Abstract
Triclosan is a broad-spectrum antimicrobial compound commonly used in oral hygiene products. Investigation of its activity against Candida albicans showed that triclosan was fungicidal at concentrations of 16 mg/L. However, at subinhibitory concentrations (0.5-2 mg/L), triclosan antagonized the activity of fluconazole. Although triclosan induced CDR1 expression in C. albicans, antagonism was still observed in cdr1Δ and cdr2Δ strains. Triclosan did not affect fluconazole uptake or alter total membrane sterol content, but did induce the expression of FAS1 and FAS2, indicating that its mode of action may involve inhibition of fatty acid synthesis, as it does in prokaryotes. However, FAS2 mutants did not exhibit increased susceptibility to triclosan, and overexpression of both FAS1 and FAS2 alleles did not alter triclosan susceptibility. Unexpectedly, the antagonistic effect was specific for C. albicans under hypha-inducing conditions and was absent in the non-filamentous efg1Δ strain. This antagonism may be due to the membranotropic activity of triclosan and the unique composition of hyphal membranes.
Keywords: triclosan, fluconazole, antagonism, Candida albicans, EFG1, hyphae
Introduction
Triclosan (5-chloro-2-[2,4-dichlorophenoxy]phenol) is a small hydrophobic bisphenolic compound that exhibits a broad spectrum of antimicrobial activity (McDonnell and Russell, 1999). It is widely used in a variety of oral healthcare products, where it has been shown to possess potent anti-plaque activity (Marsh, 1991; Bhargava and Leonard, 1996). Triclosan is also commonly incorporated into soaps and plastics as an antimicrobial in both domestic and healthcare settings. In studies with Escherichia coli, FabI encoding a component of the fatty acid synthase machinery has been identified as the primary target of triclosan inhibition (McMurry et al., 1998; Heath et al., 1999). FabI encodes an NADH-dependent enoyl reductase that catalyzes the final reaction of the fatty acid elongation cycle.
Because of the pervasiveness of triclosan in the everyday environment, concerns have been raised about the safety of this compound (Levy, 2001). In particular, the role of triclosan in selecting for bacteria resistant to multiple drugs and antibiotics has become a concern (Chuanchuen et al., 2001). However, the use of triclosan hand-washes has not been directly linked to changes in bacterial susceptibility to antibiotics (Aiello et al., 2004). Although triclosan exhibits antifungal activity, few studies have examined the effects of this agent on Candida albicans, the major fungal pathogen of humans (Giuliana et al., 1997; Yu et al., 2011). C. albicans is a cause of oral and vaginal mucosal infections, commonly referred to as thrush. In critically ill patients, C. albicans can also cause life-threatening systemic infection. Since C. albicans is a common resident of the oral cavity, daily use of oral healthcare products containing triclosan would expose this organism to significant quantities of this agent. However, the interaction between triclosan and common azole antifungal drugs has not been fully investigated. In a recent study, it has been shown that triclosan can exhibit synergy with fluconazole against fluconazole-resistant C. albicans strains (Yu et al., 2011). In this study, we investigated the activity of triclosan against azole-susceptible C. albicans and other common Candida species and identified an antagonistic interaction between triclosan and the azole antifungal drugs.
Materials & Methods
Strains and Growth Conditions
For routine strain maintenance, Candida strains (Appendix Table 1) were cultured in yeast extract peptone dextrose (YEPD) broth or agar at 37°C. Fluconazole susceptibility was determined by broth microdilution (BMD) according to EUCAST Edef 7.1 (Rodriguez-Tudela et al., 2008). The medium used for BMD was RPMI-1640 containing L-glutamine, buffered with MOPS and supplemented with 2% (w/v) glucose. To promote growth in the yeast phase, some BMD experiments were performed with yeast nitrogen base (YNB) medium without amino acids, supplemented with 2% (w/v) glucose. Where noted, media were supplemented with triclosan (TRC; Irgasan, Fluka, Buchs, Switzerland). IC50s and IC80s for fluconazole were defined as the concentration of drug that was required to inhibit growth by 50% or 80% relative to drug-free controls, respectively. The fractional inhibitory concentration index (∑FIC) was calculated to determine whether drug interactions were antagonistic or synergistic according to the formula: ∑FIC = (MIC fluconazole in triclosan / MIC fluconazole alone) + (MIC triclosan in fluconazole / MIC triclosan alone) (Te Dorsthorst et al., 2002; Rodriguez-Tudela et al., 2008).
Fluconazole Uptake by Fungal Cells
Fluconazole uptake was determined in RPMI medium in the absence and presence of triclosan (1 mg/L) with [3H]-fluconazole [final concentration 50 nM (0.015 mg/L)] as previously described (Mansfield et al., 2010). Fluconazole accumulation was also measured during batch growth with triclosan (1 mg/L) and [3H]-fluconazole, with measurements taken at 1, 3, and 24 hrs.
Analysis of Total Membrane Sterol Content in C. albicans
Total membrane sterols were isolated from cells grown in RPMI medium in the presence and absence of 1 mg/L triclosan, as described previously (Martel et al., 2010a). Derivatized sterols were analyzed by Gas Chromatography-Mass Spectrometry (GC-MS) and identified with reference to retention times and fragmentation spectra for known standards. Sterol chromatograms were analyzed with Agilent software (MSD Enhanced ChemStation, Agilent Technologies Inc., Palo Alto, CA, USA) for the derivation of integrated peak areas (Martel et al., 2010b).
RNA Isolation and Quantitation
To facilitate isolation of large amounts of RNA, we grew cells under conditions identical to those used in the BMD assays, but in 50-mL volumes in 75 cm2 polystyrene tissue culture flasks, and harvested them after 24 hrs. RNA was isolated and cDNA synthesized as described previously (O’Connor et al., 2010). qRT-PCR was carried out in an ABI FAST7500 with SYBR Green (Applied Biosystems, Warrington, UK) according to the manufacturer’s instructions. Primers used in qRT-PCR are listed in Appendix Table 2, prefixed ‘RT’. Expression of FAS1, FAS2, CDR1, and CDR2 was measured. ACT1 was included as an internal control, and all measurements were normalized against ACT1 in each sample before comparison with other conditions. 2−ΔΔCT values were calculated according to Schmittgen and Livak (2008) and are represented graphically.
Results
Triclosan Antagonizes Azole Activity against C. albicans
Triclosan was fungicidal against C. albicans at a concentration of 16 mg/L. Measurement of fluconazole MICs by the EUCAST broth microdilution assay showed that the addition of subinhibitory concentrations of triclosan (0.5-2.0 mg/L) to RPMI-1640 medium interfered with fluconazole antifungal activity against C. albicans (Fig. 1). The fluconazole IC50 in the absence of triclosan was 0.125 mg/L, which increased to 8 mg/L fluconazole in the presence of 1 mg/L triclosan. Calculation of the ∑FIC yielded a value of 64.25, which indicated an antagonistic interaction. This phenotype was confirmed with 8 additional C. albicans isolates (Appendix Table 1). In addition, the activity of other azoles (ketoconazole, itraconazole, and miconazole) against C. albicans was antagonized similarly by 1 mg/L triclosan, but the activity of amphotericin B was not affected (Appendix Fig. 1).
Figure 1.
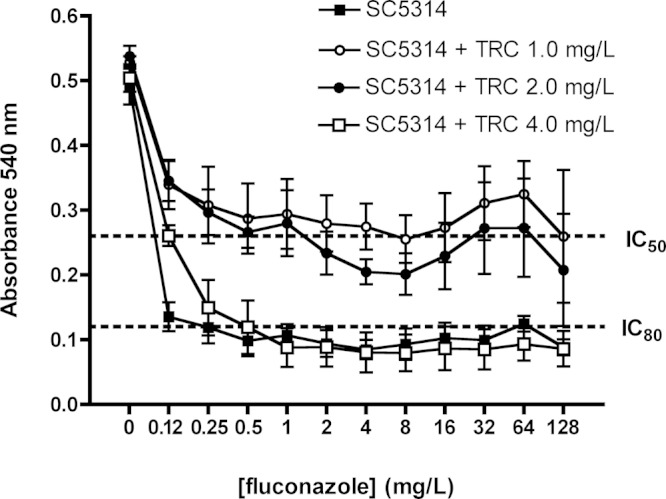
Susceptibility of C. albicans SC5314 in RPMI-1640 to fluconazole. Drug susceptibilities were tested by the EUCAST method. Fluconazole susceptibility of SC5314 was tested in RPMI-1640 medium in the absence and presence of triclosan (1, 2, and 4 mg/L). Dotted line indicates the IC50 and IC80 cut-offs as indicated. All plates were incubated at 37°C for 24 hrs, and growth was measured as absorbance at 540 nm. Results shown are the average of data generated in four separate experiments.
Expression of CDR1 and CDR2 in Response to Triclosan
Addition of 1 mg/L triclosan to the growth medium caused a significant increase in expression of CDR1 in CAF2-1 (Fig. 2A). Triclosan exposure resulted in a small but non-significant decrease in CDR2 mRNA levels in CAF2-1. To investigate whether changes in drug pump expression were directly involved in antagonism, we measured the IC80 of triclosan and the level of triclosan-mediated fluconazole antagonism in Δcdr1 and Δcdr2 mutants (Appendix Fig. 2). These mutations did not affect triclosan IC80 (Appendix Fig. 2A). Deletion of CDR1 alone or in combination with CDR2 caused increased susceptibility to fluconazole (IC80 was reduced to 0.25 mg/L compared with 0.5 mg/L in the parental strain; Appendix Fig. 2B). However, mutation of the drug efflux pumps CDR1 and CDR2 did not eliminate the antagonism (Appendix Fig. 2B).
Figure 2.
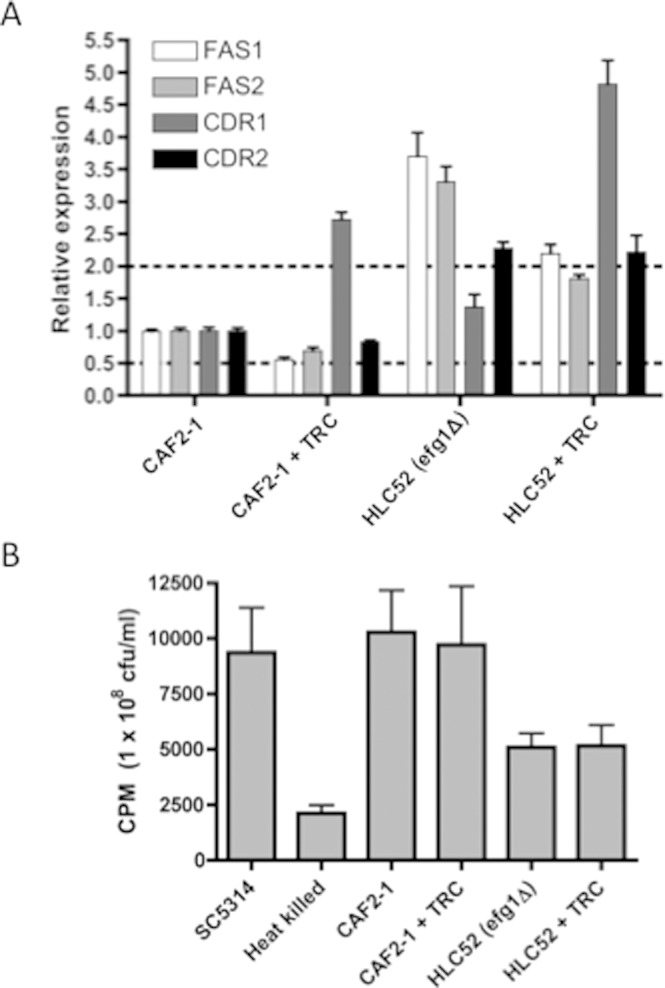
Analysis of fluconazole efflux and uptake. (A) Relative expression of FAS1, FAS2, CDR1, and CDR2 in CAF2-1 and HLC52 (Δefg1) in the presence and absence of 1 mg/L triclosan. The upper and lower dashed lines indicate 2-fold increased and 2-fold decreased expression relative to CAF2-1, respectively. Cells were grown for 24 hrs on YEPD plates at 37°C and inoculated into RPMI-1640 at 2 x 105 cfu/mL, grown for 24 hrs at 37°C, and harvested. (B) [3H]-Fluconazole accumulation by C. albicans strains, expressed as counts per min (CPM)/108 cells. Cells were grown in RPMI-1640 medium and washed, and the accumulation of [3H]-fluconazole was measured following 24-hour incubation. Live and heat-killed SC5314 were included as positive and negative controls, respectively.
Fluconazole Accumulation Is Not Influenced by Triclosan
We assessed whether triclosan antagonized fluconazole activity by altering fluconazole uptake (Mansfield et al., 2010). Addition of triclosan did not significantly affect fluconazole accumulation in CAF2-1 (Fig. 2B). Fluconazole accumulation was also measured in cells pre-grown in triclosan (1 mg/L) prior to cell starvation, or during batch growth with triclosan (1 mg/L) and [3H]-fluconazole. No significant effect was observed in any condition (data not shown).
Membrane Sterol Content Is Not Affected by Triclosan
The membrane sterol contents of C. albicans cells exposed to triclosan (1 mg/L) in RPMI-1640 medium were investigated. No significant difference in sterol profile was identified in triclosan-treated and untreated cells (Appendix Table 3).
Alteration of FAS2 Levels Does Not Influence Triclosan Sensitivity
Triclosan exposure resulted in a drop in the expression levels of the fatty acid synthase encoding genes FAS1 and FAS2 by approximately 50% and 30%, respectively (Fig. 2A). To investigate whether Fas2p, the functional orthologue of the bacterial target FabI, is a possible target of triclosan inhibition, we investigated whether a heterozygous FAS2/fas2Δ mutant (CFD1) had altered triclosan susceptibility. Despite having only one copy of FAS2, CFD1 was found to have no alteration in triclosan MIC (Appendix Fig. 3A) or in triclosan-induced fluconazole antagonism (data not shown). We also overexpressed FAS1 and FAS2 in SC5314 using the pNIM1 doxycycline-inducible expression element (see Appendix for methods) (Park and Morschhauser, 2005). Overexpression of FAS1 and FAS2 from pNIM1 was confirmed by qRT-PCR and was reproducibly at least 2.0-fold greater at doxycycline concentrations ≥ 10 mg/L. Induction of FAS1 or FAS2 from the pNIM1 element with doxycycline (10-40 mg/L) did not reduce the triclosan susceptibility of SC5314 (Appendix Fig. 3B) or the antagonism of fluconazole (Appendix Fig. 4).
Figure 3.
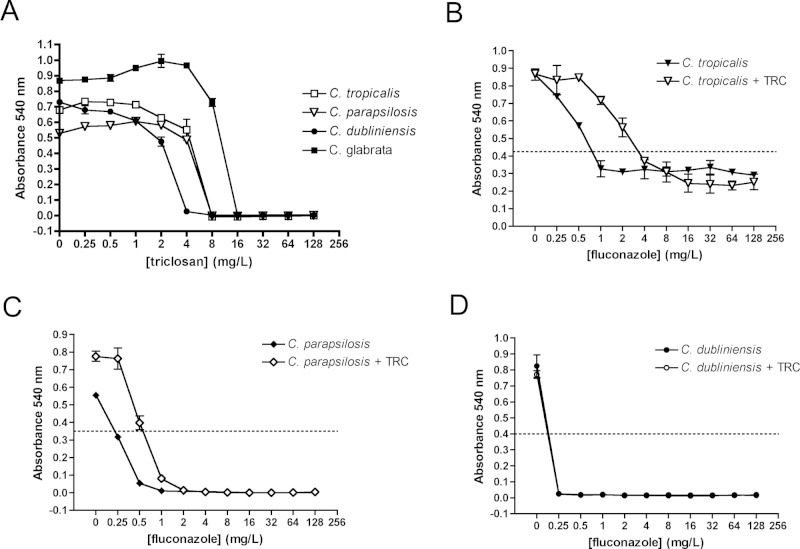
Sensitivity to triclosan (A) and fluconazole (B-D) of non-albicans Candida species. Drug susceptibilities were tested by the EUCAST method. Strains (C. tropicalis 3111 [B], C. parapsilosis HEM20 [C], and C. dubliniensis Wü284 [D]) were grown on YEPD plates at 37°C for 24 hrs before inoculation at 2 x 105 cfu/mL. Triclosan was added at 1 mg/L where indicated in panels B-D. Plates were incubated at 37°C for 24 hrs, and growth was measured as absorbance at 540 nm. Results are the average of at least 3 independent experiments. Dotted lines on Y-axes indicate IC50 values.
Figure 4.
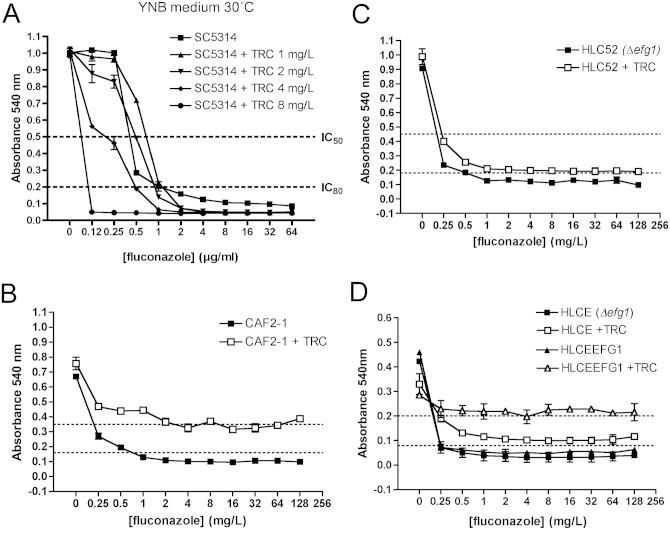
Fluconazole antagonism in C. albicans requires hypha formation. Drug susceptibilities were tested by the EUCAST method with YNB (A) or RPMI (B-D). Strains SC5314, CAF2-1, and HLC52 (Δefg1) were grown on YEPD plates at 37°C for 24 hrs before inoculation at 2 x 105 cfu/mL. Triclosan was added at 1 mg/L (+ TRC) unless otherwise indicated. Plates were incubated at 37°C for 48 hrs, and growth was measured as absorbance at 540 nm. Results are the average of at least three independent experiments. Dotted lines on Y-axes indicate IC50 or IC80 values as indicated.
Antagonism of Fluconazole Activity Is Restricted to C. albicans Hyphae
Triclosan was fungicidal against C. tropicalis, C. parapsilosis, C. glabrata, and C. dubliniensis at concentrations between 4 and 16 mg/L (Fig. 3A). Antagonism was not observed in C. glabrata (data not shown). C. tropicalis and C. parapsilosis isolates exhibited a 2- to 4-fold increase in fluconazole IC50 in the presence of 1 mg/L triclosan (Figs. 3B, 3C). However, the antagonistic effect at high fluconazole concentrations (> 8 mg/L) seen in C. albicans was not observed in either species. Unexpectedly, the closely related species C. dubliniensis (5 isolates, Appendix Table 1) exhibited a complete absence of fluconazole antagonism (Fig. 3D). Since C. dubliniensis grows exclusively in the yeast phase in RPMI medium, and C. albicans forms true hyphae, we investigated whether morphology affected antagonism (Moran et al., 2007; O’Connor et al., 2010). Antagonism assays were repeated with a growth medium that promotes growth of C. albicans in the yeast phase (YNB, pH 5.6). C. albicans exhibited an identical triclosan IC50 in RPMI and YNB (16 mg/L) but exhibited a 4-fold higher fluconazole IC50 (0.5 mg/L) in YNB medium (Fig. 4A). However, antagonism of fluconazole activity by triclosan was not observed in YNB medium. To further explore the role of morphology in antagonism, we also examined antagonism in strain HLC52, which has a homozygous deletion in EFG1 (Δefg1), a key regulator of hypha formation. Deletion of EFG1 resulted in growth in the yeast form and greatly reduced antagonism in the presence 1 mg/L triclosan and fluconazole compared with the control CAF2-1 (Figs. 4B, 4C). Complementation of Δefg1 with a single copy of EFG1 (strain HLCEFG) restored antagonism (Fig. 4D). Analysis of gene expression showed that the Δefg1 mutant exhibited a significant increase in expression of both FAS1 and FAS2 compared with CAF2-1 (Fig. 2). In addition, CDR2 expression was constitutively high compared with CAF2-1, even in the absence of triclosan. Analysis of fluconazole uptake in Δefg1 cells indicated that they accumulated less fluconazole than CAF2-1; however, the levels were not affected by the addition of triclosan (Fig. 2).
Discussion
Triclosan is commonly used as an anti-plaque agent and displays a high level of oral retention in plaque and on tooth surfaces for several days following administration (Creeth et al., 1993). We observed that, against C. albicans, subinhibitory triclosan concentrations (0.5-2 mg/L) could antagonize the activity of fluconazole. Although triclosan accumulates to high concentrations in plaque, its aqueous solubility is < 10 mg/L (Loftsson et al., 1999). Triclosan is therefore unlikely to reach IC50 concentrations in saliva or other bodily fluids for extended periods, and residual concentrations in saliva and plasma are within the range of antagonistic triclosan concentrations identified here (Creeth et al., 1993; Lin, 2000; Calafat et al., 2008).
A recent study by Yu et al. reported synergy between fluconazole and triclosan against fluconazole-resistant C. albicans isolates, but did not report the effects of this compound on azole-susceptible yeasts (Yu et al., 2011). These authors did not detect the antagonism described here because of the intrinsic fluconazole resistance of the isolates studied (IC50s >16 µg/mL). As such, the effects of triclosan on fluconazole MIC described here would not have been apparent in these isolates.
To elucidate its mechanism, we carried out a detailed study of triclosan-mediated fluconazole antagonism. The possibility that this phenomenon could be due to a physical interaction between the two drugs could be excluded, since the antagonism was not observed in non-albicans Candida species. Our data exclude changes in membrane sterol content, altered drug efflux, or altered uptake as mechanisms of triclosan-induced azole antagonism. Our data also exclude a role for the fungal orthologue of FabI, encoded by FAS2 in C. albicans, in the mode of action of triclosan. Strains exhibiting increased or decreased expression of FAS2 did not exhibit altered susceptibility to triclosan or antagonism. From these studies, we concluded that FAS2 was unlikely to be the major target of triclosan in C. albicans. Since these data were generated, the crystal structure of the Fas2 enzyme from S. cerevisiae has been determined at high resolution, and it was concluded that triclosan is unlikely to bind to the enoyl reductase active site, supporting the genetic evidence presented here (Jenni et al., 2007).
One unexpected observation from these studies was that antagonism was specific for the hyphal form of C. albicans. The hyphal form of C. albicans is highly adherent and invasive, and triclosan antagonism may therefore allow invasive fungal infections to persist. Antagonism was not observed in YNB medium, which at 30°C restricts C. albicans to the yeast morphology, or in the efg1Δ mutant HLC52, which is unable to form hyphae in RPMI-1640 medium. Although the efg1Δ mutant exhibited deregulated expression of FAS1 and FAS2, analysis of our data indicated that fatty acid synthases are unlikely to be the targets of triclosan in C. albicans.
Since our data exclude the involvement of many specific targets in the mode of action of triclosan (fatty acid synthases, sterol metabolism, CDR mechanisms), we hypothesize that triclosan may act as a non-specific membranotropic agent against C. albicans, mediating non-specific damage to the plasma membrane that accounts for its fungicidal and antagonistic activities. Recent research has re-appraised the role of membrane intercalation by triclosan as part of its biocidal action (Villalain et al., 2001; Lygre et al., 2003; Guillen et al., 2004). At low concentrations, triclosan has been shown to alter bacterial membrane fluidity and function without actually causing cell lysis, whereas at higher concentrations cell lysis may occur (Regos et al., 1979; Villalain et al., 2001). In C. albicans, subinhibitory concentrations of triclosan (≤ 8 mg/L) could also induce changes in membrane fluidity, and this could be the cause of fluconazole antagonism, perhaps by counteracting the disruptive effects of toxic sterols. Indeed, fluconazole resistance has previously been associated with increased membrane fluidity (Kohli et al., 2002). The different activities of triclosan at subinhibitory concentrations in yeast and hyphal cells may also be related to altered membrane content and fluidity (Prasad et al., 2010). The efg1Δ mutant has a significantly different lipid composition compared with the wild-type, and exhibits decreased membrane fluidity and increased fluconazole accumulation. We can only hypothesize at this stage that changes in membrane content and fluidity in hyphal cells result in a different interaction with triclosan compared with yeasts.
This study raises concerns about the concurrent use of triclosan-containing products and azole antifungals. The widespread use of triclosan in everyday hygiene and oral healthcare products makes it highly likely that infecting C. albicans strains are regularly exposed to this agent. How this affects antifungal therapy in these patients has yet to be explored, and further investigations will be required to determine whether this interaction is clinically significant.
Supplementary Material
Acknowledgments
We thank Joachim Ernst for strains HLCE and HLCEFG1; Dominique Sanglard for strains DSY447, DSY651, and DSY654; and Ronald Cihlar for strains CFD1 and CFD2. Analysis of total membrane sterols was carried out with the support of the EPSRC National Mass Spectrometry Service Centre, Swansea University.
Footnotes
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
This work was supported by the Irish Health Research Board (HRB grant RP/2002/6). HNO and TCW were supported by NIH NIDCR grant RO1 DE017078.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Aiello AE, Marshall B, Levy SB, Della-Latta P, Larson E. (2004). Relationship between triclosan and susceptibilities of bacteria isolated from hands in the community. Antimicrob Agents Chemother 48:2973-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava HN, Leonard PA. (1996). Triclosan: applications and safety. Am J Infect Control 24:209-218. [DOI] [PubMed] [Google Scholar]
- Calafat AM, Ye X, Wong LY, Reidy JA, Needham LL. (2008). Urinary concentrations of triclosan in the U.S. population: 2003-2004. Environ Health Perspect 116:303-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuanchuen R, Beinlich K, Hoang TT, Becher A, Karkhoff-Schweizer RR, Schweizer HP. (2001). Cross-resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: exposure of a susceptible mutant strain to triclosan selects nfxB mutants overexpressing MexCD-OprJ. Antimicrob Agents Chemother 45:428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creeth JE, Abraham PJ, Barlow JA, Cummins D. (1993). Oral delivery and clearance of antiplaque agents from triclosan-containing dentifrices. Int Dent J 43(4 Suppl 1):387-397. [PubMed] [Google Scholar]
- Giuliana G, Pizzo G, Milici ME, Musotto GC, Giangreco R. (1997). In vitro antifungal properties of mouthrinses containing antimicrobial agents. J Periodontol 68:729-733. [DOI] [PubMed] [Google Scholar]
- Guillen J, Bernabeu A, Shapiro S, Villalain J. (2004). Location and orientation of triclosan in phospholipid model membranes. Eur Biophys J 33:448-453. [DOI] [PubMed] [Google Scholar]
- Heath RJ, Rubin JR, Holland DR, Zhang E, Snow ME, Rock CO. (1999). Mechanism of triclosan inhibition of bacterial fatty acid synthesis. J Biol Chem 274:11110-11114. [DOI] [PubMed] [Google Scholar]
- Jenni S, Leibundgut M, Boehringer D, Frick C, Mikolasek B, Ban N. (2007). Structure of fungal fatty acid synthase and implications for iterative substrate shuttling. Science 316:254-261. [DOI] [PubMed] [Google Scholar]
- Kohli A, Smriti NFN, Mukhopadhyay K, Rattan A, Prasad R. (2002). In vitro low-level resistance to azoles in Candida albicans is associated with changes in membrane lipid fluidity and asymmetry. Antimicrob Agents Chemother 46:1046-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SB. (2001). Antibacterial household products: cause for concern. Emerg Infect Dis 7(3 Suppl):512S-515S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YJ. (2000). Buccal absorption of triclosan following topical mouthrinse application. Am J Dent 13:215-217. [PubMed] [Google Scholar]
- Loftsson T, Leeves N, Bjornsdottir B, Duffy L, Masson M. (1999). Effect of cyclodextrins and polymers on triclosan availability and substantivity in toothpastes in vivo. J Pharm Sci 88:1254-1258. [DOI] [PubMed] [Google Scholar]
- Lygre H, Moe G, Skalevik R, Holmsen H. (2003). Interaction of triclosan with eukaryotic membrane lipids. Eur J Oral Sci 111:216-222. [DOI] [PubMed] [Google Scholar]
- Mansfield BE, Oltean HN, Oliver BG, Hoot SJ, Leyde SE, Hedstrom L, et al. (2010). Azole drugs are imported by facilitated diffusion in Candida albicans and other pathogenic fungi. PLoS Pathog 6:e1001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh PD. (1991). Dentifrices containing new agents for the control of plaque and gingivitis: microbiological aspects. J Clin Periodontol 18:462-467. [DOI] [PubMed] [Google Scholar]
- Martel CM, Parker JE, Bader O, Weig M, Gross U, Warrilow AG, et al. (2010a). A clinical isolate of Candida albicans with mutations in ERG11 (encoding sterol 14alpha-demethylase) and ERG5 (encoding C22 desaturase) is cross resistant to azoles and amphotericin B. Antimicrob Agents Chemother 54:3578-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel CM, Parker JE, Bader O, Weig M, Gross U, Warrilow AG, et al. (2010b). Identification and characterization of four azole-resistant erg3 mutants of Candida albicans. Antimicrob Agents Chemother 54:4527-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell G, Russell AD. (1999). Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev 12:147-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry LM, Oethinger M, Levy SB. (1998). Triclosan targets lipid synthesis. Nature 394:531-532. [DOI] [PubMed] [Google Scholar]
- Moran GP, MacCallum DM, Spiering MJ, Coleman DC, Sullivan DJ. (2007). Differential regulation of the transcriptional repressor NRG1 accounts for altered host cell interactions in Candida albicans and Candida dubliniensis. Mol Microbiol 66:915-929. [DOI] [PubMed] [Google Scholar]
- O’Connor L, Caplice N, Coleman DC, Sullivan DJ, Moran GP. (2010). Differential filamentation of Candida albicans and C. dubliniensis is governed by nutrient regulation of UME6 expression. Eukaryot Cell 9:1383-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YN, Morschhauser J. (2005). Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryot Cell 4:1328-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad T, Hameed S, Manoharlal R, Biswas S, Mukhopadhyay CK, Goswami SK, et al. (2010). Morphogenic regulator EFG1 affects the drug susceptibilities of pathogenic Candida albicans. FEMS Yeast Res 10:587-596. [DOI] [PubMed] [Google Scholar]
- Regos J, Zak O, Solf R, Vischer WA, Weirich EG. (1979). Antimicrobial spectrum of triclosan, a broad-spectrum antimicrobial agent for topical application. II. Comparison with some other antimicrobial agents. Dermatologica 158:72-79. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Tudela JL, Arendrup MC, Barchiesi F, Bille J, Chryssanthou E, Cuenca-Estrella M, et al. (2008). EUCAST definitive document EDef 7.1: method for the determination of broth dilution MICs of antifungal agents for fermentative yeasts. Clin Microbiol Infect 14:398-405. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101-1108. [DOI] [PubMed] [Google Scholar]
- Te Dorsthorst DT, Verweij PE, Meis JF, Punt NC, Mouton JW. (2002). Comparison of fractional inhibitory concentration index with response surface modeling for characterization of in vitro interaction of antifungals against itraconazole-susceptible and -resistant Aspergillus fumigatus isolates. Antimicrob Agents Chemother 46:702-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalain J, Mateo CR, Aranda FJ, Shapiro S, Micol V. (2001). Membranotropic effects of the antibacterial agent triclosan. Arch Biochem Biophys 390:128-136. [DOI] [PubMed] [Google Scholar]
- Yu L, Ling G, Deng X, Jin J, Jin Q, Guo N. (2011). In vitro interaction between fluconazole and triclosan against clinical isolates of fluconazole-resistant Candida albicans determined by different methods. Antimicrob Agents Chemother 55:3609-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.