The structure of LsrB from Y. pestis complexed with autoinducer-2 (AI-2) has been determined at 1.75 Å resolution. Bound AI-2 adopts the (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran conformation, which is the same conformation as is observed in the S. typhimurium and S. meliloti LsrB–AI-2 structures.
Keywords: LsrB, Yersinia pestis, autoinducer-2
Abstract
The crystal structure of LsrB from Yersinia pestis complexed with autoinducer-2 (AI-2; space group P212121, unit-cell parameters a = 40.61, b = 61.03, c = 125.23 Å) has been solved by molecular replacement using the structure of LsrB from Salmonella typhimurium (PDB entry 1tjy) and refined to R = 0.180 (R free = 0.213) at 1.75 Å resolution. The electron density for bound AI-2 and the stereochemistry of the AI-2-binding site are consistent with bound AI-2 adopting the (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran conformation, just as has been observed in the crystal structures of the Salmonella typhimurium and Sinorhizobium meliloti LsrB–AI-2 complexes.
1. Introduction
Bacteria can adapt to changes in local population densities by sensing the extracellular concentration of communication signals. This regulatory mechanism is generally called ‘quorum sensing’ and the types of signals recognized are composed of diverse functional chemistry, ranging from acylhomoserine lactones in Gram-negative species to linear or modified peptides in Gram-positive species (Fuqua et al., 2001 ▶; Thoendel & Horswill, 2010 ▶). An additional molecule called autoinducer-2 (AI-2) is produced by many species spanning different phylogenetic groups and has been proposed to function as a universal quorum-sensing signal (Surette et al., 1999 ▶).
AI-2 was originally identified and characterized in the marine bacterium Vibrio harveyi (Bassler et al., 1993 ▶). The signal is released into the extracellular environment and induces bioluminescence in a concentration-dependent manner through a two-component sensory relay (Vendeville et al., 2005 ▶). The structure of AI-2 was unknown until crystallographic studies of the LuxP periplasmic receptor revealed a bound furanosyl borate diester (2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran–borate; S-THMF–borate) in the active site (Chen et al., 2002 ▶). Interestingly, the LuxP receptor and other V. harveryi sensory components are not present in many species of bacteria (Sun et al., 2004 ▶). Studies on Salmonella typhimurium were the first to unravel these differences and to identify the alterative luxS-regulated (Lsr) chromosomal locus. Unlike the V. harveyi paradigm model, the AI-2 signal is imported into the S. typhimurium cytoplasm through the action of an ABC transporter encoded in the lsr region (Taga et al., 2001 ▶, 2003 ▶). The S. typhimurium AI-2 structure is also divergent, which was revealed through crystallographic studies of the LsrB periplasmic receptor. The S. typhimurium version of AI-2 is a (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran (R-THMF) that lacks the liganded B atom (Miller et al., 2004 ▶). Subsequent studies demonstrated that both S-THMF–borate and R-THMF are derived from the common intermediate dihydroxypentanedione (DPD). DPD is produced through the activated methyl cycle, in which S-adenosylhomocysteine is converted in sequential steps to homocysteine and DPD through the action of the Pfs nucleosidase and LuxS enzymes (Miller et al., 2004 ▶). More recently, AI-2 in the crystal structure of LsrB–AI-2 from Sinorhizobium meliloti (Pereira et al., 2008 ▶; PDB entry 3ejw) has been shown to adopt the same R-THMF conformation.
Here, we report the LsrB–AI-2 structure for the pathogen Yersinia pestis, the causative agent of plague in humans. Y. pestis is known to have a luxS gene and preliminary studies indicate that it produces AI-2 (Bobrov et al., 2007 ▶). We find that AI-2 synthesized from S-adenosylhomocysteine using purified Pfs nucleosidase and LuxS enzymes from Y. pestis (Yu et al., 2011 ▶) adopts the R-THMF conformation when bound to Y. pestis LsrB.
2. Materials and methods
2.1. Cloning
Avirulent Y. pestis CO92 pgm− pCD1 (Lcr+, strain R88) was obtained from Robert Perry (University of Kentucky) and maintained in Brain Heart Infusion Broth (BHI). Genomic DNA was purified from strain R88 using a Gentra Puregene bacterial DNA-purification kit (Gentra Systems, Minneapolis, Minnesota, USA) and used as a template source for cloning of lsrB. A truncated LsrB lacking the amino-terminal leader sequence (Δ2–25) was PCR-amplified from genomic DNA using the oligonucleotides 5′-GTTGTTCATATGGCGGAACGCATCGCATTTATC-3′ (forward) and 5′-GTTGTTGGATCCCCGCTAAGATTAAAAGTCGTATTTACTG-3′ (reverse). The PCR product was digested with NdeI and BamHI and cloned into pET28a cut with the same enzymes, resulting in an N-terminal His tag. There is no C-terminal T7 tag owing to the stop codon in the reverse primer. Clones were sequenced at the University of Iowa DNA Core Facility and positive clones were transformed into Escherichia coli overexpression strain ER2566 (New England Biolabs).
2.2. Expression and purification
E. coli overexpression strain ER2566 carrying plasmid pET28a-lsrB was grown at 310 K in 8 l Luria–Bertani medium to an optical density of ∼1.0 at 600 nm. Expression of lsrB in the culture was induced with 1 mM IPTG for 4 h. The cells were harvested by centrifugation and resuspended in 90 ml equilibration buffer (50 mM sodium phosphate pH 8.0, 0.3 M sodium chloride, 10 mM imidazole). The suspension was lysed with 9 ml Bugbuster (Novagen) and incubated at 277 K with gentle rocking for 4 h. The LsrB protein was purified by nickel-affinity chromatography using His-Select resin (Sigma) by the batch method. After centrifugation at 30 000g for 30 min at 277 K, the cleared lysate was incubated with 25 ml equilibrated resin for 2 h at 277 K with gentle rocking. The resin was then centrifuged at 5000g for 5 min at 277 K and the supernatant was saved. The resin was washed twice with ten volumes of equilibration buffer. The LsrB protein was eluted from the resin with two column volumes of elution buffer (50 mM sodium phosphate pH 8.0, 0.3 M sodium chloride, 250 mM imidazole). Elution fractions were pooled and concentrated using an Amicon PM10 membrane and finally dialyzed against 10 mM sodium phosphate pH 6.5. The concentration of His-tagged LsrB was determined to be 22.1 mg ml−1 by the Bradford assay (Bio-Rad).
2.3. Crystallization
Hanging-drop vapor-diffusion crystallization screens were performed using a Mosquito robot (TTP LabTech, Cambridge, Massachusetts, USA) and pre-filled crystallization trays that were prepared using a Genesis RSP 150/8 liquid handler (Tecan, Männedorf, Switzerland). The drop and reservoir volumes were 0.8 and 100 µl, respectively, with the initial protein concentration in the drops being 11 mg ml−1 (1:1 ratio of protein:well solution). Four different 96-condition screens were utilized: Crystal Screen/Crystal Screen 2 and SaltRx from Hampton Research (Aliso Viejo, California, USA), Wizard I and II from Emerald BioSystems (Bainbridge Island, Washington, USA) and the PEGs and PEGs II Suite from Qiagen (Germantown, Maryland, USA). Crystallization trays were stored at 291 K and the drops were monitored for crystal growth using RockMaker software and a RockImager 2 (Formulatrix, Waltham, Massachusetts, USA). Within 2 d, crystals began to form in numerous crystallization conditions, with the most promising crystals growing from drops that had lower molecular-weight PEGs as the precipitating agent. Large (∼0.3 × 0.3 × 0.2 mm) well formed single crystals that grew from drops containing 20% PEG 3350 and 200 mM diammonium hydrogen phosphate (Qiagen PEGs Suite condition No. 92) were used for data collection after 6 d of growth.
2.4. Diffraction data collection
A single LsrB crystal that was grown from 20% PEG 3350, 200 mM diammonium hydrogen phosphate was transferred into 200 µl mother liquor in which the concentration of PEG 3350 was increased to 35%. The mother liquor also contained 145 µM AI-2 that had been synthesized from S-adenosylhomocysteine using purified Pfs nucleosidase and LuxS enzymes from Y. pestis (Yu et al., 2011 ▶). The crystal was soaked at 291 K for approximately 24 h and flash-cooled in a nylon loop for data collection. Diffraction data for the AI-2-soaked crystal were collected at the University of Iowa Protein Crystallography Facility using a Rigaku RUH3R rotating-anode generator that was fitted with Osmic Blue Optics and an R-AXIS IV++ area detector (Rigaku, The Woodlands, Texas, USA). The crystal was maintained at 100 K using an Oxford CryoStream 700. Diffraction data were collected using StructureStudio and processed using XDS (Kabsch, 2010 ▶) and SCALA (Evans, 2006 ▶) software. Crystal parameters and data-processing statistics are presented in Table 1 ▶.
Table 1. Crystal and refinement parameters for LsrB from Y. pestis (PDB entry 3t95).
Values in parentheses are for the highest resolution shell.
| Data processing | |
| Wavelength (Å) | 1.5418 |
| Unit-cell parameters (Å) | a = 40.61, b = 61.03, c = 125.23 |
| Space group | P212121 |
| Resolution range (Å) | 62.62–1.75 (1.84–1.75) |
| No. of observations | 207868 |
| No. of unique reflections | 31947 (4339) |
| Data completeness (%) | 98.9 (93.7) |
| Multiplicity | 6.5 (5.7) |
| 〈I/σ(I)〉 | 23.3 (6.5) |
| Rmerge | 0.049 (0.261) |
| REFMAC5 refinement | |
| Resolution range (Å) | 19.75–1.75 (1.80–1.75) |
| No. of reflections for Rwork | 30271 (1973) |
| No. of reflections for Rfree | 1618 (93) |
| No. of protein atoms | 2398 |
| No. of solvent atoms | 380 |
| No. of ligand atoms | 10 |
| Mean overall B factor (Å2) | 17.4 |
| Mean protein B factor (Å2) | 15.6 |
| Mean solvent B factor (Å2) | 29.3 |
| Mean ligand B factor (Å2) | 9.4 |
| Final Rwork | 0.180 (0.306) |
| Final Rfree | 0.213 (0.415) |
| R.m.s.d. bond lengths (Å) | 0.013 |
| R.m.s.d. bond angles (°) | 1.361 |
| R.m.s.d. B factors (Å2) | |
| Main-chain bond-related | 0.701 |
| Main-chain angle-related | 1.296 |
| Side-chain bond-related | 2.168 |
| Side-chain angle-related | 3.614 |
2.5. Structure solution and refinement
The structure of the LsrB–AI-2 complex was solved by molecular replacement using Phaser (McCoy et al., 2007 ▶) in the CCP4 (v.6.1.13) software package (Winn et al., 2011 ▶). When the structure of S. typhimurium apo LsrB (PDB entry 1tm2; Miller et al., 2004 ▶) was used as the probe structure no molecular-replacement solution was obtained, but using the structure of the S. typhimurium LsrB–AI2 complex (PDB entry 1tjy; Miller et al., 2004 ▶) as a probe did yield a unique molecular-replacement solution. The AI-2 ligand was not included in this probe structure in order to minimize any bias with regard to the molecular form of the AI-2 bound to Y. pestis LsrB. Strong well defined electron density was found in the Y. pestis LsrB AI-2 binding site, but AI-2 was not added to the atomic model right away. The amino-acid differences (37 positions) between S. typhimurium and Y. pestis LsrB were incorporated into the model using the Coot software package (Emsley et al., 2010 ▶) and the model was subjected to maximum-likelihood refinement using REFMAC5 (Murshudov et al., 2011 ▶). There was interpretable electron density for the entire LsrB sequence (Ala26–Phe339) and some electron density for the last two residues, His24 and Met25, of the N-terminal His tag, but these residues were not added to the atomic model because the weak electron density could not be interpreted unambiguously. A total of 380 water molecules were added to the model in iterations of Coot real-space refinement followed by maximum-likelihood refinement using REFMAC5. Finally, the AI-2 molecule was added to the model and one round of refinement with REFMAC5 resulted in R and R free values of 0.1800 and 0.2128, respectively. The final model has good stereochemistry, as indicated by the refinement statistics reported in Table 1 ▶. The final model and diffraction data have been deposited in the Protein Data Bank (Berman et al., 2000 ▶; PDB code 3t95).
3. Results and discussion
3.1. LsrB domain structure
With the similarities of the Y. pestis lsr region to those of other enteric bacteria, we hypothesized that Y. pestis recognizes the non-boron-containing form of AI-2 (R-THMF). To investigate this question, the Y. pestis CO92 LsrB protein was overexpressed and purified using nickel-affinity chromatography. The LsrB protein was cocrystallized with AI-2 that had been biosynthesized using recombinant Y. pestis LuxS and Pfs proteins. The structure was solved at 1.75 Å resolution by molecular replacement using the structure of the LsrB–AI-2 complex (PDB entry 1tjy) from S. typhimurium (Miller et al., 2004 ▶).
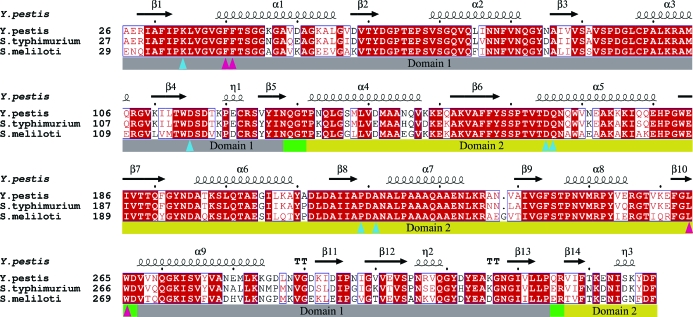
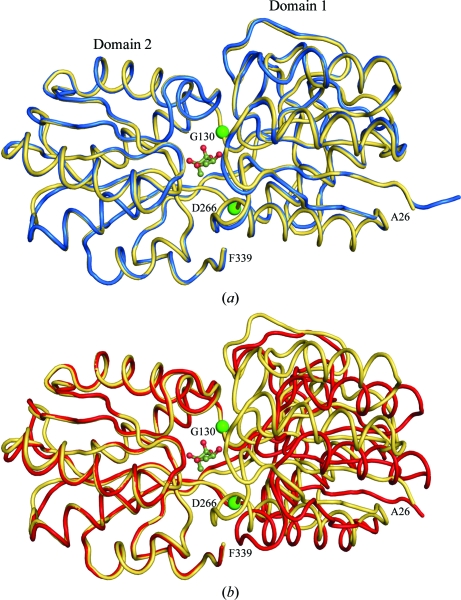
The LsrBs from Y. pestis, S. typhimurium and Si. meliloti share high sequence similarity (Fig. 1 ▶), with the Y. pestis sequence being 87% and 76% identical to the S. typhimurium and Si. meliloti sequences, respectively. All three receptors adopt a class 1 periplasmic binding-protein fold (Quiocho & Ledvina, 1996 ▶; Dwyer & Hellinga, 2004 ▶), in which two α/β-domains are connected through three short connecting linkers or hinges (Fig. 1 ▶) and each α/β-domain is composed of two segments, one coming from the first half of the polypeptide chain and the other from the second half of the chain. The Y. pestis LsrB–AI-2 complex shows striking structural similarity to the LsrB–AI-2 complexes from S. typhimurium (Miller et al., 2004 ▶) and Si. meliloti (Pereira et al., 2008 ▶), with Cα root-mean-square deviations (r.m.s.d.) of 0.279 Å (269 Cα) for S. typhimurium and 0.346 Å (267 Cα) for chain A and 0.271 Å (261 Cα) for chain B of the Si. meliloti asymmetric unit. There is a significant tertiary-structure conformation change associated with binding of AI-2 (Fig. 2 ▶), which is evident when one superimposes domain 2 residues Gly130–Asp266 and calculates the r.m.s.d. for the residues that were not superimposed. When the domain 2 residues of the Y. pestis LsrB–AI2 complex are superimposed on the domain 2 residues of the S. typhimurium LsrB–AI2 complex, the r.m.s.d. is 0.213 Å for the superimposed residues and 0.278 Å for the residues not included in the superposition, indicating there is very little (if any) difference in the tertiary structures of the two LsrB–AI-2 complexes. In contrast, an analogous superposition of the Y. pestis LsrB–AI-2 complex on S. typhimurium apo LsrB yields r.m.s.d.s of 0.274 and 2.525 Å for the superimposed and non-superimposed residues, respectively. This is indicative of a significant ligand-associated conformational change and explains why molecular replacement using the S. typhimurium apo LsrB structure failed to provide a solution for the Y. pestis LsrB–AI-2 complex.
Figure 1.
Sequence alignment of LsrB from Y. pestis, S. typhimurium and Si. meliloti, in which residues that are conserved in all three proteins are highlighted with a red background and residues that are conserved in two of the proteins are displayed in red text. Secondary-structural elements of Y. pestis LsrB are indicated at the top of each panel. Bars below the sequences indicate domain structure, with hinge residues indicated by green bars and domain 1 and domain 2 residues indicated by grey and yellow bars, respectively. Residues that interact with AI-2 are indicated by triangles: blue for residues that form hydrogen bonds and magenta for residues that form van der Waals interactions. This figure was generated using ClustalW (Larkin et al., 2007 ▶) and ESPript/ENDscript (Gouet et al., 2003 ▶).
Figure 2.
Ribbon drawing showing AI-2 bound in the cleft between α/β-domains 1 and 2. (a) shows the LsrB–AI-2 complex from Y. pestis (yellow) and the LsrB–AI-2 complex from S. typhimurium (blue) after domain 2 residues Gly130–Asp166 have been superimposed. (b) shows the LsrB–AI-2 complex from Y. pestis (yellow) and apo LsrB from S. typhimurium (red) after domain 2 residues Gly130–Asp166 have been superimposed. The locations of Gly130 and Asp166 are labelled and indicated by green spheres.
3.2. AI-2 conformation and binding site
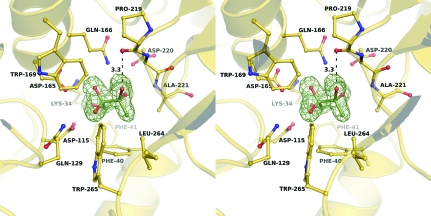
Strong electron density in the central cleft between the two α/β-domains was easily interpretable and was modelled as the R-THMF conformation of the AI-2 ligand (Fig. 3 ▶). These data are consistent with the Y. pestis LuxS and Pfs proteins synthesizing the non-boron-containing form of AI-2 and the Y. pestis LsrB recognizing this form of AI-2.
Figure 3.
F o − F c electron-density map calculated before the AI-2 molecule was added to the LsrB atomic model. Contours within 2.8 Å of the AI-2 molecule are drawn at 3σ. Residues that form hydrogen bonds or van der Waals interactions with the AI-2 molecule are shown as ball-and-stick models. The hydrogen bond between the C2 hydroxyl group of the AI-2 ring and the carbonyl O atom of Pro218 is indicated.
The AI-2 binding site in Y. pestis LsrB is highly conserved when compared with the binding sites of S. typhimurium and Si. meliloti LsrBs. The residues that form hydrogen bonds and van der Waals interactions with AI-2 are completely conserved in the three proteins (Fig. 1 ▶). Of particular note is the hydrogen bond between the C2 hydroxyl group of the AI-2 ring and the carbonyl O atom of Pro218 (Fig. 3 ▶), as this interaction, together with van der Waals interactions between the C2 methyl group and Phe40, Ala221, Leu264 and Trp265, may be necessary to differentially bind the 2R enantiomer over the 2S enantiomer. If AI-2 were to bind in the 2S conformation, both the carbonyl O atom of Pro218 and the C2 hydroxyl of AI-2 would be buried without hydrogen-bonding partners, a scenario that would be energetically unfavorable.
4. Conclusions
The structure of LsrB from Y. pestis complexed with autoinducer-2 (AI-2) has been solved by molecular replacement using the structure of LsrB from S. typhimurium (PDB entry 1tjy) and refined at 1.75 Å resolution. The electron density of the bound AI-2 matched the (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran (R-THMF) conformation, which is the same conformation that is bound by LsrBs from S. typhimurium and Si. meliloti.
Supplementary Material
PDB reference: LsrB–AI-2 complex, 3t95
Acknowledgments
We thank R. Perry for Y. pestis strain R88. We also thank the staff of the Protein Crystallography Facility at the University of Iowa for assistance with this project. This work was supported by Office of Naval Research Grant N00014-06-1-1176 to ARH.
References
- Bassler, B. L., Wright, M., Showalter, R. E. & Silverman, M. R. (1993). Mol. Microbiol. 9, 773–786. [DOI] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed]
- Bobrov, A. G., Bearden, S. W., Fetherston, J. D., Khweek, A. A., Parrish, K. D. & Perry, R. D. (2007). Adv. Exp. Med. Biol. 603, 178–191. [DOI] [PubMed]
- Chen, X., Schauder, S., Potier, N., Van Dorsselaer, A., Pelczer, I., Bassler, B. L. & Hughson, F. M. (2002). Nature (London), 415, 545–549. [DOI] [PubMed]
- Dwyer, M. A. & Hellinga, H. W. (2004). Curr. Opin. Struct. Biol. 14, 495–504. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Fuqua, C., Parsek, M. R. & Greenberg, E. P. (2001). Annu. Rev. Genet. 35, 439–468. [DOI] [PubMed]
- Gouet, P., Robert, X. & Courcelle, E. (2003). Nucleic Acids Res. 31, 3320–3323. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J. & Higgins, D. G. (2007). Bioinformatics, 23, 2947–2948. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Miller, S. T., Xavier, K. B., Campagna, S. R., Taga, M. E., Semmelhack, M. F., Bassler, B. L. & Hughson, F. M. (2004). Mol. Cell, 15, 677–687. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Pereira, C. S., McAuley, J. R., Taga, M. E., Xavier, K. B. & Miller, S. T. (2008). Mol. Microbiol. 70, 1223–1235. [DOI] [PMC free article] [PubMed]
- Quiocho, F. A. & Ledvina, P. S. (1996). Mol. Microbiol. 20, 17–25. [DOI] [PubMed]
- Sun, J., Daniel, R., Wagner-Döbler, I. & Zeng, A.-P. (2004). BMC Evol. Biol. 4, 36. [DOI] [PMC free article] [PubMed]
- Surette, M. G., Miller, M. B. & Bassler, B. L. (1999). Proc. Natl Acad. Sci. USA, 96, 1639–1644. [DOI] [PMC free article] [PubMed]
- Taga, M. E., Miller, S. T. & Bassler, B. L. (2003). Mol. Microbiol. 50, 1411–1427. [DOI] [PubMed]
- Taga, M. E., Semmelhack, J. L. & Bassler, B. L. (2001). Mol. Microbiol. 42, 777–793. [DOI] [PubMed]
- Thoendel, M. & Horswill, A. R. (2010). Adv. Appl. Microbiol. 71, 91–112. [DOI] [PMC free article] [PubMed]
- Vendeville, A., Winzer, K., Heurlier, K., Tang, C. M. & Hardie, K. R. (2005). Nature Rev. Microbiol. 3, 383–396. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yu, J., Kavanaugh, J. S., Madsen, M. L., Carruthers, M. D., Phillips, G. J., Boyd, J. M., Horswill, A. R. & Minion, F. C. (2011). Submitted.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: LsrB–AI-2 complex, 3t95