The structure of NFeoB from S. thermophilus harbouring an N11A mutation has been determined to 1.85 Å resolution.
Keywords: FeoB, G-protein domain, Streptococcus thermophilus
Abstract
The uptake of ferrous iron in prokaryotes is mediated by the G-protein-coupled membrane protein FeoB. The protein contains two N-terminal soluble domains that are together called ‘NFeoB’. One of these is a G-protein domain, and GTP hydrolysis by this domain is essential for iron transport. The GTPase activity of NFeoB is accelerated in the presence of potassium ions, which bind at a site adjacent to the nucleotide. One of the ligands at the potassium-binding site is a conserved asparagine residue, which corresponds to Asn11 in Streptococcus thermophilus NFeoB. The structure of an N11A S. thermophilus NFeoB mutant has been determined and refined to a resolution of 1.85 Å; the crystals contained a mixture of mant-GDP-bound and mant-GMP-bound protein. The structure demonstrates how the use of a derivatized nucleotide in cocrystallization experiments can facilitate the growth of diffraction-quality crystals.
1. Introduction
Iron is an essential element for all forms of life, and in prokaryotes acquisition of ferrous iron is achieved through the G-protein-coupled membrane protein FeoB. This three-domain protein contains two cytoplasmic globular domains at its N-terminus followed by a large polytopic membrane-spanning domain. The soluble N-terminal domains are together named ‘NFeoB’ and are comprised of a G-protein domain (G-domain) followed by an α-helical domain. NFeoB is approximately 30 kDa in mass and numerous studies have investigated the structure of NFeoB in its apo form and also bound to various nucleotides (Guilfoyle et al., 2009 ▶; Hattori et al., 2009 ▶; Köster et al., 2009 ▶; Ash et al., 2010 ▶, 2011 ▶; Hung et al., 2010 ▶; Petermann et al., 2010 ▶). These studies found that the G-domain adopts the classical G-protein fold and the helical domain interacts with one face of the G-domain. Like other small G-proteins, the G-domain of FeoB possesses two Switch regions that undergo conformational changes in response to nucleotide binding and hydrolysis (Ash et al., 2010 ▶, 2011 ▶).
Biophysical studies on NFeoB initially demonstrated that the protein exhibits slow intrinsic GTPase activity. For example, the k cat values for Escherichia coli and Streptococcus thermophilus NFeoB were found to be 0.0015 and 0.0028 s−1, respectively (Ash et al., 2010 ▶; Eng et al., 2008 ▶; Marlovits et al., 2002 ▶). However, the solution of the structure of NFeoB from S. thermophilus (NFeoBSt) bound to a nonhydrolyzable GTP analogue (PDB entry 3lx5; Ash et al., 2010 ▶) led to the finding that the GTPase activity of the protein is accelerated more than 15-fold in the presence of potassium ions, in a manner similar to the potassium-activated G-protein MnmE (Scrima & Wittinghofer, 2006 ▶). A subsequent structure of NFeoBSt bound to GDP–AlF4 − and K+ showed that the potassium ion binds at a site adjacent to the nucleotide in the GTP-bound form of the protein (PDB entry 3ss8; Ash et al., 2011 ▶). The six-coordinate potassium-binding site in NFeoBSt consists of the backbone carbonyl groups of Gly29 and Trp31 from the Switch I region, three O atoms from the nucleotide phosphates and the side-chain amide of Asn11.
Coordination of the potassium ion by the Switch I region is accompanied by a large conformational change in the protein, in which the Switch adopts a distinctive structure that differs from that found in, for example, the Ras or Gα families of G-proteins. This GTP-bound Switch I conformation is a conserved structural feature of members of the TrmE-Era-EngA-YihA-septin-like (TEES) superfamily of G-proteins (Ash et al., 2010 ▶), which all contain a conserved asparagine residue in the equivalent position to Asn11 of NFeoBSt. It was thus predicted that other members of this family would similarly possess potassium-dependent GTPase activity (Ash et al., 2010 ▶; Scrima & Wittinghofer, 2006 ▶). Numerous studies have now surfaced in support of this hypothesis and other potassium-activated G-proteins have been identified (Anand et al., 2010 ▶; Hwang & Inouye, 2001 ▶; Scrima & Wittinghofer, 2006 ▶; Tomar et al., 2011 ▶).
As part of our studies investigating the potassium-activated GTPase activity of NFeoBSt, an N11A mutant was created to verify the location of the potassium ion-binding site and to explore the role of this amino acid in potassium binding (Ash et al., 2010 ▶). Mutation of Asn11 to alanine abolished potassium binding and potassium-dependent activation of the enzyme, highlighting the importance of this residue in the coordination sphere of the ion. Here, a high-resolution structure of this NFeoBSt N11A mutant protein (referred to subsequently as N11A) has been determined in complex with a fluorescently-labelled nucleotide.
2. Materials and methods
2.1. Protein purification
In an earlier study, an N11A mutation was introduced into wild-type NFeoBSt (residues 1–270; Q5M586; Ash et al., 2010 ▶). Mutated NFeoBSt was expressed as an N-terminally tagged GST-fusion protein from a pGEX-4T-1 vector. Details of the expression and purification of N11A have been described previously (Ash et al., 2010 ▶). The protein was purified, stored and crystallized in buffer consisting of 20 mM Tris pH 8.0, 100 mM NaCl, 5%(v/v) glycerol. After purification, concentrated protein (∼15 mg ml−1) was aliquoted and stored at 193 K until use in crystallization trials. Because N11A was expressed as a fusion protein, two additional amino acids, Gly-Ser, remained at the N-terminus after cleavage of the tag and are present in the final crystallized product.
2.2. Crystallization
For crystallization of N11A, protein (7.0 mg ml−1 final concentration) was mixed with 10 mM MgCl2 and either GDP (1 mM), the nonhydrolyzable GTP analogue GMPPNP (0.8 mM; Jena Bioscience), or the fluorescently labelled GDP nucleotide mant-GDP (0.5 mM; Invitrogen). Crystallization conditions were tested for each sample at 293 K using the hanging-drop vapour-diffusion method with commercially available screens (PACT and JCSG+; Qiagen). Drops containing equal volumes (200 nl) of reservoir and protein solutions were set up using a Mosquito nanolitre liquid-handling robot (TTP LabTech). Diffraction-quality crystals of N11A were only obtained from the cocrystallization experiment with mant-GDP. Crystals grew after three weeks from condition B9 of the PACT screen [0.2 M lithium chloride, 0.1 M MES pH 6.0, 20%(w/v) PEG 6000]. Crystals obtained in the initial screen were used in diffraction studies without further optimization. Crystals were cryoprotected by brief immersion in mother liquor containing 20%(v/v) glycerol and then directly frozen in an N2-gas stream (100 K) for data collection.
2.3. Data collection and refinement
Diffraction data were recorded on a MAR 345 image-plate detector using Cu Kα X-rays from a Rigaku RU-200 rotating-anode X-ray generator. Integration and scaling of diffraction intensities was performed using the HKL-2000 software suite (Otwinowski & Minor, 1997 ▶). The structure of the N11A mutant protein was solved by molecular replacement using the program Phaser (McCoy et al., 2007 ▶) from the CCP4 suite (Winn et al., 2011 ▶). The structure of wild-type NFeoBSt bound to GDP (PDB code 3lx8; Ash et al., 2010 ▶) was used as a search model after removal of all nonprotein atoms. Refinement was carried out using REFMAC v.5.5.0109 (Murshudov et al., 2011 ▶) and manual model building was performed in Coot (Emsley & Cowtan, 2004 ▶). The model was refined using isotropic temperature factors, and a total of 18 964 parameters were refined in the final model.
3. Results and discussion
3.1. Crystallization of the N11A mutant protein
Crystals of N11A grew after three weeks from a cocrystallization experiment with mant-GDP (hereafter referred to as mGDP). mGDP is a derivatized nucleotide in which a fluorescent N-methylanthraniloyl (mant) group is attached to either the 2′-hydroxy or 3′-hydroxy group of the ribose ring. Commercially available mGDP solutions contain a mixture of 2′-labelled and 3′-labelled nucleotides. Such modified nucleotides are frequently used in experiments which monitor nucleotide binding and release, since the fluorescence of the mant group is highly sensitive to the nature of its chemical environment. Importantly, efforts to crystallize N11A bound to unlabelled nucleotides proved unfruitful, so crystallization was attempted in the presence of mGDP. Diffraction-quality crystals were then obtained directly from initial screening trials.
N11A crystallized in space group C2221 and the crystals diffracted to 1.85 Å resolution. The data-collection and refinement statistics are presented in Table 1 ▶. The asymmetric unit contains two protein molecules and the final model consists of residues 0–259 in chain A (residue 0 remains from the expression tag), residues −1 to 262 in chain B (residues −1 and 0 remain from the expression tag), two nucleotide molecules, 416 water molecules, three glycerol molecules and one Mg atom.
Table 1. Data-processing and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Data processing | |
| Wavelength (Å) | 1.5418 |
| Space group | C2221 |
| Unit-cell parameters (Å) | a = 60.1, b = 63.8, c = 265.2 |
| Resolution (Å) | 50–1.85 (1.88–1.85) |
| Total reflections | 272067 |
| Unique reflections | 43892 (2060) |
| Completeness (%) | 99.3 (93.4) |
| 〈I/σ(I)〉 | 24.9 (2.5) |
| 〈Multiplicity〉 | 6.2 (3.3) |
| Rmerge† | 0.068 (0.379) |
| Refinement | |
| No. of molecules in asymmetric unit | 2 |
| No. of atoms | 4741 |
| No. of water molecules | 416 |
| Resolution | 50–1.85 (1.90–1.85) |
| Unique reflections | 43738 (3020) |
| Completeness (%) | 99.2 (92.8) |
| R.m.s.d. bonds (Å) | 0.010 |
| R.m.s.d. angles (°) | 1.50 |
| 〈Isotropic B factor〉 (Å2) | |
| Protein | 20.8 |
| Ligand | 20.3 |
| Solvent | 28.8 |
| Ramachandran plot‡ | |
| Favoured (%) | 98.3 |
| Allowed (%) | 100 |
| Rwork§ | 17.6 |
| Rfree¶ | 21.6 |
| PDB code | 3tah |
R
merge = 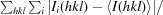
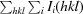 .
.
As calculated by MolProbity (Chen et al., 2010 ▶).
R
work = 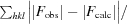
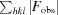 .
.
Calculated as for R work using 5% of the diffraction data that was excluded during refinement (2197 reflections).
3.2. The overall structure of N11A
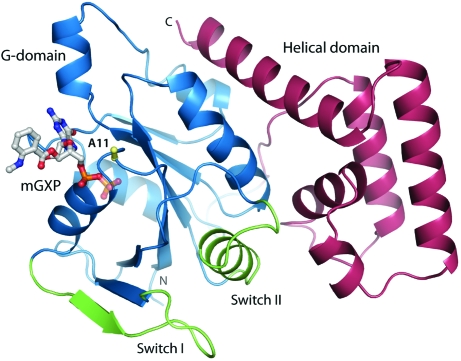
The structure of N11A is comprised of a G-domain (residues 1–160) followed by a helical domain (residues 171–270) (Fig. 1 ▶). The G-domain protein possesses the canonical small G-protein fold, consisting of a six-stranded β-sheet and six α-helices. The helical domain consists of five α-helices, which interact with Switch II and one face of the G-domain. There are no significant conformational differences between the two molecules in the asymmetric unit, and the two chains superpose with an r.m.s. deviation of 0.4 Å over 254 Cα atoms.
Figure 1.
Structure of N11A bound to mGXP. Cα trace of the N11A mutant protein showing the G-domain (blue), the Switch regions (green) and the helical domain (pink). The nucleotide and the side chain of the mutated Ala11 residue (yellow) are shown in stick representation.
In the N11A structure, the Switch I region (residues 24–37) is oriented away from the nucleotide-binding site and residues 31–40 form an additional β-strand as part of the β-sheet core of the G-domain. The Switch II region consists of a loop and an α-helix (residues 61–77), which lie at the interface between the G-domain and the helical domain. The conformations of both Switch regions are the same as observed in the structure of wild-type GDP-bound NFeoBSt (PDB code 3lx8; Ash et al., 2010 ▶). The overall folds of wild-type GDP-bound NFeoBSt and the N11A protein agree very closely, with an r.m.s. deviation of only 0.6 Å over 250 Cα atoms.
A magnesium ion is coordinated between chains A and A* (symmetry operation x + ½, y + ½, z). During refinement, an unexplained 11σ sphere of positive F o − F c electron density became apparent and the octahedral geometry and short bond lengths (∼2.1 Å) to its ligands were characteristic of a magnesium coordination site (Harding, 2001 ▶). Indeed, Mg2+ was present at 5 mM concentration in the crystallization drop. The magnesium is coordinated by Asn126 and Asp128 from chain A, Glu211 from chain A* and three water molecules.
3.3. Details of the nucleotide-binding site
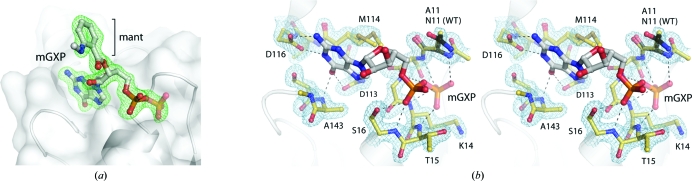
As shown in Fig. 2 ▶(a), the active site in the crystals of N11A is occupied by a mant-labelled nucleotide, consistent with the use of mGDP for cocrystallization. However, refinement with an mGDP nucleotide modelled at full occupancy in the active site produced a peak of negative F o − F c electron density over the β-phosphate group. In addition, the temperature factors of the β-phosphate were significantly higher (∼33 Å2) than those of the rest of the nucleotide (∼23 Å2). These observations were the same for both mGDP molecules in the asymmetric unit. Because the β-phosphate is the recipient of five hydrogen bonds and also possesses limited degrees of freedom, we may exclude the possibility that the higher temperature factors are indicative of its disorder. Rather, the data suggest that the crystals of N11A contained both mGDP-bound and mGMP-bound protein, and that the electron density at the nucleotide-binding site arises from an average of the two forms. The mGMP nucleotide results from the hydrolysis of mGDP throughout the three-week crystallization experiment: a spontaneous reaction which is estimated to occur at a rate of 1–2% per day (Invitrogen). To account for the contribution of mGMP-bound protein in the crystals, the β-phosphate group was assigned a fractional occupancy of 0.6 in the final model. This satisfied the electron density and gave a refined temperature factor approximately equal to those of the surrounding protein residues and the rest of the nucleotide. We will therefore refer to the bound nucleotide as ‘mGXP’ in further discussions, indicative of the presence of both mGDP-bound and mGMP-bound protein in the crystal.
Figure 2.
Details of the nucleotide-binding site. (a) OMIT electron-density map around the nucleotide contoured at 3σ. (b) Interactions with the mGXP nucleotide. The position of the Asn11 side chain in the wild-type GDP-bound protein (PDB entry 3lx8; Ash et al., 2010 ▶) is indicated by black sticks. A 2F o − F c electron-density map around the active-site residues is shown as a blue mesh contoured at 1.5σ. In this and other figures the β-phosphate from the nucleotide is shown at 60% opacity, indicative of the 60% contribution from mGDP and the 40% contribution from mGMP in the crystals.
The interactions at the nucleotide-binding site in mGXP-bound N11A (Fig. 2 ▶ b) agree with those observed for wild-type GDP-bound NFeoB proteins from various organisms. These interactions have been analyzed in detail in a host of earlier publications (Hattori et al., 2009 ▶; Köster et al., 2009 ▶; Ash et al., 2010 ▶; Hung et al., 2010 ▶; Guilfoyle et al., 2009 ▶) and will not be described again here. It is however important to note that the structural integrity of the active site is maintained in the current mGXP-bound N11A structure, consistent with the observation that the Asn11 side chain does not form any interactions with the protein or the nucleotide in the structure of GDP-bound wild-type protein (shown in black sticks in Fig. 2 ▶ b). Furthermore, the mant fluorophore from the derivatized mGDP nucleotide does not make any contacts with the protein chain to which it is bound. Instead, it forms a crystal-packing contact with an adjacent molecule, thereby mediating the assembly of the N11A crystals.
3.4. The importance of mGXP for crystal growth
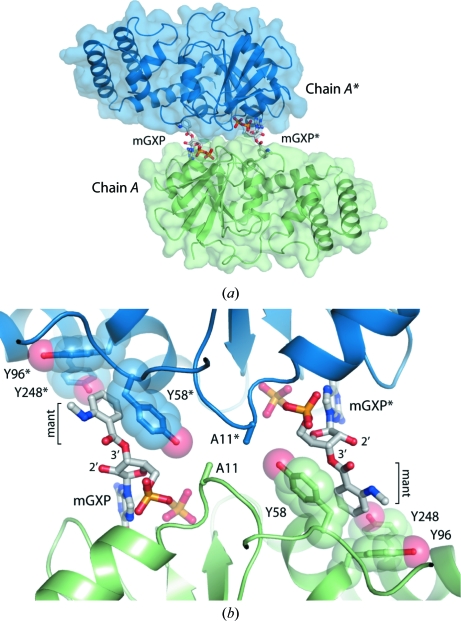
Cocrystallization of the N11A protein with mant-labelled GDP was instrumental in producing crystals, and examination of the crystal packing reveals the nature of this relationship. As shown in Fig. 3 ▶(a), the mant group from each nucleotide makes contact with a symmetry-related molecule across a crystallographic twofold axis, such that the resultant interface consists of two mGXP ‘bridges’. This is observed for both chains (symmetry operations x, −y, −z and −x, y, −z + ½) and the entire crystal is assembled from mGXP-linked dimers. The interface is comprised entirely of π-stacking interactions between the mant group and a cluster of three tyrosine residues (Tyr58, Tyr96 and Tyr248) from the adjacent protein chain (Fig. 3 ▶ b). These interactions form almost the entire contact surface at the crystal-packing interface. The only protein–protein interaction at the interface is between the mutated Ala11 residues from each chain, which form a small buried hydrophobic patch (Fig. 3 ▶ b).
Figure 3.
The importance of the mant fluorophore for crystal assembly. (a) Overview of the crystal-packing interface mediated by mGXP. (b) Structural basis for mant-dependent crystal formation and the selective incorporation of 3′-hydroxy-labelled nucleotide. The nucleotide is shown in stick representation and the interacting tyrosine cluster is shown as transparent spheres.
Although the mGDP solution used in crystallization contained a mixture of 2′- and 3′-hydroxy-labelled nucleotides, the active site is fully occupied by 3′-labelled mGXP. Examination of the structure indicates that for protein molecules bound to 2′-hydroxy-labelled mGXP, the mant group would be positioned too far from the adjacent molecule to interact with the tyrosine cluster and mediate crystal growth (Fig. 3 ▶ b). Therefore, proteins bound to 3′-labelled nucleotides were selectively incorporated into the crystals.
4. Conclusions
The structure of N11A bound to mGXP illustrates that mutation of Asn11 to alanine in NFeoBSt did not compromise the overall structural integrity of the protein nor impact upon interactions with GDP. Therefore, the structure supports the suggestion that the observed loss of potassium-dependent GTPase activity in the N11A mutant protein is solely a consequence of disruption of the potassium-binding site that is assembled upon GTP binding (Ash et al., 2010 ▶). The successful growth of N11A crystals through cocrystallization with mGDP provides an example in which a small chemical modification mediated crystal assembly in an otherwise intractable system. There are numerous structures in the Protein Data Bank bound to mant-labelled nucleotides and the mant group participates in a crystal-packing interface in at least one of these (PDB code 1gnp; Scheidig et al., 1995 ▶). If necessary, the use of derivatized nucleotides could therefore be exploited when attempting crystallization of nucleotide-binding proteins.
Supplementary Material
PDB reference: N11A mutant NFeoB, 3tah
Acknowledgments
This study was supported by the National Health and Medical Research Council (grant Nos. 570784 and 632703). M-RA is the recipient of an Australian Postgraduate Award.
References
- Anand, B., Surana, P. & Prakash, B. (2010). PLoS One, 5, e9944. [DOI] [PMC free article] [PubMed]
- Ash, M.-R., Guilfoyle, A., Clarke, R. J., Guss, J. M., Maher, M. J. & Jormakka, M. (2010). J. Biol. Chem. 285, 14594–14602. [DOI] [PMC free article] [PubMed]
- Ash, M.-R., Maher, M. J., Guss, J. M. & Jormakka, M. (2011). PLoS One, 6, e23355. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Eng, E. T., Jalilian, A. R., Spasov, K. A. & Unger, V. M. (2008). J. Mol. Biol. 375, 1086–1097. [DOI] [PMC free article] [PubMed]
- Guilfoyle, A., Maher, M. J., Rapp, M., Clarke, R., Harrop, S. & Jormakka, M. (2009). EMBO J. 28, 2677–2685. [DOI] [PMC free article] [PubMed]
- Harding, M. M. (2001). Acta Cryst. D57, 401–411. [DOI] [PubMed]
- Hattori, M., Jin, Y., Nishimasu, H., Tanaka, Y., Mochizuki, M., Uchiumi, T., Ishitani, R., Ito, K. & Nureki, O. (2009). Structure, 17, 1345–1355. [DOI] [PubMed]
- Hung, K.-W., Chang, Y.-W., Eng, E. T., Chen, J.-H., Chen, Y.-C., Sun, Y.-J., Hsiao, C.-D., Dong, G., Spasov, K. A., Unger, V. M. & Huang, T. (2010). J. Struct. Biol. 170, 501–512. [DOI] [PMC free article] [PubMed]
- Hwang, J. & Inouye, M. (2001). J. Biol. Chem. 276, 31415–31421. [DOI] [PubMed]
- Köster, S., Wehner, M., Herrmann, C., Kühlbrandt, W. & Yildiz, O. (2009). J. Mol. Biol. 392, 405–419. [DOI] [PubMed]
- Marlovits, T. C., Haase, W., Herrmann, C., Aller, S. G. & Unger, V. M. (2002). Proc. Natl Acad. Sci. USA, 99, 16243–16248. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Petermann, N., Hansen, G., Schmidt, C. L. & Hilgenfeld, R. (2010). FEBS Lett. 584, 733–738. [DOI] [PubMed]
- Scheidig, A. J., Franken, S. M., Corrie, J. E., Reid, G. P., Wittinghofer, A., Pai, E. F. & Goody, R. S. (1995). J. Mol. Biol. 253, 132–150. [DOI] [PubMed]
- Scrima, A. & Wittinghofer, A. (2006). EMBO J. 25, 2940–2951. [DOI] [PMC free article] [PubMed]
- Tomar, S. K., Kumar, P. & Prakash, B. (2011). Biochem. Biophys. Res. Commun. 408, 459–464. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: N11A mutant NFeoB, 3tah