplementary Figure 11 for a summary of changes). The MD simulation of the pre-transfer state was used as a control in this study to provide the baseline behavior of the system.
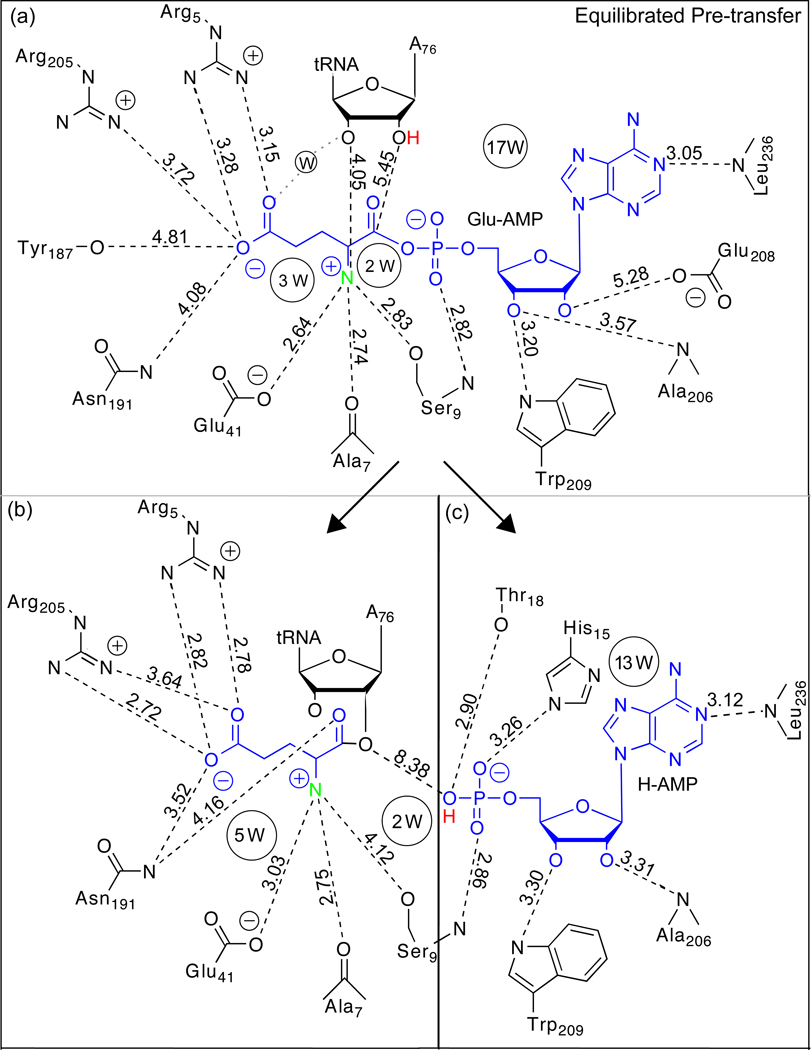
Figure 3.
Molecular interactions with the small molecule substrate/product in the GluRS active site. In panel (a) Glu-AMP interacts with A76 on the tRNA, multiple residue sidechains, and water. Panels (b) and (c) show the active site residues around the (b) charging glutamate and (c) H-AMP in the equilibrated Post (H-AMP) state. The Glu-AMP, charging glutamate, and H-AMP atoms are colored blue while the H-AMP proton is colored red and the glutamate α–nitrogen is colored green for emphasis. All hydrogen atoms other than the H-AMP proton have been removed for clarity. The distances shown were measured between the heavy atoms and averaged over the last 16 ns of the 20 ns trajectories. Residues in the HIGH or KMSK sequence motifs are denoted accordingly.
Assignment of the protonation state of residues at the GluRS·tRNAGlu interface and within the active site is a key consideration when developing models for protein-RNA dissociation. The pKa values of all titratable sidechains on GluRS in the Pre-transfer state were estimated using the PROPKA program at the end of the simulation (see Methods), and the values for residues within the active site are reported in Table 1. In the Pre-transfer state, all histidine residues (including His15 in the HIGH motif) are predicted to have pKa < 7, indicating that the neutral form of histidine is dominant at pH 7.0. In addition, all lysine and arginine residues are predicted to have pKa > 7 while the acidic aspartate and glutamate residues are predicted to have pKa < 7, suggesting that they exist as charged residues in the Pre-transfer state.
Table 1.
Predicted pKa values of residues within 5 Å of the small molecule substrate/products, the (H-)AMP, and the α–amino group of the charging glutamate in the Pre-transfer state and several post-transfer states. Residues with pKa values both above and below 7 are shown in bold.
| pKa Calculations after 20 ns Simulations | |||||
|---|---|---|---|---|---|
| Residue | Pre-transfer | Post (H-AMP) | Post (AMP) | Post (AMP/GluNH2 /E41COOH) |
Post (no AMP/GluNH2) |
| (H-)AMP | - | 5.51 | 7.70 | 6.44 | - |
| Glu amino | 9.86 | 7.23 | 7.69 | 5.12 | 9.47 |
| Glu carboxy | 2.11 | 0.89 | 2.50 | 0.38 | 4.50 |
| Arg5 | 11.91 | 11.09 | 10.35 | 10.07 | 9.29 |
| His15 | 2.79 | 3.52 | 2.93 | 3.48 | 2.24 |
| Glu41 | 1.86 | 8.06 | 8.90 | 4.50 | 6.03 |
| Tyr187 | 13.11 | 12.08 | 11.18 | 11.39 | 15.75 |
| Arg205 | 11.45 | 9.54 | 11.15 | 12.27 | 12.98 |
| Glu208 | 3.94 | −4.91 | 3.39 | 2.99 | 3.25 |
| Lys243 | 11.10 | 10.94 | 11.18 | 11.03 | 9.76 |
Charging mechanism and post-transfer states
A concerted mechanism has been proposed for tRNA aminoacylation of the homologous GlnRS·tRNAGln system [8] which was later experimentally tested on TyrRS·tRNATyr [32]. In the transfer step of the proposed aminoacylation reaction mechanism, the 2′-hydroxyl oxygen from A76 nucleophilically attacks the α-carbonyl carbon of Glu-AMP while, simultaneously, the A76 2′-hydroxyl proton shifts to the Glu-AMP α-phosphate resulting in Glu-tRNAGlu and H-AMP [8]. Assuming a similar concerted mechanism holds for GluRS·tRNAGlu, we created in silico a series of relevant post-transfer states (see Methods).
Figure 2 provides a summary of the terminology and differences in initial setup between the simulations. In the first post-transfer state (Post (H-AMP)), the amino acid moiety on the adenylate is transferred to the tRNA, and the proton previously on the 2′-OH of A76 is transferred to the AMP phosphate, which is the only base in close proximity to the ribose hydroxyl group. As this state models the system immediately following the transition state, it is expected to exhibit binding affinities similar to the Pre-transfer state. Separation of H-AMP from the amino acid moiety causes a few significant rearrangements in the active site (see Figure 3b and c). The H-AMP α-phosphate shifts closer to the HIGH motif and forms hydrogen bonds with His15 and Thr18. Both residue sidechains rotate to accommodate the new interactions. In solution, H-AMP has a pKa of 6.9 [33], but in the protein, interactions with His15 and Ser9 reduce the pKa of the H-AMP phosphate to 5.51 (see Table 1). This suggests that the proton could be further transferred to another residue on the protein or to a water molecule, forming the Post (AMP) state. Because its sidechain is buried within the protein, neutral His15 is predicted to have a pKa of 3.52 and therefore is unlikely to serve as a general base for abstracting the H-AMP proton. As a check, a system containing AMP and protonated His15 was simulated (Post (AMP/H15h) state), but the free energy analysis showed that this state has tighter binding at the complex interface than the Pre-transfer state (see Supplementary Table 8) and therefore does not lead to tRNA dissociation.
On the side of the active site containing the amino acid moiety, the electrostatic and hydrogen bonding environment around the Glu41 sidechain causes its local pKa to rise to 8.06 at the end of the Post (H-AMP) simulation and 8.90 after 20 ns in the Post (AMP) state (see Table 1). Because these simulations were performed at pH 7.0, Glu41 most likely exists in the neutral glutamic acid form in the Post (AMP) state. The α-ammonium group of the charging glutamate has a predicted pKa of 7.23 and therefore can be in either the cationic or neutral form. This suggests that the nearby negatively charged Glu41 sidechain could deprotonate the α-ammonium group and presumably participate in further proton transfer events. A Post (AMP/GluNH2/E41h) simulation, similar to the Post (AMP) state in all respects except that a proton from the α-ammonium group was transferred to the Glu41 sidechain resulting in a neutral α-amino group and a glutamic acid, was also run. The pKa values at the end of this simulation suggest that the neutral Glu41 (4.50) becomes deprotonated after exposure to solvent, but the α-amino group (5.12) remains neutral.
The next set of states was modeled under the assumption that AMP had already undocked. Initially, eight water molecules were placed in the vacant AMP binding site. This site becomes further hydrated as the increasingly mobile KMSK loop allows water molecules to enter. These waters penetrate deep into the active site, weakening contacts between the charging amino acid and multiple active site residues. Simulations of the Post (no AMP/GluNH2) state, in which the Glu41 sidechain is charged again and the α–amine of the charging glutamate is neutral, are expected to show the fast release of the tRNA as the water molecules that replace the AMP moiety assist in solvating and weakening the interactions between the charged tRNA and GluRS. The interactions of the charging glutamate sidechain with Arg5 and Arg205 are solvated, leading to greater motion that loosens the contacts between the α–ammonium group and its triad of conserved binding residues. Once the charging amino acid breaks its contact with the Glu41 sidechain in the Post (no AMP/GluNH2) state, the pKa of the α-amino group increases to 9.47 (see Table 1). Hence, the α-amine of the charging amino acid is likely to be reprotonated while the Glu41 sidechain displays a propensity to remain charged upon losing contact. The motion of the CCA hairpin as it undocks over 80 ns is shown in Supplementary Figure 12.
Universal handle for aa-tRNA release
Glu41 binds the α–ammonium group, which is a fundamental part of all charging amino acids. Figure 4b shows that Glu41 (amino acid 3 or AA3) is conserved as either glutamate or aspartate in GluRS enzymes across all domains of life. Further, comparison of ten available crystal structures of Class I aaRSs [7, 34, 35, 36, 37, 38, 39, 40, 41, 42] reveals that in nearly all specificities a conserved aspartate or glutamate is in the same orientation in the active site and binds the α–ammonium group of the charging amino acid, confirming previous studies [11]. This residue is analogous to the “universal aspartate” found in the editing domain of LeuRS and IleRS that interacts with the same α–ammonium [43, 44]. The pKa calculations and sequence conservation studies suggest that Glu41 may represent a universal handle in Class I aaRSs to stabilize the charging amino acid during aminoacylation. The transfer of a proton from the α–ammonium group to the universal handle may be a standard strategy for facilitating release of the charged tRNA following the transfer reaction. The other residues forming the conserved triad binding the α–ammonium group are usually conserved in structure but not in sequence over all Class I aaRSs. They are conserved in sequence within each specificity over all three domains of life, possibly indicating coevolution with the charging amino acid. The first (amino acid 1 or AA1) includes Ala7 in bacterial GluRS and commonly binds the α–ammonium group through its main chain carbonyl oxygen. This is analogous to the methionine in the LeuRS editing domain that stabilizes the charging amino acid for hydrolysis through a hydrogen bond between the methionine main chain carbonyl oxygen and the charging amino acid α–amino group [44]. The second (amino acid 2 or AA2) includes Ser9 and generally forms a hydrogen bond through the sidechain. This residue, which is a glutamine in TyrRS, was previously shown to stabilize the transition state of the second step of aminoacylation [32]. Interestingly, IleRS, LeuRS, and MetRS each contain an aromatic residue in the same structural position, which does not form a hydrogen bond, but instead stacks on the backbone of the charging amino acid. This difference may be necessary to stabilize these hydrophobic charging amino acids.
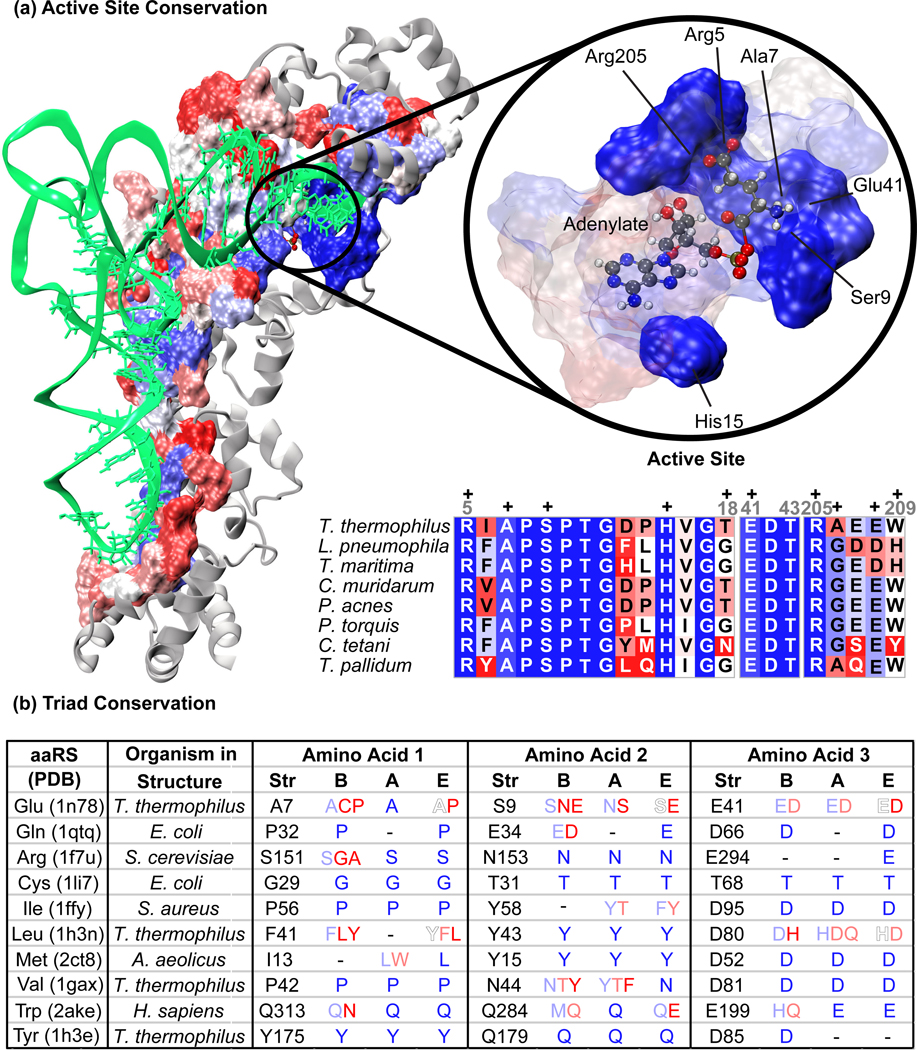
Figure 4.
Conservation of GluRS at the tRNA binding interface and active site. The structure on the left shows the residues within 5 Å of tRNA colored by their sequence conservation (percent identity). Blue indicates highly conserved, white is nearly 50% conserved, and red is not conserved. The structure on the right shows the highly conserved residues in the active site making contacts with the adenylate in solid blue with nearby residues of varying conservation as transparent. A subset of sequences in the discriminating GluRS alignment is shown, representing several of the major bacterial phyla. The ‘+’ symbols indicate columns of residues interacting with the adenylate either in the right panel or in Figure 3. In panel (b), the structures used to create the Class I overlap are shown along with the identity of each residue in the conserved triad (amino acid or AA1, AA2, AA3) for each particular structure. AA3 is the hypothesized universal handle. The adjacent columns list the residues present in the same position in the sequence alignment for a given domain of life (B = Bacteria, A = Archaea, and E = Eucarya). Conservation is indicated by the same color scheme as above. Residues that are not conserved within a domain of life are denoted by a dash.
The conservation of AA3 in both sequence and structure is nearly universal in Class I aaRSs, with three exceptions. Based on the available crystal structure, CysRS uses a threonine to form a hydrogen bond rather than an aspartate for a salt bridge. The close proximity of a zinc ion in the CysRS active site changes the electrostatics to control binding of the sidechain of the charging cysteine. The CysRS·tRNACys·Cys-AMP complex was modeled and simulated, and after equilibration, the charging cysteine α–amino group remained in contact with threonine, not interacting with the nearby aspartate (model available at http://www.scs.uiuc.edu/~schulten/publications.html). Bacterial TyrRS contains the conserved aspartate in a domain specific insertion, but recent crystal structures of the archaeal and eukaryal TyrRS show that AA3 may be a conserved glutamine in these domains of life [45, 46]. Experimental studies will be needed to verify this difference. The bacterial/archaeal types of TyrRS are known to be structurally divergent [2]. The aspartate in ArgRS is in a poorly conserved part of the structure, which is expected given the wide variety of ArgRS subtypes within each domain of life [2]. Due to a lack of crystal structures containing appropriate substrates, no additional interpretation can be offered.
The RMSD of the macromolecular backbones (Cα and P atoms) relative to the starting structure was used to monitor the equilibration of each state. There is an initial increase in the protein RMSD which levels off at 1.7 Å after 4 ns and remains approximately constant throughout all the runs. The RMSD of the entire tRNA in the Pre-transfer state remains around 3 Å with the largest fluctuations occuring in the anticodon arm. The motion of the CCA hairpin was monitored by measuring the distance between the charging amino acid and active site residues (Supplementary Figure 13), and it shows the large structural change in the Post (no AMP/GluNH2) simulation as it undocks.
Magnesium ion perturbations
Inclusion of Mg2+ ions in the simulations helps capture more physiologically realistic states but also introduces structural perturbations. These random fluctuations are independent of the active site modifications and can hinder the identification of resulting system changes, complicating the comparison of structural and free energy results across different system states. To maintain comparable distribution of Mg2+ across the various simulations, a metric was developed using Mg2+ ion residency times and locations to identify runs in which there were substantial deviations. A resident ion was defined as deviating less than 3 Å from its location for at least 4 ns (see Methods). A comparison of two Pre-transfer runs provides an illustrative example of how different distributions of Mg2+ ions cause large fluctuations in tRNA structure. In a replicate run, the complex interface showed substantially weaker interactions near the tRNA acceptor stem and stronger binding near the anticodon arm compared to the first run. A Mg2+ ion had entered the deep groove of the acceptor stem and brought phosphates on either side of the groove closer together. The tightening of this helix pulled the first two base pairs of the acceptor stem away from the catalytic domain, resulting in a tRNA conformation that was significantly different from the crystal structure (the frame of reference used for comparing the different system states). Although this new conformation could be physiologically realistic, the equilibration time required to return the tRNA to a conformation comparable to the crystal structure would have been too computationally expensive. The first run featured seven resident Mg2+ ions while the second had nine with the additional two Mg2+ ions near the acceptor stem and anticodon arm (see Supplementary Figure 14). To ensure that the system states had similar ion behavior, multiple replicates were performed, and simulations with approximately seven resident Mg2+ ions were accepted for further analysis.
Communication decreases in interaction network as tRNA dissociates
Analysis of the dynamical network of interactions within the aaRS·tRNA complexes was used to identify signs of dissociation during the course of the 20 ns simulations (which are relatively short compared to the biological timescale of tRNA release) [25]. This type of network analysis uses local coupled motions between pairs of residues to track allosteric signaling in biomolecules. In these networks, a node represents either a nucleotide, an amino acid, or the adenylate present in the pre-transfer complex, and edges connect nodes that are in contact for a majority of the last 5 ns of the trajectory (see Methods). The cross-correlations calculated from the atomic fluctuations during the same time window are used to weight each edge such that as the correlation reduces, the edge distance lengthens. Greater edge distances indicate lower communication between the connected pair of nodes since the corresponding residues are moving more independently of one another. Signaling pathways are the dominant routes for communication between important interfacial contacts and the active site, and they can be found by tracing the most optimal path through the weighted edges in the network. Shorter optimal path lengths have larger correlation along the path. The sum of all the path lengths from the interfacial interactions to the active site can therefore be used to quantitatively compare changes in correlation during tRNA release. The reduction of correlation with time during the release of the charged tRNA from the protein may also be observable experimentally through mutagenesis studies.
The shortest paths shown in Table 2 are calculated between A76 (site of aminoacylation) and the identity elements in different parts of the tRNA. Identity elements are specificity-dependent nucleotides that have been experimentally shown to affect the efficiency of aminoacylation by the cognate aaRS [47, 48, 49]. The identity elements for tRNAGlu are divided into three groups according to their position on the nucleic acid: acceptor stem (G1·C72, G2·U71, and C4·G69), D arm (U11·A24, C12·G23··C9, and U13·G22··A46), and anticodon arm (C34, U35, C36, and A37). The path distances are summed over all identity elements in a group. The sum of all path lengths is comparable for the Pre-transfer and the Post (H-AMP) states, indicating that the correlation in the post-transfer state immediately following the transition state is nearly the same as that in the pre-transfer state. In all subsequent post-transfer states, the correlations reduce along the signaling pathways, demonstrating that the aaRS·tRNA interfacial contacts are weakening. In the Post (AMP) and Post (no AMP) states, the signaling pathways exhibit comparable distances with the largest change occurring at the acceptor stem, where the tRNA is starting to undock. There is no significant change in the D arm and the anticodon arm, suggesting that the acceptor stem is released first, as has been hypothesized from crystal structures of Trp-tRNATrp binding to TrpRS [6]. The Post (no AMP/GluNH2) system has the largest increase in the distances along the communication pathways, indicating that the aaRS·tRNA complex is the closest to dissociation in this system. The acceptor stem distances show the largest loss in correlation to the enzyme because many of the interactions between these nucleotides and GluRS have broken (see Supplementary Figure 15). The identity elements in the anticodon and D arms remain in contact with GluRS residues, but because the number of contacts between the partially undocked CCA hairpin and the protein is significantly reduced, longer paths are required to pass communication to A76.
Table 2.
Comparison of optimal path distances between different system states. The shortest distances between A76 and identity elements distal from the active site are listed for each system state. The identity elements are divided into groups based on their position in the tRNA structure: acceptor stem, core, and anticodon arm. The distances within each group are summed with the final total at the bottom. Shorter distances indicate stronger signaling. The dash for G1 in the Post (H-AMP) state indicates that it has lost all contacts with its neighbors and therefore is not connected to the rest of the network.
| Source | Pre-transfer | Post (H-AMP) | Post (AMP) | Post (no AMP) | Post (no AMP/GluNH2) |
|---|---|---|---|---|---|
| G1 | 4.80 | - | 7.67 | 7.05 | 15.42 |
| C72 | 1.35 | 2.24 | 3.59 | 5.90 | 13.86 |
| G2 | 2.50 | 5.51 | 5.92 | 4.51 | 12.26 |
| U71 | 1.94 | 2.48 | 2.98 | 2.63 | 12.97 |
| C4 | 2.25 | 3.69 | 3.97 | 3.03 | 9.85 |
| G69 | 3.30 | 2.87 | 2.78 | 2.78 | 8.26 |
| Acceptor Stem Subtotal |
16.14 | 16.79 | 26.91 | 25.90 | 72.62 |
| U11 | 4.55 | 3.09 | 3.16 | 2.88 | 9.15 |
| A24 | 3.79 | 3.35 | 3.66 | 3.36 | 10.25 |
| C12 | 3.30 | 2.65 | 2.63 | 2.59 | 8.61 |
| G23 | 4.07 | 3.47 | 4.46 | 3.68 | 9.62 |
| C9 | 4.29 | 3.06 | 3.15 | 3.15 | 9.42 |
| U13 | 3.32 | 2.80 | 3.19 | 2.75 | 8.84 |
| G22 | 4.22 | 4.22 | 3.99 | 4.11 | 10.06 |
| A46 | 4.80 | 3.68 | 3.96 | 3.85 | 9.81 |
| Core Subtotal |
32.34 | 26.32 | 28.20 | 26.37 | 75.76 |
| C34 | 5.44 | 5.25 | 5.19 | 6.09 | 12.34 |
| U35 | 4.71 | 4.33 | 4.18 | 5.72 | 11.18 |
| C36 | 4.25 | 4.00 | 6.40 | 5.93 | 10.69 |
| A37 | 3.96 | 3.60 | 5.16 | 4.71 | 10.58 |
| Anticodon Arm Subtotal |
18.36 | 17.18 | 20.93 | 22.45 | 44.79 |
| Total | 66.84 | 60.29 | 76.04 | 74.72 | 193.17 |
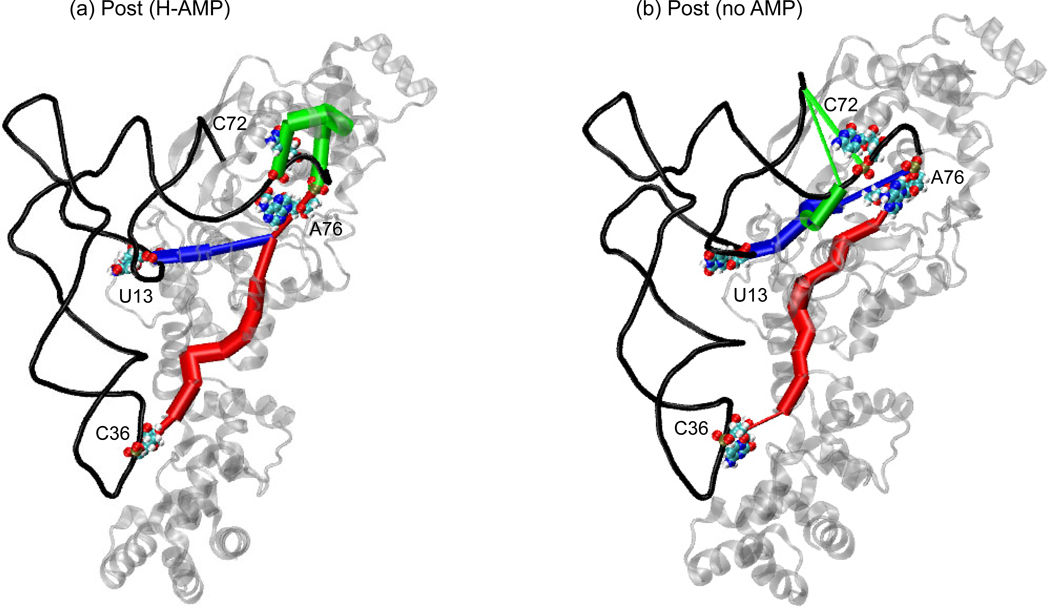
In Figure 5, the shortest paths between A76 and the identity elements C72, U13, and C36 are compared for the Post (H-AMP) and Post (no AMP) states. Paths are traced along the nodes, and the thickness of each edge is scaled by the correlation weights (thicker edges show greater correlation). As seen in the representative optimal path between C72 and A76, in the Post (H-AMP) state, the acceptor stem communicates with the active site through the CP1 insertion of the protein. In the Post (no AMP) state, the contacts between the CP1 insertion and the acceptor stem have broken as the CCA hairpin unbends and begins to unbind. Instead, the optimal path length increases as it passes through the tRNA before merging with the shortest path from U13 to A76 that passes through the catalytic domain of the protein. The shortest paths from U13 and C36 merge as they pass through the AMP before reaching A76 in the Post (H-AMP) state. The absence of the AMP causes a shift in these shortest paths in the Post (no AMP) state even though the anticodon loop and the D arm do not undock significantly. As discussed in the next section, the dissociation of the charged tRNA involves a conformational twist that brings the charged glutamate and CCA hairpin out of the active site while the D and anticodon stems are driven slightly further into the GluRS. A similar picture is presented by the interface edges connecting the tRNA and protein subnetworks (Supplementary Table 5).
Figure 5.
Comparison between the dynamical networks for the Post (H-AMP) and Post (no AMP) states. The correlations from the last 5 ns of the 20 ns simulation of the Post (H-AMP) and Post (no AMP) states were used to calculate the dynamical network (see Methods). The shortest or optimal paths are shown between A76 and identity elements C72 (green), U13 (blue), and C36 (red). Thicker edges indicate higher correlation between two nodes. The green and red paths are longer and thinner in (b) than in (a) revealing that the Post (no AMP) state complex has lost correlation between the charging amino acid on A76 and nucleotides C36 and C72 (see Table 2). The blue path, however, is only slightly thinner in (b) indicating that signaling is comparable for U13 between the two states.
Energetics of GluRS·tRNAGlu binding interface
GluRS specifically recognizes and binds tRNAGlu through contacts spanning the acceptor stem to the anticodon, and these interactions must break during tRNA dissociation. The residues and nucleotides making significant energetic contributions at the interface in the Pre-transfer state shown in Figure 6 were identified by averaging the electrostatic and van der Waals interaction energies over the last 5 ns of each simulation. Additional post-transfer states are provided in Supplementary Figure 16. These energies were calculated with a cutoff of 21 Å to fully include bound tRNA helices while limiting the analysis to the GluRS binding interface. The evolutionary importance of these interface residues is shown by masking the contact energy values by percent sequence identity across the bacterial GluRS evolutionary profile.
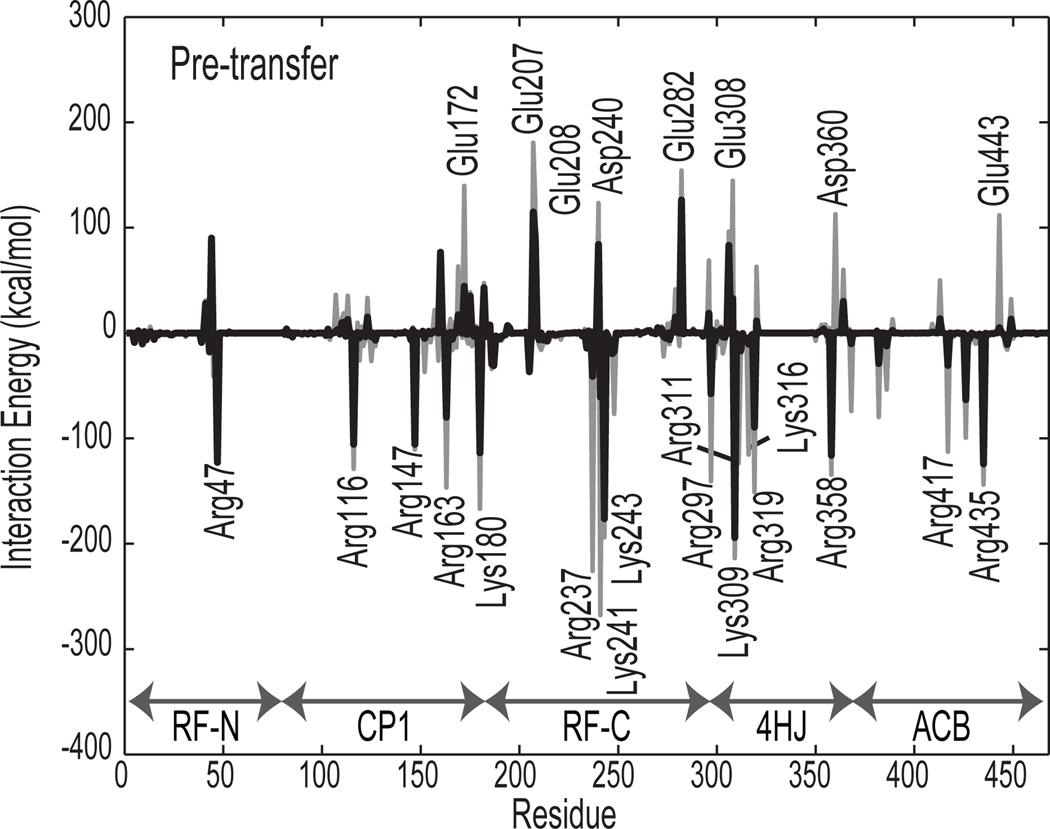
Figure 6.
Mean nonbonded interaction energies between tRNAGlu and GluRS. The energies are averaged over the last 5 ns of the 20 ns simulation of the Pre-transfer state (GluRS·Glu-AMP·tRNAGlu). Gray peaks show the full energetic interaction while black peaks show the energy scaled by percent sequence identity (see Methods). Labeled peaks have absolute value greater than 100 kcal/mol.
Because tRNA is highly electronegative, positively charged residues on GluRS generally form attractive contacts while negatively charged residues repel the tRNA. Figure 6 shows twice as many significantly attractive interactions as repulsive for the Pre-transfer complex, and all major non-bonded interactions involve charged amino acids near the highly negative tRNA backbone phosphates. The attractive interactions are generally stronger than the repulsive because positively charged residues can form direct contacts with the tRNA while repulsive forces keep the negatively charged residues further away. Most of the strongly attractive residues are close in sequence to repulsive residues, energetically balancing the interactions such that the tRNA is not bound too tightly to dissociate from GluRS. The strongly attractive residues at the tRNA interface are also more highly conserved than the repulsive residues (59% conservation vs. 43% with background GluRS conservation at 41%). Almost half of these attractive amino acids have greater than 75% sequence identity whereas only one of the repulsive residues is similarly well conserved. Six of the strongly interacting residues contact identity elements on the tRNA through salt bridges to backbone phosphates or hydrogen bonds to polar groups on the ribose or base (see Supplementary Table 6).
The strongest interactions occur in the second half of the Rossman fold, especially near the KMSK motif (residues 243–246), which binds the 3′ tRNA acceptor stem, and in the 4HJ near the D stem and the 5′ end of the anticodon stem. The stronger attractive interactions limit the rate at which dissociation can occur. Finally, the anticodon and its neighboring nucleotides are bound by Arg358 (86% with 14% lysine), Arg417, and Arg435 (86%) primarily through interactions with functional groups on the nucleobases while being counterbalanced by repulsion from nearby Asp360 and Glu443. Arg358 (Lys358) is notable because the presence of this positively charged residue is used by bacterial D-GluRS to discriminate between tRNAGlu and tRNAGln [18]. Comparison between interface energies for the Pre-transfer and post-transfer states showed a general picture of the beginning of tRNA dissociation in agreement with the network analysis (see Supplementary Information). While the charged CCA end leaves the GluRS catalytic site, the tRNA elbow region undergoes a rocking motion that moves the acceptor stem further away from GluRS and the D stem closer.
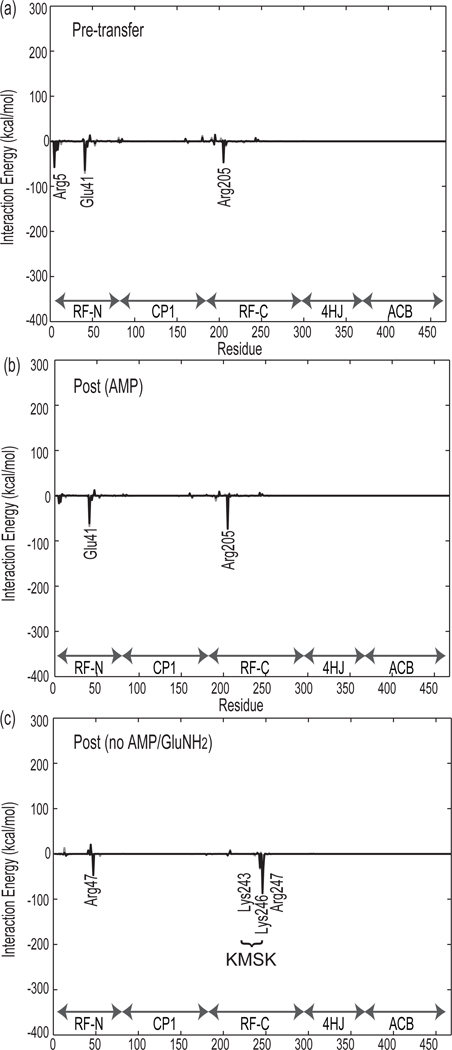
Nonbonded interactions to charging glutamate
A similar analysis was performed in the active site between GluRS and the charging glutamate moiety attached to the tRNA (see Figure 7). In the Pre-transfer state, the three most attractive contacts to the charging glutamate are through salt bridges from the glutamate sidechain to Arg5 and Arg205 and from its α–ammonium group to the conserved Glu41 (see Figure 4 for structural reference). In the Post (H-AMP) state (not shown), the interactions are almost identical, but the contact to Arg5 has been lost in the Post (AMP) state. Although Arg5 and Arg205 interact strongly with the glutamate during transfer, both of their sidechains are extended and accessible to water. Water molecules intervene between Arg5 and the charging glutamate, breaking the corresponding salt bridge. This state is likely to be in the process of dissociating, as seen in the previous section, but with the glutamate remaining bound to two of its three primary contacts, it does not progress at the same rate as the Post (no AMP/GluNH2) state. The glutamate contacts differ significantly in the partially undocked Post (no AMP/GluNH2) state. As the charging glutamate leaves the active site, water molecules come between it and the active site residues, and all three original salt bridges are broken. As the CCA end pivots around the phosphoester linkage between nucleotides 70 and 71, the glutamate forms new contacts to Arg47 and positively charged residues Lys243, Lys246, and Arg247 in the KMSK loop. During this simulation, the sidechain of the charging glutamate makes transient salt bridges with all four of these residues. The longest-lived salt bridges are made between the glutamate and Lys246.
Figure 7.
Mean nonbonded interaction energies between the charging amino acid (glutamate) and GluRS in the Pre-transfer, Post (AMP), and Post (no AMP/GluNH2) states. Labeled residues have interaction energies greater than 25 kcal/mol. These values are averaged over the last 5 ns of the 20 ns trajectories.
Free energies of binding predict tRNA dissociation irrespective of AMP
Free energies of binding
The technique of Molecular Mechanics–Poisson-Boltzmann Surface Area (MM-PBSA) is used to calculate both the average free energy of complex formation and binding of ligands [50, 51], 〈ΔGbinding〉 = 〈GAB〉 − (〈GA〉 + (GB〉) where AB denotes a macromolecular complex formed from components A and B. This can be reformulated so that the free energy difference is expressed as a sum of terms that can be calculated from MD trajectories: 〈ΔGbinding〉 = 〈ΔEVdW + ΔEelec + ΔGpolar + ΔGnonpolar〉 − T〈ΔS〉. The 〈·〉 indicate time averages over the last 5 ns of each 20 ns trajectory. The first two terms are used to measure the van der Waals and electrostatic interaction energies between the two molecules and are derived from molecular mechanics. Gpolar, the polar solvation energy, is the energy required to move the system from a dielectric of εin = 1.00 to εout = 78.54. The polar solvation energy is efficiently calculated using a Poisson-Boltzmann implicit continuum solvent model [52]. The nonpolar solvation energy, Gnonpolar, is the energy required to create a cavity in the solvent for a given system and is proportional to the solvent accessible surface area. The last term refers to the change in entropy on complex formation and was calculated from the MD simulations using Schlitter’s formula [53]. Since the free energies are calculated using the complex and system components from the same trajectory, free energies associated with any conformational changes in the isolated components are neglected in this analysis. Also, as MM-PBSA is not generally successful at reproducing absolute binding free energy values accurately, the focus here is on the trends in binding free energies between different system states.
The network analysis indicated that the loss of correlation is comparable in the Post (AMP) or Post (no AMP) states. In order to address the question: which is energetically more feasible to occur first - the dissociation of AMP (GluRS·Glu-tRNAGlu·AMP ⇌ GluRS·Glu-tRNAGlu + AMP) or charged tRNA (GluRS·Glu-tRNAGlu·AMP ⇌ GluRS·AMP + Glu-tRNAGlu) - two sets of calculations were performed and reported in Tables 3 and 4, respectively. In the first, the free energy of binding was estimated for the small molecule substrate/products (AMP or H-AMP) in the Pre-transfer, Post (H-AMP), and Post (AMP) states, and in the second, the free energy of binding was estimated for charged tRNA to GluRS in the presence and absence of the appropriate small molecule. The experimentally measured free energy of binding of tRNA to homologous class I aaRS enzymes in the absence of any cofactors is −8 to −9 kcal/mol [54, 4]. The interior dielectric was chosen to make the Pre-transfer have a comparable value (see Supplementary Table 7 for the detailed calculation).
Table 3.
MM-PBSA free energy estimates in kcal/mol for the adenylate/(H-)AMP substrate binding GluRS·tRNAGlu. The 95% confidence interval range for each value is ± the number shown below in parentheses. Standard deviations for the ΔGbinding were all 6–7 kcal/mol.
| Small Molecule Substrate Free Energies of Binding | |||
|---|---|---|---|
| Pre-transfer | Post (H-AMP) | Post (AMP) | |
| 〈ΔEVdW〉 | −41.34 (0.20) |
−41.79 (0.13) |
−36.77 (0.13) |
| 〈ΔEcoulomb + ΔGpolar〉 | −3.90 (0.38) |
39.47 (0.32) |
38.47 (0.29) |
| 〈ΔGnonpolar〉 | −5.81 (0.00) |
−4.85 (0.00) |
−4.72 (0.00) |
| −TΔS | 11.70 | 8.40 | 8.40 |
| 〈ΔGbinding〉 | −39.15 (0.29) |
1.23 (0.31) |
5.38 (0.27) |
Table 4.
MM-PBSA free energy differences in kcal/mol for tRNAGlu binding to GluRS (with small molecule substrate/products). The 95% confidence interval range for each value is ± the number shown below in parentheses. Standard deviations for the ΔGbinding were all 14–21 kcal/mol
| tRNA Free Energies of Binding | |||||
|---|---|---|---|---|---|
| Pre-transfer | Post (H-AMP) | Post (AMP) | Post (no AMP) | Post (no AMP/GluNH2) | |
| 〈ΔEVdW〉 | −244.49 (0.34) |
−234.03 (0.45) |
−209.52 (0.31) |
−215.70 (0.50) |
−206.53 (0.35) |
| 〈ΔEcoulomb + ΔGpolar〉 | 228.57 (0.74) |
213.26 (1.07) |
214.16 (0.95) |
213.06 (1.09) |
204.70 (0.79) |
| 〈ΔGnonpolar〉 | −29.07 (0.02) |
−30.74 (0.04) |
−28.20 (0.03) |
−28.05 (0.03) |
−26.34 (0.03) |
| −TΔS | 36.20 | 37.10 | 38.60 | 37.20 | 37.40 |
| 〈ΔGbinding〉 | −8.79 (0.65) |
−14.41 (0.91) |
15.04 (0.84) |
6.51 (0.85) |
9.23 (0.65) |
Small molecule product undocking
The small molecule product binding free energies indicate that H-AMP/AMP can dissociate from the GluRS·Glu-tRNAGlu complex after the reaction (See Table 3). The adenylate binds tightly to the GluRS·tRNAGlu component while both H-AMP and AMP have a significantly reduced affinity for complex formation. This results from a change in the ΔGpolar + ΔEcoulomb because of the transfer of the glutamate moiety to the tRNA. The charging glutamate backbone forms a salt bridge with Glu41 while its sidechain carboxylate forms two salt bridges with Arg5 and Arg205, accounting for the strong binding of the adenylate to active site residues. In comparison, the AMP moiety forms relatively weaker hydrogen bonds with protein residues in the active site (see Figure 3). In addition, the transfer of glutamate to tRNA reduces the surface area (also leading to smaller ΔGnonpolar) as well as the charge distribution on the interaction surface of the complex. Conformational and charge differences on the small molecule product account for the AMP being more likely to exit the active site than H-AMP. The 5 kcal/mol difference in the ΔEvdW between the Post (H-AMP) and Post (AMP) states is due to the AMP shifting in the pocket and making contact with fewer residues, particularly His15. The difference in charge from H-AMP to AMP increases the contribution of the ΔGpolar term. The proton transfer causes the charge of H-AMP/AMP to change from −1 to −2 while the charge of the GluRS:Glu-tRNAGlu complex remains −71. This results in a larger coulombic repulsion after proton transfer which compensates for the change in ΔGpolar.
tRNA dissociation
The overall trends in binding energies between tRNA and GluRS in the various system states seen in Table 4 are similar to those exhibited by the reduction in correlation measured by the network analysis. The Pre-transfer and Post (H-AMP) states have strongly negative values that indicate tighter binding than in the other post-transfer states. The main contribution to the decrease of the binding free energy comes from the change in the van der Waals interaction, indicating a decrease in the contacts between the tRNA and the GluRS. Notably, this value is the least attractive in the Post (AMP) and Post (no AMP/GluNH2) states, both of which were identified as having the weakest interface contacts by both the network and local energetics analyses. With the exception of the Post (H-AMP) state, our results are in contradiction to the experiments on CysRS [4] that report a larger binding affinity to the charged tRNACys than to the uncharged tRNA. The results indicate that the charged tRNA can dissociate from GluRS in the presence or absence of AMP. Given that the AMP can also undock on transfer of the charging amino acid to tRNA, the free energy calculations suggest that the undocking of the products from GluRS can proceed along competitive thermodynamic pathways, which could lead to kinetic partitioning. The only 20 ns simulation in which significant undocking of the tRNA acceptor stem from GluRS was actually observed was in the Post (no AMP/GluNH2) state.
Free energies of binding as a function of time
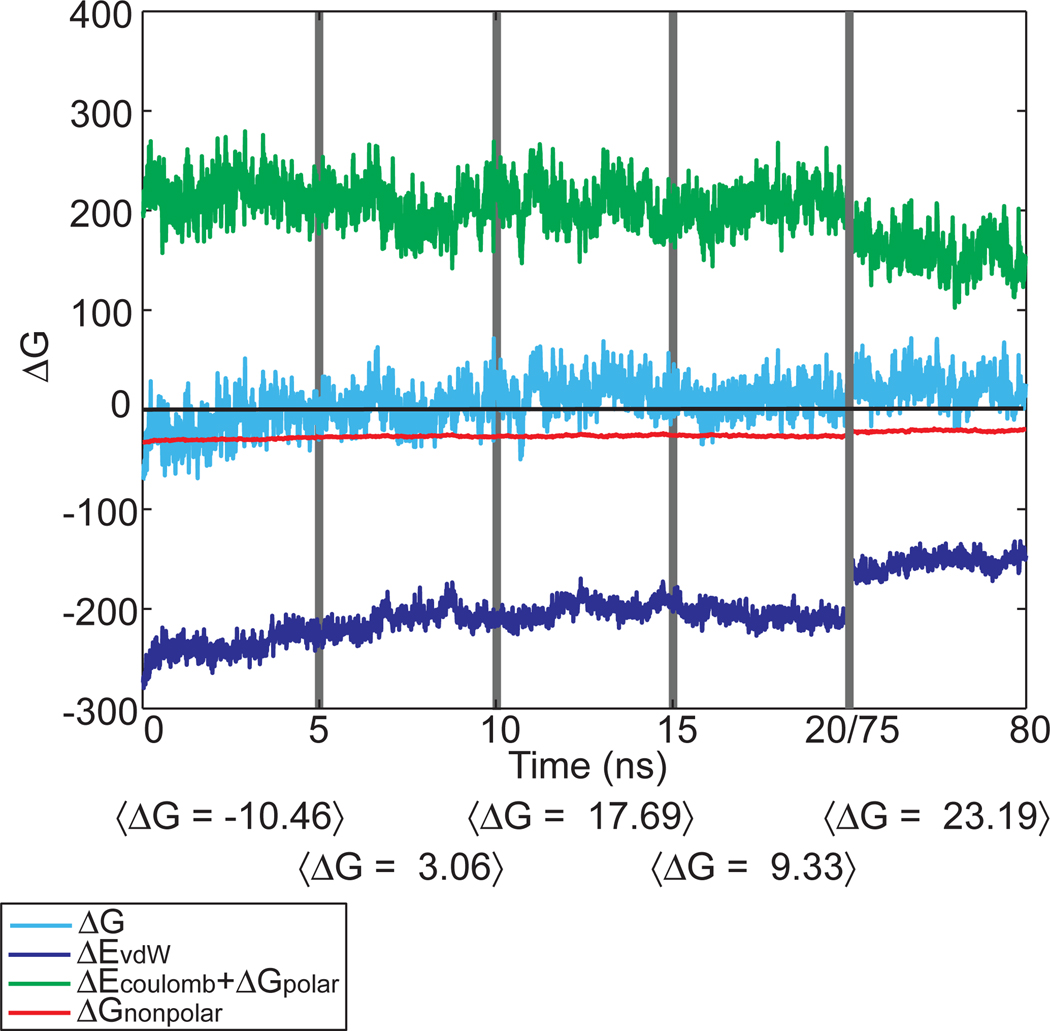
The free energies of binding were calculated from the last 5 ns of the 20 ns simulations to capture the short time binding characteristic of each system state that can be easily accessed computationally. The total ΔG(t) as a function of time is shown in Figure 8 for the first 20 ns and the last 5 ns of the 80 ns Post (no AMP/GluNH2) state simulation. The average free energy over the first 5 ns of this simulation is roughly equivalent to the final 5 ns of the Pre-transfer state, indicating that the Post (no AMP/GluNH2) state is still inhabiting the ensemble of states similar to the Pre-transfer state. However, the free energy of binding rapidly rises as the system equilibrates with the modeled active site changes, peaking at 17.69 kcal/mol between 10 and 15 ns. During this time, the charging amino acid exits the active site and the CCA hairpin begins to undock (see Supplementary Figure 13). The free energy becomes less repulsive during the next 5 ns as the charging amino acid makes surface interactions with the KMSK loop, stabilizing the CCA hairpin. During the last 5 ns of the 80 ns run, each of the individual components has moved closer to zero as expected. As the interface opens, ΔEvdW and ΔGnonpolar weaken because the surface of each molecule becomes more exposed to solvent. There may be increasing error in the electrostatic components related to the treatment of the bulk dielectric as water enters the interface and creates a cleft between the two macromolecules, but the generally positive free energies indicate that Glu-tRNAGlu is dissociating from GluRS.
Figure 8.
ΔG(t) averaged over 5 ns windows from the 80 ns Post (no AMP/GluNH2) simulation for charged tRNA dissociation from GluRS. The components of the total ΔG(t) sampled every 40 ps are given over the first 20 ns and the last 5 ns of the 80 ns simulation. The entropy averaged over 5 ns windows is included in ΔG(t) as well as 〈ΔG〉 computed over 5 ns windows.
Exit strategies for dissociation of aa-tRNA
Figure 9 depicts several of the representative states as a series of events leading to tRNA dissociation. The first event is aminoacylation, the transition between the pre- and post-transfer states. The pre-transfer state (b) can be well approximated from the crystal structure containing the analog, and the Post (H-AMP) state (c) previously postulated to exist [8] following transfer of the amino acid moiety is also in agreement with results from quantum chemistry calculations on a model system (data not shown). Our network analysis and free energy calculations indicate that the Post (H-AMP) state complex is stable and has dynamic and energetic properties similar to the Pre-transfer state complex. Transfer of the 2′-hydroxyl proton to AMP causes significant conformational changes in the active site with the HIGH motif providing stability for the H-AMP. The formation of hydrogen bonds between H-AMP and both His15 and Ser9 reduces the pKa of H-AMP to 5.51, which sets up the next transition with the transfer of the proton to a general base.
Figure 9.
Proposed tRNA exit strategies. The various pathways can be constructed from the states considered in this study: each begins after the tRNA has associated with GluRS and the adenylate has been formed (a), and ends with the formation of a proposed migration complex with EF-Tu (g). Dotted arrows imply additional dissociation events that could occur.
While the HIGH motif would seemingly be a good candidate for this transfer, pKa calculations show that the histidine has a strong propensity for remaining neutral. The only other general base in the vicinity is water, and the Post (AMP) state (d) models this. In the Post (AMP) state, both the AMP and the charged tRNA can dissociate along competitive pathways. However, the CCA hairpin, which has been hypothesized to dissociate prior to the rest of the tRNA, remains strongly bound to the active site residues during the 20 ns simulation.
The complete release of the charged CCA hairpin is prevented by contacts between the α-ammonium group of the charging amino acid and the conserved residue Glu41. On the transfer of the charging amino acid to the tRNA, the pKa of the α–ammonium group decreases to 7.23, while the pKa of Glu41 increases to 8.06, indicating that the proton could be transferred from the charging amino acid to Glu41, reducing the interactions between the two (Post (AMP/GluNH2/E41h) state) (e). The removal of AMP creates a channel for water molecules to interact with Glu41COOH and solvate contacts between the charging amino acid and other active site residues. The proton can be transferred subsequently to the water molecules entering through the vacant AMP binding site.
The Post (no AMP) state models the complex in the case where AMP has undocked before tRNA. The affinity of GluRS for the charged tRNA is similar to its affinity in the presence of AMP. The CCA hairpin becomes highly solvated by additional water molecules entering through the mobile KMSK loop and vacant AMP binding site. This helps reduce interactions between the charged CCA hairpin of the tRNA molecule and GluRS. However, the rocking motion that assists in weakening the contacts between the acceptor stem and the CP1 insertion also causes a conformational twist that strengthens the interactions between GluRS and the tRNA core. This results in a energetic trade-off which causes the Post (no AMP) and the Post (AMP) states to have comparable shortest path lengths.
Removal of the proton and AMP in the Post (no AMP/GluNH2) state (f) allows the CCA hairpin and charged amino acid to move out of the active site within 20 ns (Figure 7). The anticodon loop and D arm remain bound, while fluctuations in the acceptor stem increase.
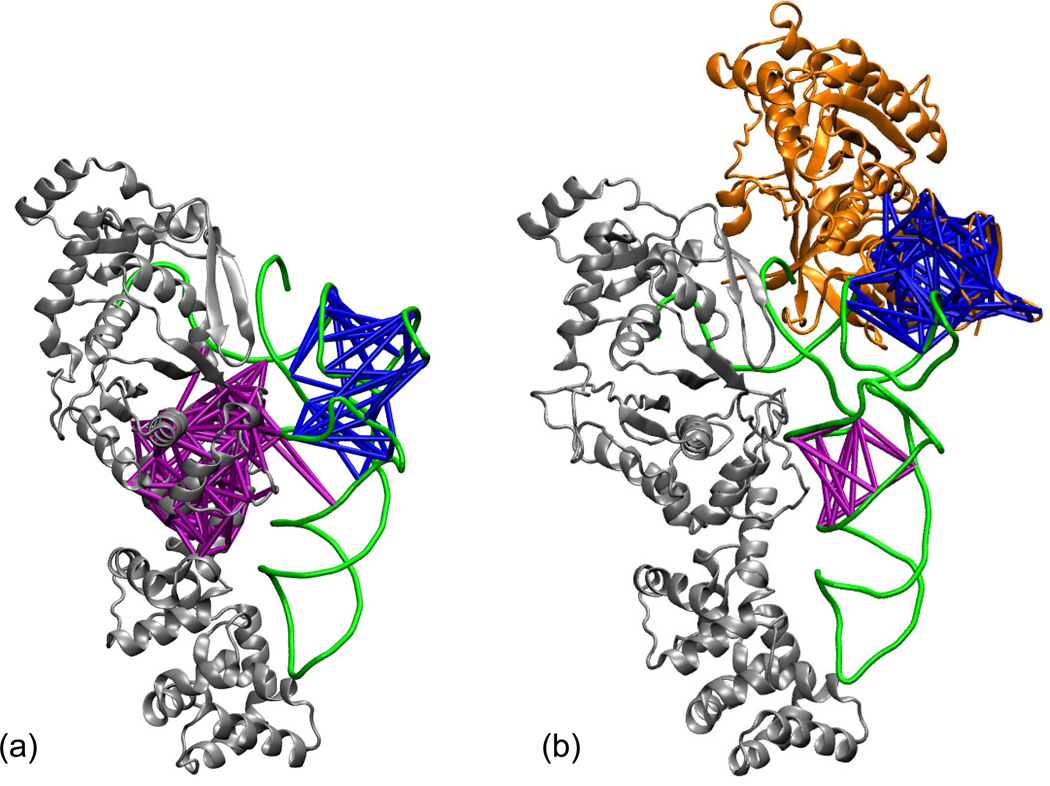
The final state in Figure 9 is the proposed migration complex consisting of GluRS·tRNAGlu·EF-Tu·GTP. The structural details of the docking of EF-Tu to the GluRS·tRNAGlu complex are unknown, but recent studies have suggested that EF-Tu can bind the tRNA while it is still bound to the aaRS [5, 4]. The steric clashes reported upon docking of the crystal structures [4] are eliminated by selecting configurations from the MD simulations in which the CP1 has rocked away from EF-Tu. Because the binding affinity of tRNA for EF-Tu is 300-fold higher than the aaRS [4], this might also serve as a powerful stimulant for tRNA release. We modeled the initial migration complex using equilibrated EF-Tu from T. thermophilus (from [55]) and the Post (no AMP) state complex after 20 ns. After 20 ns of equilibration, the dynamical network and community partitioning in the migration complex were calculated from the last 5 ns and compared to the network from the Post (no AMP) state simulation. Shown in Figure 10 are network communities comprised of the tRNA core and T arm. In the left panel showing the GluRS·tRNAGlu in the absence of EF-Tu, four nucleotides in the tRNA core remain strongly correlated with the GluRS. However, in the presence of EF-Tu (right panel), the tRNA core gradually loses its correlations with the GluRS while the T arm becomes strongly correlated with the EF-Tu. While this simulated model of the migration complex is only one representative of the ensemble of partially docked states, it indicates that EF-Tu can have a strong stimulating effect on the release of the tRNA from the aaRS. Subsequent deprotonation of the α–ammonium group, which is still bound to conserved active site residues, results in release of the CCA hairpin towards the binding site in EF-Tu (see Supplementary Figure 17). Open questions remain with regards to the initial approach and binding of EF-Tu to the GluRS:tRNA complex. When and how EF-Tu binds could affect interactions between GluRS and tRNA both before and after aminoacylation.
Figure 10.
Community analysis of the (a) Post (no AMP) complex and (b) the GluRS·tRNAGlu·EF-Tu·GTP complex formed from the Post (no AMP) system. Two communities are displayed for each complex: one containing nucleotides in the the D stem (purple) and one containing the base of the T stem (blue). In the Post (no AMP) state, a community with four D stem nucleotides includes amino acids from the C-terminal half of the GluRS Rossman fold, and the T stem community contains the whole T arm. When EF-Tu is placed so that it binds the tRNA, the T stem community merges with the third domain of EF-Tu, and the D stem peels away from GluRS, resulting in a community containing only nucleotides. Dynamical networks were created from the final 5 ns of 20 ns trajectories.
Conclusion
Experiments reveal that the complete dissociation of the charged tRNA from the class I aaRSs takes place in the millisecond-second timescale and is stimulated by the presence of EF-Tu [4, 5], but our calculations indicate that there can be initial signs of tRNA release even at timescales of tens of nanoseconds. An important factor affecting the release of charged tRNA is the protonation state of residues in the active site of the aaRS. Results from network analysis, local nonbonded interaction energies, and free energies of binding all show that the Pre-transfer and Post (H-AMP) states form stronger GluRS·tRNA interactions than all other post-transfer states regardless of the presence or absence of AMP. The pKa calculations suggest that one of the α-ammonium hydrogens on the charging glutamate can transfer to the Glu41 sidechain carboxylate while in the active site. Glu41 is predicted to be a nearly universal handle that acts as a general base to facilitate tRNA release from the active site upon transfer of the amino acid. If Glu41 becomes solvated, either through the removal of AMP or the dissociation of the CCA hairpin, it would return to its charged state. Similarly, the pKa measured at the beginning and end of the Post (no AMP/GluNH2) simulation shows that once the CCA end leaves the active site, breaking contacts with Glu41, the α-amino group on the charging glutamate can become reprotonated. From binding free energies it appears that the pathways for AMP and tRNA dissociation are thermodynamically competitive, but once AMP has left the active site and the α-ammonium group is deprotonated, dissociation of the CCA hairpin can occur on the nanosecond timescale. Binding of EF-Tu to the GluRS·tRNAGlu can stimulate tRNA release. Further studies will be needed to determine the molecular details of the migration of the tRNA to the EF-Tu and whether its complete dissociation from the aaRS occurs prior to or during delivery of the tRNA to the ribosome A-site.
Methods
Bioinformatics
Evolutionary analyses of the structures and sequences of the Class I aaRS catalytic domains have already been conducted [56, 57] and are in good agreement with phylogenetic analyses of the complete sequences [2]. The evolutionary analyses in this study were performed to measure the conservation of the residues and nucleotides important for binding either within the active site or along the GluRS·tRNAGlu interface. Because the ACB domain is nonhomologous between the bacterial version of GluRS and the archaeal and eukaryotic versions, the evolutionary profile was limited to exclusively bacterial sequences. Further, because the bacterial GluRS has diverged into discriminating and non-discriminating versions, the set of sequences used to build the evolutionary profile was filtered to include only discriminating GluRS sequences containing the characteristic residue Arg358 [18].
D-GluRS
The GluRS sequence from T. thermophilus was used to perform a BLAST search [58] over the NCBI non-redundant database [59] with an E-value cutoff of 10−5. The alignment of all bacterial GluRS sequences was performed with ClustalW [60] and manually improved (particularly in the highly divergent 4HJ). Only sequences with arginine or the similar lysine in corresponding positions of the characteristic residue Arg358 in the discriminating GluRS from T. thermophilus were retained. The evolutionary profile was prepared by applying SeqQR [57] with a 50% sequence identity cutoff, yielding a set of 23 sequences which are listed in Supplementary Table 9, representing 12 of the major bacterial phyla.
The SeqQR algorithm is used to determine the set of the most linearly independent sequences to form a statistically well-balanced profile that represents the phylogenetic diversity while minimizing the number of sequences.
tRNA
The sequences for bacterial tRNAGlu (which have CUC and UUC anticodons) were downloaded from the tRNA Compilation 2000 [61] and the Integrated Microbial Genome database at the Joint Genome Institute [62]. The tRNA sequence alignment was performed using ClustalW with manual improvement. SeqQR was used with a sequence identity cutoff of 85%, yielding 56 sequences in the final evolutionary profile. The MultiSeq plugin [63] in VMD [64] was used to construct and analyze the evolutionary profiles. The organisms represented in the evolutionary profiles are provided in Supplementary Table 9.
Class I alignment
The Class I structural alignment was prepared by aligning the catalytic domains of several aaRS·substrate structures in MultiSeq (1N78 [7], 1QTQ [34], 1F7U [35], 1LI7 [36], 1FFY [37], 1H3N [38], 2CT8 [39], 1GAX [40], 2AKE [41], and 1H3E [42]). The triad residues were located by their proximity and orientation relative to the α-amino group on the adenylate analog or charging amino acid. The individual sequence alignments were prepared by downloading all Class I aaRS sequences from the Swiss-Prot database [65] and grouping them by domain of life and specificity. These were aligned with ClustalW and manually improved. Evolutionary profiles were created using SeQR with a sequence identity cutoff of 70%, which yielded statistically well-balanced sets for both domain/specifities with many sequences (bacterial) and few sequences (eukaryotic). The sequence of the aaRS structure in a given specificity was aligned to each evolutionary profile to locate the columns containing the triad residues. In columns containing multiple residue types, the residues are listed in Figure 4b in order from those appearing in the most sequences to the least and colored by their percent sequence identity (blue = most conserved, white = 50% conserved, and red = minimally conserved). Columns containing more than four residues in the evolutionary profile are considered unconserved.
Molecular Modeling
Pre-transfer complex
The Pre-transfer system state was modeled from the GluRS·tRNAGlu·analog crystal structure [Protein Data Bank (PDB) code 1N78] [7], which contains the T. thermophilus GluRS, transcribed tRNAGlu, and the adenylate analog glutamol-AMP. A carbonyl oxygen was added to the glutamol-AMP in the crystal structure using the Psfgen plugin in VMD [64], modifying it to glutamyl-AMP. Histidine protonation states were checked with the Whatif server [66] and verified through visual inspection.
Post-transfer complexes
All of the post-transfer complexes were modeled from the crystal structure. The backbone carbonyl carbon was detached from the AMP and reconnected to the 2′ ribose oxygen of A76 to create Glu-tRNAGlu. This new bond was 4.00 Å in the model and relaxed to 1.35 Å during minimization. In the Post (H-AMP) state, a proton was attached to the AMP phosphate oxygen that previously had been bonded to the charged glutamate. The capture of the proton by water was modeled in the Post (AMP) state by removing the 2′-OH proton from the system and compensating for the charge difference by adding a monovalent ion to the bulk solution. To test the validity of the Post (H-AMP) and Post (AMP) states, an intermediate state was modeled from the Post (H-AMP) system after 5 ns of equilibration wherein the proton on the H-AMP was removed and a potassium ion was added to bulk solution. This system was then equilibrated for 20 ns and showed that active site residues relaxed to positions similar to those in the modeled Post (AMP) state (data not shown).
In the Post (AMP/H15h) state, His15 in the HIGH motif was protonated (residue type HSP in the CHARMM27 force field). In the Post (AMP/GluNH2/E41h) state, the α-ammonium group was deprotonated and Glu41 was protonated using the glutamic acid patch in CHARMM. In the Post (no AMP/GluNH2) state, a proton from the α-ammonium group on the charged glutamate was removed.
Migration complex
The migration complex was assembled from proteins and tRNA in two different simulations. The EF-Tu structure was taken from an EF-Tu·GTP·Cys-tRNACys trajectory in [55], and the bound GluRS and Glu-tRNAGlu structures were taken from the Post (no AMP) trajectory. The structures in the first frames of each simulation were overlapped based on a structural alignment of their respective tRNA backbones, excluding the anticodon arms. Then each trajectory was aligned back to its first frame based on the same tRNA backbone selection. A pair of trajectory frames was selected in which GluRS and EF-Tu had no steric clashes. Structures from these frames were merged to create the final migration complex. Water and ions within 10 Å of GluRS·tRNAGlu and EF-Tu were included with the migration complex unless they produced steric clashes with molecules in the system. Finally, the remaining solvent box was generated and neutralizing K+ atoms were added.
Another system was created based on the migration complex after 10 ns of simulation. A proton from the α-ammonium group on the charged glutamate was removed and one of the bulk water molecules was replaced by a K+ atom to maintain charge neutrality. After the next 20 ns, this group was reprotonated, and a K+ atom was removed from the system.
Molecular dynamics
System setup
Psfgen was used to add hydrogen atoms to the molecules. Each system state was neutralized by placing magnesium and potassium ions with ionize 3, a program that calculates the Coulombic interaction energy for the placement of an ion on a uniform grid. Each ion is added to the system at the energetic minimum, and the process is repeated until all ions have been placed. The Mg2+ placement protocol developed previously was used to place three Mg2+ on the primary solvation shell of the tRNA (at 2 A) and 14 Mg2+ and 44 K+ at 6.5 Å from the complex in its ionic cloud [55]. For states with changes in charge within the solute, the number of neutralizing K+ ions was modified accordingly. The Mg2+ ion primary solvation shells were filled with a maximum of six TIP3 water molecules [67].
The concentration and motion of Mg2+ ions have powerful effects on nucleic acid structure and dynamics (for a review see [68]). As demonstrated previously with EF-Tu·Cys-tRNACys studies [55], Mg2+ ions disrupt RNA backbone conformations leading to structural fluctuations when they move in and out of tRNA hairpin loops. However, by interacting with the RNA deep groove, cations can stabilize helices by shielding the repulsive electrostatic interactions between phosphates on either side of the groove. An RNA helix with Mg2+ ions in its deep groove becomes more rigid and less able to conform to an induced fit at a protein binding interface. In this study, several replicate simulations were performed for each system state, and the Mg2+ ion distribution after 5 ns was assessed through ion occupancy and residency calculations to determine if there were too many (> 8) or too few (< 7) resident Mg2+ atoms associated with the tRNA. This is a check to ensure that the initial movement of Mg2+ ions did not lead to unusual structural fluctuations. The residency locations were determined by tracking the positions of the Mg2+ ions over time. Frames from the last 16 ns of each simulation (40 ps intervals) were aligned by the tRNAGlu backbone. Mg2+ ions remaining within 3 Å of a starting position for 4 ns were classified as resident ions. Occupancy maps were calculated using the VolMap plugin to VMD. The occupancy was calculated at a 1 Å resolution and averaged over all frames.
To ensure full solvation of the active site and GluRS·tRNAGlu binding interface, Solvate 1.04 was used with two gaussians to add two layers of water molecules to the system. This resulted in the placement of eight additional water molecules in the active site of the Pre-transfer system. The Solvate 1.2 plugin in VMD was used to place the bulk water, reaching a system size of approximately 120 × 80 × 120 Å and an average of 110,436 atoms.
The NAMD2 software [69] and CHARMM27 force field [70] were used to perform the MD simulations. The simulations were calculated in the NPT ensemble with periodic boundary conditions; the Langevin piston method [71] was applied to maintain pressure at 1.01325 bar and Langevin dynamics were used to maintain temperature at 298.15 K. Electrostatics were efficiently treated with the Particle Mesh Ewald summation [72]. A multiple time-stepping algorithm was used to evaluate bonded interactions at 1 fs, vdW interactions every 2 fs, and electrostatic forces every 4 fs. A cutoff of 12 Å and a switching distance of 10 Å were used for the vdW force calculations.
Minimization
Minimization was carried out according to the protocol previously established for protein·RNA complexes [55] to ensure that water molecules were associated with the macromolecules prior to allowing the macromolecules to move. The system was minimized in four stages: 2,000 steps with heavy atoms fixed, 3,000 steps with heavy atoms fixed excluding water, 5,000 steps with macromolecule backbone atoms fixed, and 20,000 steps of unconstrained minimization.
Equilibration
Equilibration was conducted using a temperature jump protocol similar to that developed by Auffinger and Westhof [73]. This allowed cations to enter the deep groove of the negatively charged nucleic acid by systematically raising the temperature and freeing harmonic constraints in five steps [55]. These steps were: a temperature of 100 K with heavy atoms and ions constrained (25,000 fs), 200 K with hydrogens and waters freed (25,000 fs), 250 K with all hydrogens, waters, ions, and small molecules unconstrained (25,000 fs), 250 K and the protein and nucleic acid backbones constrained (25,000 fs), and 298.15 K with no constraints (19.9 ns). The harmonic constraints in all steps were set to 1kcal mol−1 Å2.
Parameters
The parameters for glutamyl-AMP were derived by analogy to AMP and glutamate in the CHARMM27 force field. The parameters for the ester bond connecting the charged glutamate to the 2′ hydroxyl group of A76 were taken from those developed previously from the ester bond in fatty acids [55]. The H-AMP parameters were derived by analogy to pyrophosphate, protonated pyrophosphate, and AMP. The parameters for the deprotonated α-amino group on the charging glutamate were the same as those used previously in the docking of Cys-tRNACys to EF-Tu [55].
RMSD calculations
The average RMSD for the protein and nucleic acid was calculated separately using the Cα and phosphorous atoms at 40 ps intervals over the 20 ns simulations. The Cα and phosphorous atoms of the complex backbone in the crystal structure were used as the reference for the standard Kabsch method [74] implemented in VMD.
pKa calculations
Protonation states for titratable protein and ligand groups in the active site were assigned using PROPKA 2.0 [75]. PROPKA uses a fast heuristic method to compute the pKa perturbations of the titratable sidechains due to desolvation, hydrogen bonding, and charge-charge interactions in the environment of the protein. A well equilibrated frame at the end of 20 ns of simulation was used in the pKa calculation for each of the system states. The bound magnesium ions in the primary solvation shell of the tRNA were found to have minimal effect on the pKa of the protein sidechains and were therefore not included in the calculation. Input files for PROPKA were prepared so that the bulk pKa values were set to 6.9 for the AMP/H-AMP phosphate and 9.47 for the amino acid moiety on the AMP adenylate and A76.
Correlation calculations
To calculate correlations within and between GluRS residues and tRNAGlu nucleotides, the backbone atoms from the last 5 ns (10,000 frames) of each 20 ns trajectory were used to compute the normalized covariance (correlation) matrix with the program Carma [76]. The elements of this matrix are the correlations in motion of the residues and nucleotides. Values are close to one if two residues/nucleotides are moving in the same direction in a majority of frames (correlation), approaching negative one if they move in opposite directions (anticorrelation), and near zero if their motion is uncorrelated.
Dynamical network construction and community analysis
A weighted GluRS·tRNAGlu network was constructed for each system state based on dynamical contacts as outlined previously [25]. Each amino acid, nucleotide, or small molecule substrate/product is represented by a node, and edges between nodes are defined by dynamical contacts. A contact is present when the heavy atoms of two residues/nucleotides are within 4.5 Å of each other in 75% of the trajectory frames analyzed (50 ps intervals from the last 5 ns of each simulation). Covalently bonded neighbors were neglected. The edge weights wij were calculated from the correlation values (Cij) which define the probability of information transfer across each edge: wij = −log(|Cij|).
Shortest paths through the network represent the dominant mode of communication between two nodes. Path lengths are the sum of the edge weights between consecutive nodes (k,l) along the path, Dij = Σk,lWkl, and the shortest paths were determined using the Floyd-Warshall algorithm [77, 78]. Betweenness of an edge is defined as the number of shortest paths traversing that edge, and high betweenness values identify contacts that are important for communication across the complex.
The Girvan-Newman algorithm was used to identify tightly interconnected modules or communities of nodes [79]. This algorithm iteratively removes edges with the highest betweenness values and recalculates the betweenness of all remaining edges to determine the optimal community structure as measured by the modularity score. This score is a measure of the difference between the probability of inter- and intracommunity edges and is maximized during the calculation.
Local energetics analysis
The nonbonded energetics were calculated using the NAMDEnergy plugin in VMD with the CHARMM27 force field. The vdW and electrostatics interaction energies were calculated between each protein residue and either the tRNA (minus any atoms from the charging glutamate) or the charging amino acid. Energies were calculated at 10 ps intervals and averaged over the last 5 ns of each trajectory. The switching and cutoff distances of 18 Å and 21 Å ensured that electrostatic energy was calculated relative to both members of a nucleic acid base pair. To determine if the residues/nucleotides with strong interaction energies were conserved, a mask was calculated based on the multiple sequence alignment of each molecule (see Bioin-formatics in Methods). The percent identity over the bacterial sequences was determined based on the T. thermophilus sequence. The average interaction energies per residue were scaled by percent identity.
MM-PBSA Calculation of Free Energies
Molecular mechanics-Poisson-Boltzmann Surface Area (MM-PBSA) is a method for calculating binding free energies from a simple thermodynamic cycle [80, 50, 51, 81, 82, 83, 55, 84]. The difference in energy between the complex and two components or unbound docking partners is calculated from a single trajectory of the complex according to 〈ΔGbinding〉 = 〈ΔEVdW + ΔEcoul + ΔGnonpolar + ΔGpolar〉−T〈ΔS〉. Each of the values in the averaged terms is the difference for either a small molecule substrate/product undocking from the protein·tRNA complex , or the tRNA dissociating from the protein·substrate complex . The three bound Mg2+ ions placed during system setup and the water molecules in their first solvation shell were included with the tRNA, all of which bind the tRNA core and are stable elements of the nucleic acid structure. The values were averaged over 500 frames from the final 5 ns of each 20 ns simulation (10,000 frames for the entropy calculation). Calculations for the individual components were performed over the same trajectory as the complex [51, 55], which neglects any contributions from changes in conformation that could exist were the individual components simulated separately.
The bonded interactions (bonds, angles, dihedrals, and impropers) cancel in each difference calculation and therefore are not shown. The EVdW term is the sum of all pairwise VdW interaction energies and was calculated using the NamdEnergy plugin in VMD. The Coulombic energy (ΔEcoul), which is the sum of all pairwise interactions in the system scaled by εin, was calculated with Coulomb, a program distributed with APBS [52].
The nonpolar solvation free energy (ΔGnonpolar) is the energetic cost of creating a cavity in the solvent for a given system. Gnonpolar = γSASA + b was calculated for the complex and components with SASA set to the solvent accessible surface area using a solvent radius of 1.4 Å, γ = 0.00542kcal mor−1 Å−2, and b = 0.92kcal/mol. This term was calculated in VMD. The polar solvation (ΔGpolar), the energy required to move the charged solute from a vacuum dielectric (εin = 1.00) to an aqueous dielectric (εout = 78.54), was computed for the complex and individual components. The 1.00 interior dielectric was chosen to scale the final free energy of the Pre-transfer system state to approximately −8 kcal/mol, consistent with experiment (see Results). A temperature of 298.15 K and CHARMM27 radii/charges were used on a grid of 193 × 129 × 193 with automated focussing. The implicit salt concentrations of 117 mM KCl and 38 mM MgCl2 were used to model the explicit concentrations in the system states. Each frame was aligned to the crystal structure based on the Cα and P atoms to limit error from changes in orientation. The box size was based on the crystal structure with 15% padding in each dimension. The grid was centered on the complex for all components. This term was numerically calculated with the Poisson-Boltzmann solver APBS [52].
The entropy terms (ΔS) were estimated from the covariance matrix of atomic position fluctuation using Schlitter’s formula [53, 85, 86], which relates the entropy of a solute molecule to the sum of decoupled simple harmonic oscillators obtained from the principal component modes in the molecular dynamics simulation. The coordinates of the backbone atoms (Cα and phosphorous) in 10,000 frames from the last 5 ns of each trajectory were input to the program Carma [76] to generate and diagonalize the mass-weighted covariance matrix. The resulting eigenvalues were then used to calculate the determinant and substituted into Schlitter’s formula to calculate the entropy of the solute [55]. Because the small molecule substrate/products had only one or two backbone atoms, which is an insufficient number to calculate the covariance matrix, the entropy was estimated using the Sakur-Tetrode equation for dimolecular and unimolecular ideal gases [87].
Supplementary Material
Acknowledgements
The authors thank ZLS group members, particularly Li Li and Elijah Roberts, for many helpful discussions. They also wish to thank Nathan Baker for APBS assistance, Jan Jensen for help with PROPKA 2.0, Susan Martinis for experimental interpretations, and John Stone for VMD graphics suggestions. Funding for ABP, JE, and AS was provided by NSF (MCB04-46227), NSF (MCB08-44670), NSF (PHY08-22613), and NIH Chemical Biology Training Grant (5T32GM070421). Supercomputer and local computing time was provided by NCSA LRAC (MCA03T027) and and NSF CRIF (0541659).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Grubmueller, H. 1996 Solvate 1.0. http://www.mpibpc.gwdg.de/abteilungen/071/solvate/docu.html
Contributor Information
Alexis Black Pyrkosz, Email: ablack2@illinois.edu.
John Eargle, Email: eargle@illinois.edu.
Anurag Sethi, Email: sethi2@illinois.edu.
Zaida Luthey-Schulten, Email: zan@illinois.edu.
References
- 1.Ibba I, S¨oll D. Aminoacyl-tRNA synthesis. Ann. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Woese CR, Olsen GJ, Ibba M, S¨oll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ibba M, Francklyn C, Cusack S, editors. The Aminoacyl-tRNA synthetases. Georgetown, Texas: Landes Bio-science; 2005. [Google Scholar]
- 4.Zhang CM, Perona JJ, Ryu K, Francklyn C, Hou YM. Distinct kinetic mechanisms of the two classes of Aminoacyl-tRNA synthetases. J. Mol. Biol. 2006;361:300–311. doi: 10.1016/j.jmb.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 5.Hausmann CD, Praetorius-Ibba M, Ibba M. An aminoacyl-tRNA synthetase:elongation factor complex for substrate channeling in archaeal translation. Nucl. Acids Res. 2007;35:6094–6102. doi: 10.1093/nar/gkm534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang XL, Otero FJ, Ewalt KL, Liu J, Swairjo MA, Kohrer C, RajBhandary UL, Skene RJ, McRee DE, Schimmel P. Two conformations of a crystalline human tRNA synthetase-tRNA complex: implications for protein synthesis. EMBO J. 2006;25:2919–2929. doi: 10.1038/sj.emboj.7601154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sekine SI, Nureki O, Dubois DY, Bernier S, Chenevert R, Lapointe J, Vassylyev DG, Yokoyama S. ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding. EMBO J. 2003;22:676–688. doi: 10.1093/emboj/cdg053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perona JJ, Rould MA, Steitz TA. Structural basis for transfer RNA aminoacylation by Escherichia coli glutaminyl-tRNA synthetase. Biochemistry. 1993;32:8758–8771. doi: 10.1021/bi00085a006. [DOI] [PubMed] [Google Scholar]
- 9.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 10.Cusack S. Eleven down and nine to go. Nat. Str. Biol. 1995;2:824–831. doi: 10.1038/nsb1095-824. [DOI] [PubMed] [Google Scholar]
- 11.Landes C, Perona JJ, Brunie S, Rould MA, Zelwer C, Steitz TA, Risler JL. A structure-based multiple sequence alignment of all class I aminoacyl-tRNA synthetases. Biochimie. 1995;77:194–203. doi: 10.1016/0300-9084(96)88125-9. [DOI] [PubMed] [Google Scholar]
- 12.O’Donoghue P, Luthey-Schulten Z. Evolutionary profiles derived from the QR factorization of multiple strucutral alignments gives an economy of information. J. Mol. Biol. 2005;346:875–894. doi: 10.1016/j.jmb.2004.11.053. [DOI] [PubMed] [Google Scholar]
- 13.Nureki O, Vassylyev DG, Katayanagi K, Shimizu T, Sekine S, Kigawa T, Miyazawa T, Yokoyama S, Morikawa K. Architectures of class-defining and specific domains of glutamyl-tRNA synthetase. Science. 1995;267:1958–1965. doi: 10.1126/science.7701318. [DOI] [PubMed] [Google Scholar]
- 14.Tateno M, Nureki O, Sekine S, Kaneda K, Go M, Yokoyama S. A three-dimensional structure model of the complex of glutamyl-tRNA synthetase and its cognate tRNA. FEBS Lett. 1995;377:77–81. doi: 10.1016/0014-5793(95)01295-8. [DOI] [PubMed] [Google Scholar]
- 15.Sekine S, Nureki O, Shimada A, Vassylyev DG, Yokoyama S. Structural basis for anticodon recognition by discriminating glutamyl-tRNA synthetase. Nature Struct. Biol. 2001;8:203–206. doi: 10.1038/84927. [DOI] [PubMed] [Google Scholar]
- 16.Siatecka M, Rozek M, Barciszewski J, Mirande M. Modular evolution of the Glx-tRNA synthetase family-rooting of the evolutionary tree between the bacteria and archaea/eukarya branches. Eur. Biophys. J. 1998;256:80–87. doi: 10.1046/j.1432-1327.1998.2560080.x. [DOI] [PubMed] [Google Scholar]
- 17.Dubois DY, Blais SP, Huot JL, Lapointe J. A C-truncated glutamyl-tRNA syn-thetase specific for tRNA(Glu) is stimulated by its free complementary distal domain: mechanistic and evolutionary implications. Biochemistry. 2009;48:6012–6021. doi: 10.1021/bi801690f. [DOI] [PubMed] [Google Scholar]
- 18.Lee J, Hendrickson TL. Divergent anticodon recognition in contrasting glutamyl-tRNA synthetases. J. Mol. Biol. 2004;344:1167–1174. doi: 10.1016/j.jmb.2004.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamour V, Quevillon S, Diriong S, N’Guyen VC, Lipinski M, Mirande M. Evolution of the Glx-tRNA synthetase family: the glutaminyl enzyme as a case of horizontal gene transfer. Proc. Natl. Acad. Sci. USA. 1994;91:8670–8674. doi: 10.1073/pnas.91.18.8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salazar JC, Ahel I, Orellana O, Tumbula-Hansen D, Krieger R, Daniels L, Sll D. Coevolution of an aminoacyl-tRNA synthetase with its tRNA substrates. Proc. Natl. Acad. Sci. USA. 2003;100:13863–13868. doi: 10.1073/pnas.1936123100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skouloubris S, Ribas de Pouplana L, De Reuse H, Hendrickson TL. A noncognate aminoacyl-tRNA synthetase that may resolve a missing link in protein evolution. Proc. Natl. Acad. Sci. USA. 2003;100:11297–11302. doi: 10.1073/pnas.1932482100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang KM, Hendrickson TL. Recognition of tRNAGln by Helicobacter pylori GluRS2-a tRNAGln-specific glutamyl-tRNA synthetase. Nucl. Acids Res. 2009;37:6942–6949. doi: 10.1093/nar/gkp754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Budiman ME, Knaggs MH, Fetrow JS, Alexander RW. Using molecular dynamics to map interaction networks in an aminoacyl-tRNA synthetase. PROTEINS: Structure, Function, and Genetics. 2007;68:670–689. doi: 10.1002/prot.21426. [DOI] [PubMed] [Google Scholar]
- 24.Hansia P, Ghosh A, Vishveshwara S. Ligand dependent intra and inter subunit communication in human tryptophanyl tRNA synthetase as deduced from the dynamics of structure networks. Mol. Biosyst. 2009;5:1860–1872. doi: 10.1039/b903807h. [DOI] [PubMed] [Google Scholar]
- 25.Sethi A, Eargle J, Black A, Luthey-Schulten ZA. Dynamical Networks in tRNA:Protein Complexes. Proc. Natl. Acad. Sci. USA. 2009;106:6620–6625. doi: 10.1073/pnas.0810961106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bharatham N, Bharatham K, Lee Y, Woo Lee K. Molecular dynamics simulation study of valyl-tRNA synthetase with its pre- and post-transfer editing substrates. Biophys. Chem. 2009;143:34–43. doi: 10.1016/j.bpc.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Archontis G, Simonson T, Karplus M. Binding free energies and free energy components from molecular dynamics and Poisson-Boltzmann calculations. Application to amino acid recognition by aspartyl-tRNA synthetase. J. Mol. Biol. 2001;306:307–327. doi: 10.1006/jmbi.2000.4285. [DOI] [PubMed] [Google Scholar]
- 28.Hughes SJ, Tanner JA, Hindley AD, Miller AD, Gould IR. Functional asymmetry in the lysyl-tRNA synthetase explored by molecular dynamics, free energy calculations and experiment. BMC Structural Biology. 2003 doi: 10.1186/1472-6807-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kapustina M, Carter CW. Computational studies of tryptophanyl-tRNA synthetase: activation of ATP by induced-fit. J. Mol. Biol. 2006;362:1159–1180. doi: 10.1016/j.jmb.2006.06.078. [DOI] [PubMed] [Google Scholar]
- 30.Yamasaki S, Nakamura S, Terada T, Shimizu K. Mechanism of the difference in the binding affinity of E. coli tRNAGln to glutaminyl-tRNA synthetase caused by noninterface nucleotides in variable loop. Biophys. J. 2007;92:192–200. doi: 10.1529/biophysj.106.093351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson D, Lazennec C, Plateau P, Simonson T. Probing electrostatic interactions and ligand binding in aspartyl-tRNA synthetase through site-directed mutagenesis and computer simulations. PROTEINS: Structure, Function, and Genetics. 2008;71:1450–1460. doi: 10.1002/prot.21834. [DOI] [PubMed] [Google Scholar]
- 32.Xin Y, Li W, First EA. Stabilization of the Transition State for the Transfer of Tyrosine to tRNATyr by Tyrosyl-tRNA Synthetase. J. Mol. Biol. 2000;303:299–310. doi: 10.1006/jmbi.2000.4126. [DOI] [PubMed] [Google Scholar]
- 33.Ganguly S, Kundu KK. Deprotonation energetics of adenine, 5′-adenosine monophosphate and adenosine triphosphate in water from EMF and spectrophotometic measurements. J. Sol. Chem. 1994;23:1227–1246. [Google Scholar]
- 34.Rath VL, Silvian LF, Beijer B, Sproat BS, Steitz TA. How glutaminyl-tRNA synthetase selects glutamine. Structure. 1998;6:439–449. doi: 10.1016/s0969-2126(98)00046-x. [DOI] [PubMed] [Google Scholar]
- 35.Delagoutte B, Moras D, Cavarelli J. tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrates binding. EMBO J. 2000;19:5599–5610. doi: 10.1093/emboj/19.21.5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newberry KJ, Hou YM, Perona JJ. Structural origins of amino acid selection without editing by cysteinyl-tRNA synthetase. EMBO J. 2002;21:2778–2787. doi: 10.1093/emboj/21.11.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silvian LF, Wang J, Steitz TA. Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin. Science. 1999;285:1074–1077. [PubMed] [Google Scholar]
- 38.Cusack S, Yaremchuk A, Tukalo M. The 2 A crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J. 2000;19:2351–2361. doi: 10.1093/emboj/19.10.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakanishi K, Ogiso Y, Nakama T, Fukai S, Nureki O. Structural basis for anticodon recognition by methionyl-tRNA synthetase. Nat. Struct. Mol. Biol. 2005;12:931–932. doi: 10.1038/nsmb988. [DOI] [PubMed] [Google Scholar]
- 40.Fukai S, Nureki O, Sekine S, Shimada A, Tao J, Vassylyev DG, Yokoyama S. Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNA(Val) and valyl-tRNA synthetase. Cell. 2000;103:793–803. doi: 10.1016/s0092-8674(00)00182-3. [DOI] [PubMed] [Google Scholar]
- 41.Shen N, Guo L, Yang B, Jin Y, Ding J. Structure of human tryptophanyl-tRNA synthetase in complex with tRNATrp reveals the molecular basis of tRNA recognition and specificity. Nucl. Acids Res. 2006;34:3246–3258. doi: 10.1093/nar/gkl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yaremchuk A, Kriklivyi I, Tukalo M, Cusack S. Class I tyrosyl-tRNA synthetase has a class II mode of cognate tRNA recognition. EMBO J. 2002;21:3829–3840. doi: 10.1093/emboj/cdf373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bishop AC, Nomanbhoy TK, Schimmel P. Blocking site-to-site translocation of a misactivated amino acid by mutation of a class I tRNA synthetase. Proc. Natl. Acad. Sci. U.S.A. 2002;99:585–590. doi: 10.1073/pnas.012611299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lincecum TL, Tukalo M, Yaremchuk A, Mursinna RS, Williams AM, Sproat BS, Van Den Eynde W, Link A, Van Calenbergh S, Grøtli M, Martinis SA, Cusack S. Structural and mechanistic basis of pre- and posttransfer editing by leucyl-tRNA synthetase. Mol. Cell. 2003;11:951–963. doi: 10.1016/s1097-2765(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 45.Kobayashi T, Nureki O, Ishitani R, Yaremchuk A, Tukalo M, Cusack S, Sakamoto K, Yokoyama S. Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion. Nat. Struct. Biol. 2003;10:425–432. doi: 10.1038/nsb934. [DOI] [PubMed] [Google Scholar]
- 46.Tsunoda M, Kusakabe Y, Tanaka N, Ohno S, Nakamura M, Senda T, Moriguchi T, Asai N, Sekine M, Yokogawa T, Nishikawa K, Nakamura KT. Structural basis for recognition of cognate tRNA by tyrosyl-tRNA synthetase from three kingdoms. Nucl. Acids Res. 2007;35:4289–4300. doi: 10.1093/nar/gkm417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sekine S, Nureki O, Sakamoto K, Niimi T, Tateno M, Gō M, Kohno T, Brisson A, Lapointe J, Yokoyama S. Major identity determinants in the “Augmented D Helix” of tRNAGlu from Escherichia coli. J. Mol. Biol. 1996;256:685–700. doi: 10.1006/jmbi.1996.0118. [DOI] [PubMed] [Google Scholar]
- 48.Giege R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucl. Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sekine S, Nureki O, Tateno M, Yokoyama S. The identity determinants required for the discrimination between tRNAGlu and tRNAAsp by glutamyl-tRNA synthetase from Escherichia coli. Eur. Biophys. J. 1999;261:354. doi: 10.1046/j.1432-1327.1999.00293.x. [DOI] [PubMed] [Google Scholar]
- 50.Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE., 3rd Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 51.Reyes CM, Kollman PA. Structure and thermodynamics of RNA-protein binding: using molecular dynamics and free energy analyses to calculate the free energies of binding and conformational change. J. Mol. Biol. 2000;297:1145–1158. doi: 10.1006/jmbi.2000.3629. [DOI] [PubMed] [Google Scholar]
- 52.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosys-tems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schlitter J. Estimation of absolute and relative entropies of macromolecules using the covariance matrix. Chem. Phys. Lett. 1993;215:617–621. [Google Scholar]
- 54.Hong KW, Ibba M, Weygand-Durasevic I, Rogers MJ, Thomann HU, Söll D. Transfer RNA-dependent cognate amino acid recognition by an aminoacyl-tRNA synthetase. EMBO J. 1996;15:1983–1991. [PMC free article] [PubMed] [Google Scholar]
- 55.Eargle J, Black AA, Sethi A, Trabuco LG, Luthey-Schulten ZA. Dynamics of Recognition between tRNA and Elongation Factor Tu. J. Mol. Biol. 2008;377:1382–1405. doi: 10.1016/j.jmb.2008.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Donoghue P, Luthey-Schulten Z. On the evolution of structure in the aminocyl-tRNA synthetases. Microbiol. Mol. Bio. Rev. 2003;67:550–573. doi: 10.1128/MMBR.67.4.550-573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sethi A, O’Donoghue P, Luthey-Schulten Z. Evolutionary profiles from the QR factorization of multiple sequence alignments. Proc. Natl. Acad. Sci. USA. 2005;102:4045–4050. doi: 10.1073/pnas.0409715102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucl. Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Geer LY, Helmberg W, Kapustin Y, Kenton DL, Khovayko O, Lipman DJ, Madden TL, Maglott DR, Ostell J, Pruitt KD, Schuler GD, Schriml LM, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Suzek TO, Tatusov R, Tatusova TA, Wagner L, Yaschenko E. Database resources of the National Center for Biotechnology Information. Nucl. Acids Res. 2006;34:173–180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucl. Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Markowitz VM, Korzeniewski F, Palaniappan K, Szeto E, Werner G, Padki A, Zhao X, Dubchak I, Hugenholtz P, Anderson I, Lykidis A, Mavromatis K, Ivanova N, Kyrpides NC. The integrated microbial genomes (IMG) system. Nucl. Acids Res. 2006;34:344–348. doi: 10.1093/nar/gkj024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roberts E, Eargle J, Wright D, Luthey-Schulten Z. MultiSeq: unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics. 2006;7:382. doi: 10.1186/1471-2105-7-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 65.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O’Donovan C, Redaschi N, Yeh LSL. The Universal Protein Resource (UniProt) Nucl. Acids Res. 2005;33:154–159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vriend G. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 67.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 68.Draper DE. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J. Comp. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Foloppe N, MacKerrell AD., Jr All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comp. Chem. 2000;21:86–104. [Google Scholar]
- 71.Feller S, Zhang Y, Pastor R, Brooks B. Constant pressure molecular dynamics simulation: The Langevin piston method. JCP. 1995;103:4613–4621. [Google Scholar]
- 72.Darden T, York D, Pedersen L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;89:10089–10092. [Google Scholar]
- 73.Auffinger P, Westhof E. RNA hydration: three nanoseconds of multiple molecular dynamics simulations of the solvated tRNA(Asp) anticodon hairpin. J. Mol. Biol. 1997;269:326–341. doi: 10.1006/jmbi.1997.1022. [DOI] [PubMed] [Google Scholar]
- 74.Kabsch W. A discussion of the solution for the best rotation to relate two sets of vectors. Acta Cryst. 1978;A34:827–828. [Google Scholar]
- 75.Bas DC, Rogers DM, Jensen JH. Very fast prediction and rationalization of pKa values for protein-ligand complexes. PROTEINS: Structure, Function, and Genetics. 2008;73:765–783. doi: 10.1002/prot.22102. [DOI] [PubMed] [Google Scholar]
- 76.Glykos NM. Software news and updates. Carma: a molecular dynamics analysis program. J. Comp. Chem. 2006;27:1765–1768. doi: 10.1002/jcc.20482. [DOI] [PubMed] [Google Scholar]
- 77.Floyd RW. Algorithm 97: Shortest Path. Comm. ACM. 1962;5:345. [Google Scholar]
- 78.Cormen TH, Leiserson CE, Rivest RL, Stein C, editors. Introduction to Algorithms. 2nd edition. Cambridge: MIT Press; 2001. [Google Scholar]
- 79.Girvan M, Newman M. Community structure in social and biological networks. Proc. Natl. Acad. Sci. USA. 2002;99:7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Froloff N, Windemuth A, Honig B. On the calculation of binding free energies using continuum methods: application to MHC class I protein-peptide interactions. Protein Science. 1997;6:1293–1301. doi: 10.1002/pro.5560060617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rocchia W, Alexov E, Honig B. Extending the Applicability of the Nonlinear Poisson-Boltzmann Equation: Multiple Dielectric Constants and Multivalent Ions. J. of Parallel Computation. 2001;105:6507–6514. [Google Scholar]
- 82.Gohlke H, Kiel C, Case DA. Insights into protein-protein binding by binding free energy calculation and free energy decomposition for the Ras-Raf and Ras-RalGDS complexes. J. Mol. Biol. 2003;330:891–913. doi: 10.1016/s0022-2836(03)00610-7. [DOI] [PubMed] [Google Scholar]
- 83.Pogorelov TV, Autenrieth F, Roberts E, Luthey-Schulten ZA. Cytochrome c(2) Exit Strategy: Dissociation Studies and Evolutionary Implications. J. Phys. Chem. B. 2007;111:618–634. doi: 10.1021/jp064973i. [DOI] [PubMed] [Google Scholar]
- 84.Wong S, Amaro RE, McCammon JA. MM-PBSA captures a key role of intercalating water molecules at a protein:protein interface. J. Chem. Theory Comp. 2009;5:422–429. doi: 10.1021/ct8003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sch¨afer A, Mark AE, van Gunsteren WF. Absolute entropies from molecular dynamics simulation trajectories. J. Chem. Phys. 2000;113:7809–7817. [Google Scholar]
- 86.Anricrioaei I, Karplus M. On the calculation of entropy from covariance matrices of the atomic fluctuations. J. Chem. Phys. 2001;115:6289–6292. [Google Scholar]
- 87.Gupta MC. Statistical Thermodynamics. Sydney: Halsted Press; 1991. [Google Scholar]
- 88.Nissen P, Thirup S, Kjeldgaard M, Nyborg J. The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure. 1999;7:143–156. doi: 10.1016/s0969-2126(99)80021-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.