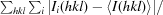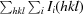Structural analysis of the catalytic module of Caldicellulosiruptor bescii family 3 pectate lyase shows that this new structure is very similar to the previously solved structure of a family 3 pectate lyase from Bacillus sp. strain KSM-P15.
Keywords: pectate lyases, PL3, Caldicellulosiruptor bescii
Abstract
A 1.5 Å resolution X-ray structure of the catalytic module of Caldicellulosiruptor bescii family 3 pectate lyase is reported (PDB entry 3t9g). The resulting structure was refined to an R factor of 0.143 and an R free of 0.178. Structural analysis shows that this new structure is very similar to the previously solved structure of a family 3 pectate lyase from Bacillus sp. strain KSM-P15 (PDB entry 1ee6), with a root-mean-square deviation of 0.93 Å and a sequence identity of 53%. This structural similarity is significant considering that C. bescii is a hyperthermophile and Bacillus sp. is a mesophile.
1. Introduction
Pectate lyases are important contributors to the plant cell-wall deconstruction mechanisms of bacterial and fungal microorganisms. Pectic polysaccharides consist of α-1,4-linked polygalactosyluronic acid residues with regions of alternating galactosyluronic acid and rhamnosyl residues (Willats et al., 2001 ▶). Pectate lyases catalyze the degradation of mixed pectins, which are a major component of the primary cell walls of higher plants, in the presence of calcium ions (Marín-Rodríguez et al., 2002 ▶). They were first discovered in plant-pathogenic bacteria that cause diseases involving the maceration of parenchymatous tissues of various dicot plants (Collmer & Keen, 1986 ▶).
With their ability to break down pectic polysaccharides, pectate lyases are clearly necessary for the efficient microbial deconstruction of the plant cell wall. These enzymes are thus an important target for detailed studies of their structure–function relationships and biochemical roles. Caldicellulosiruptor bescii family 3 pectate lyase (PL3) is especially interesting because its host organism is the most thermophilic cellulose-degrading organism known (Yang et al., 2010 ▶). Here, we report the X-ray structure of the PL3 catalytic module from the hyperthermophilic and cellulolytic bacterium C. bescii (Madigan et al., 1997 ▶; Yang et al., 2010 ▶). This report is a starting point for further structural and biochemical studies of the role of pectate lyases in plant cell-wall deconstruction.
2. Materials and methods
2.1. Cloning, expression and purification
The PL3 catalytic module encoded by C. bescii (Cbes_1854; 193 amino acids) was cloned into pET-45b (Novagen) between KpnI and XhoI sites, so that the construct contained an N-terminal 6×His affinity tag. The forward and reverse primers were GGGGTACCAATACGGGGTGGTGTTTTAGTTATTACAG and CCGCTCGAGTTAGTATTGATGTATCTGTGATTGGG, respectively. The 20.9 kDa protein was expressed in Escherichia coli BL21 (DE3) cells (10 l culture) induced with 1 mM IPTG for 16 h at 310 K. The cell pellet was collected and resuspended in 20 mM sodium phosphate pH 7.0 containing 0.3 M NaCl (buffer A) and the cells were disrupted using a French press. Cell debris was removed by centrifugation at 10 000 rev min−1 for 30 min. The supernatant was heated at 343 K for 20 min, and after removing denatured protein by centrifugation at 10 000g for 30 min the supernatant was applied onto a HisTrap FF affinity column (5 ml, GE Healthcare) equilibrated with buffer A. After washing the column, the PL3 module was eluted using an imidazole gradient (0–0.5 M) in buffer A. Based on SDS–PAGE analysis, fractions containing PL3 were combined, concentrated using ultrafiltration (Amicon, 10 kDa cutoff) and dialyzed against 20 mM sodium phosphate pH 7.0. Finally, size-exclusion chromatography was used to remove the remaining impurities using a HiLoad Superdex 75 (26/60) column (GE Healthcare, Piscataway, New Jersey, USA) in 20 mM acetic acid pH 5.0 containing 100 mM NaCl and 5 mM CaCl2. The purified protein was concentrated with a Vivaspin 20 5 kDa concentrator (GE Healthcare UK Ltd, Little Chalfont, Buckinghamshire, England) and the protein concentration was measured using a Pierce BCA Protein Assay kit (Pierce Biotechnology, Rockford, llinois, USA).
2.2. Crystallization
PL3 crystals were obtained by sitting-drop vapor diffusion using a 96-well plate with Crystal Screen HT from Hampton Research (Aliso Viejo, California, USA). 50 µl well solution was added to the reservoir and drops were made up of 0.2 µl well solution and 0.2 µl protein solution using a Phoenix crystallization robot (Art Robbins Instruments, Sunnyvale, California, USA). The crystals were grown at 293 K using 0.2 M ammonium phosphate monobasic, 0.1 M Tris pH 8.5 and 50%(v/v) (±)-2-methyl-2,4-pentanediol as the well solution. The protein solution consisted of 15 mg ml−1 protein, 20 mM acetic acid pH 5, 100 mM NaCl and 5 mM CaCl2.
2.3. Data collection and processing
Before flash-cooling in a nitrogen-gas stream at 100 K, the PL3 crystal was soaked for 20 s in a 2 µl drop of well solution with an additional 5%(v/v) glycerol and 5%(v/v) ethylene glycol. Home-source data collection was performed using a Bruker X8 MICROSTAR X-ray generator with Helios mirrors and a Bruker PLATINUM135 CCD detector. Data were indexed and processed with the Bruker suite of programs v.2011.2-0 (Bruker AXS, Madison, Wisconsin, USA).
2.4. Structure solution and refinement
Intensities were converted into structure factors and 5% of the reflections were flagged for R free calculations using the programs F2MTZ, TRUNCATE, CAD and UNIQUE from the CCP4 package of programs (Winn et al., 2011 ▶). The program MOLREP v.10.2.23 (Vagin & Teplyakov, 2010 ▶) was used for molecular replacement, using the structure of a family 3 pectate lyase from Bacillus sp. strain KSM-P15 (PDB entry 1ee6; Akita et al., 2001 ▶) as the search model. PDB entry 1ee6 was selected as the molecular-replacement model because it is the only known structure of a family 3 pectate lyase and it has the highest sequence identity to our enzyme. Refinement and manual correction were performed using REFMAC5 v.5.5.0109 (Murshudov et al., 2011 ▶) and Coot v.0.6 (Emsley & Cowtan, 2004 ▶). The MolProbity method (Chen et al., 2010 ▶) was used to analyze the Ramachandran plot, and root-mean-square deviations (r.m.s.d.s) of bond lengths and angles were calculated from the Engh and Huber ideal values of stereochemical parameters (Engh & Huber, 1991 ▶). The Wilson B factor was calculated using CTRUNCATE v.1.0.11 (Winn et al., 2011 ▶). Average B factors were calculated using ICM v.3.7-2a (Molsoft LLC, La Jolla, California, USA). The figures were created using PyMOL (http://www.pymol.org). Data-collection and refinement statistics are given in Table 1 ▶.
Table 1. X-ray data-collection and refinement statistics.
Values in parentheses are for the highest resolution bin.
| Space group | C2221 |
| Unit-cell parameters (Å, °) | a = 36.514, b = 146.685, c = 162.159, α = β = γ = 90.0 |
| Wavelength (Å) | 1.54178 |
| Temperature (K) | 100 |
| Resolution (Å) | 25–1.5 (1.6–1.5) |
| Unique reflections | 70692 (12550) |
| Observed reflections | 434755 (49823) |
| Rint† | 0.0446 (0.1495) |
| Average multiplicity | 6.2 (4.0) |
| 〈I/σ(I)〉 | 24.7 (24.7) |
| Completeness (%) | 99.6 (98.5) |
| R/Rfree | 0.143 (0.198)/0.178 (0.251) |
| No. of protein atoms | 3286 |
| No. of water molecules | 561 |
| No. of other atoms | 133 |
| R.m.s.d. from ideal bond lengths‡ (Å) | 0.029 |
| R.m.s.d. from ideal bond angles‡ (°) | 2.422 |
| Wilson B factor (Å2) | 12.5 |
| Average B factor for protein atoms (Å2) | 11.9 |
| Average B factor for water molecules (Å2) | 27.5 |
| Ramachandran plot statistics§ (%) | |
| Allowed | 99.8 |
| Favored | 95.1 |
| Outliers | 1 [Asn153, chain B] |
3. Results and discussion
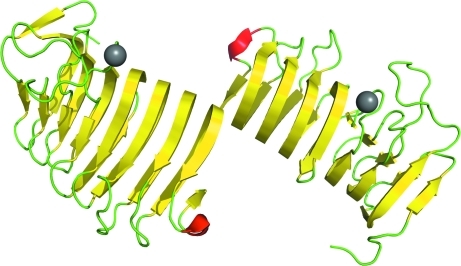
We have solved the X-ray structure of the C. bescii family 3 pectate lyase catalytic module to 1.5 Å resolution. It has two molecules in the asymmetric unit accompanied by various solvent ions and molecules, including the structurally important calcium ions of family 3 pectate lyases (Akita et al., 2001 ▶; Fig. 1 ▶). The overall fold is the parallel β-helix typical of pectate lyases (Marín-Rodríguez et al., 2002 ▶). This structure has been deposited in the PDB as PDB entry 3t9g.
Figure 1.
The overall structure of the C. bescii family 3 pectate lyase catalytic module with two molecules in the asymmetric unit. β-Strands are shown in yellow, α-helices in red and loops in green. Calcium ions are shown as gray balls.
There is one outlier in the Ramachandran plot: Asn153 of chain B (Table 1 ▶). Closer inspection of this residue shows that it is well defined in the electron density and is actually very close to being in the favored region of the Ramachandran plot. Together with Leu151, Asn152 and Val154, Asn153 forms a turn that is firmly anchored by the inward-pointing side chains of Leu151 and Val154. As it was not possible to remove this strained conformation by creating alternative conformations for Asn153 and the neighboring residues, we decided that it is a feature of this structure.
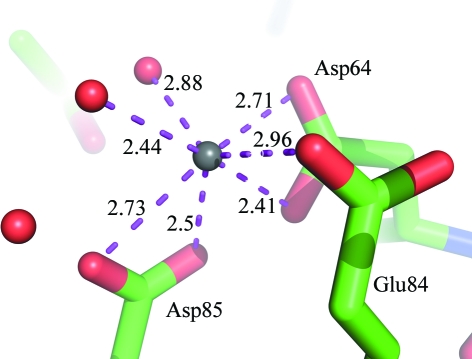
Pairwise secondary-structure matching using PDBeFold (Krissinel & Henrick, 2004 ▶) found eight unique structures with a secondary-structure similarity of between 50 and 60%. The most similar structure was that of the family 3 pectate lyase from Bacillus sp. strain KSM-P15 (PDB entry 1ee6), with an r.m.s.d. of 0.93 Å and a sequence identity of 53%. The other hits had sequence similarities of below 10% and r.m.s.d. values of greater than 4 Å, indicating that they are only similar in fold and are not of interest for further comparisons. A closer comparison with the Bacillus family 3 pectate lyase (Bacillus PL3) revealed that the first calcium ion of PL3 is in the same position as the structurally important calcium ion in Bacillus PL3 (Akita et al., 2001 ▶). Also, PL3 has an Na atom surrounded by an acidic cluster formed by Asp64, Asp85 and Glu84 in the same position and with the same conformation as those observed by Akita and coworkers for a catalytically important calcium ion when they soaked their crystals in a pH 9.5 solution. This calcium ion is believed to mediate substrate binding. We modeled an Na atom at this position because the density was too weak for calcium; in fact, the sodium we have positioned here has only 70% occupancy and the distances to the coordinating atoms are too short for a water molecule (Fig. 2 ▶). The active-site residues of PL3 (Lys108, Lys130 and Arg133) are also identical to those of Bacillus PL3, with identical or very similar residues surrounding the rest of the active site.
Figure 2.
The Na atom coordinated by the acidic cluster. The Na atom is shown as a gray ball, with waters as red dots and amino acids as sticks with red O atoms, blue N atoms and green C atoms.
Supplementary Material
PDB reference: family 3 pectate lyase catalytic module, 3t9g
Acknowledgments
This work was supported by the DOE Office of Science, Office of Biological and Environmental Research through the BioEnergy Science Center (BESC), a DOE Bioenergy Research Center.
References
- Akita, M., Suzuki, A., Kobayashi, T., Ito, S. & Yamane, T. (2001). Acta Cryst. D57, 1786–1792. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Collmer, A. & Keen, N. T. (1986). Annu. Rev. Phytopathol. 24, 383–409.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Engh, R. A. & Huber, R. (1991). Acta Cryst. A47, 392–400.
- Krissinel, E. & Henrick, K. (2004). Acta Cryst. D60, 2256–2268. [DOI] [PubMed]
- Madigan, M. T., Martinko, J. M. & Parker, J. (1997). In Brock Biology of Microorganisms, edited by M. T. Madigan, J. M. Martinko & J. Parker. Upper Saddle River, New Jersey, USA: Prentice Hall International Editions.
- Marín-Rodríguez, M. C., Orchard, J. & Seymour, G. B. (2002). J. Exp. Bot. 53, 2115–2119. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Willats, W. G., McCartney, L., Mackie, W. & Knox, J. P. (2001). Plant Mol. Biol. 47, 9–27. [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yang, S.-J., Kataeva, I., Wiegel, J., Yin, Y., Dam, P., Xu, Y., Westpheling, J. & Adams, M. W. (2010). Int. J. Syst. Evol. Microbiol. 60, 2011–2015. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: family 3 pectate lyase catalytic module, 3t9g