The purification, crystallization and X-ray structure analysis of human L30e are presented here.
Keywords: L30e, ribosomal proteins
Abstract
Many functions have been reported for the eukaryotic ribosomal protein L30e. L30e makes several inter-subunit and intra-subunit interactions with protein or RNA components of the 80S ribosome. Yeast L30e has been shown to bind to its own transcript to autoregulate expression at both the transcriptional and the translational levels. Furthermore, it has been reported that mammalian L30e is a component of the selenocysteine-incorporation machinery by binding to the selenocysteine-insertion sequence on mRNA. As high-resolution crystal structures of mammalian L30e are not available, the purification, crystallization and X-ray structure analysis of human L30e are presented here.
1. Introduction
The ribosomal protein L30e is an indispensable component of the eukaryotic 80S ribosome, in which it is part of the large (60S) ribosomal subunit. The location of L30e was first identified by a cryo-EM study of the wheat 80S ribosome (Halic et al., 2005 ▶) and further details of interactions have been determined based on the recently solved high-resolution X-ray structure of the yeast 80S ribosome (Ben-Shem et al., 2010 ▶). These studies revealed that L30e functions as a bridge for components of the 40S subunit, such as helix 22 of 18S rRNA and ribosomal protein S13e. In the 60S subunit, L30e interacts with helix 34 and helix 58 of 25S rRNA as well as with L37Ae and an unidentified protein (Halic et al., 2005 ▶). On the other hand, yeast L30e has the ability to bind to its own transcript RNA if it exists in excess and interferes with spliceosome function for the control of gene expression (Eng & Warner, 1991 ▶; Vilardell & Warner, 1994 ▶). Autoregulation during translation by the interaction between L30e and L30e mRNA has also been reported (Li et al., 1996 ▶). The molecular-recognition mechanism of the L30e transcript by L30e has been reported based on X-ray and NMR studies (Mao et al., 1999 ▶; Mao & Williamson, 1999 ▶; Chao & Williamson, 2004 ▶). L30e possesses a distinct L7Ae RNA-binding motif, originally found in the archaeal ribosomal protein L7Ae, which interacts with mRNA helical structures called kink-turns (K-turns; Klein et al., 2001 ▶). A great deal of information regarding the mechanism of interaction between L7Ae proteins and their target K-turns, which are components of the ribosomal large subunit or spliceosome ribonucleoprotein (RNP) complexes, is available at atomic resolution.
Interestingly, Chavatte and coworkers reported that L30e also makes a contribution to the UGA-selenocysteine recoding machinery in eukaryotes (Chavatte et al., 2005 ▶). They clearly showed interaction of rat L30e with the selenocysteine-insertion sequence (SECIS) in the 3′-untranslated region (UTR) of eukaryotic selenoprotein mRNAs. This interaction of the SECIS with L30e is competitive with the interaction of the SECIS with SBP2 (SECIS binding protein 2), which tethers and conveys the information of the SECIS to selenocysteine-specific elongation factor (eEFSec) on the ribosome (Fagegaltier et al., 2000 ▶; Tujebajeva et al., 2000 ▶). Both L30e and SBP2 have an L7Ae motif and they are responsible for recognizing the K-turn believed to be formed in the middle of the SECIS stem (Allmang et al., 2002 ▶). A great deal of work has been performed to define the K-turn binding site of SBP2 (Cléry et al., 2007 ▶; Copeland & Driscoll, 1999 ▶; Copeland et al., 2000 ▶; Donovan et al., 2008 ▶; Latrèche et al., 2009 ▶; Takeuchi et al., 2009 ▶), but the crystal structure of SBP2 has yet to be determined. We wished to analyze the competitive binding mechanism of SBP2 and L30e towards SECIS, which prompted us to initiate an X-ray structural study of human L30e (hL30e).
2. Expression and purification
The gene encoding hL30e (NCBI Reference NP_000980.1) was cloned into the expression plasmid pET-30b(+) (Stratagene) modified by the addition of glutathione S-transferase (GST) in fusion with an N-terminal His×10 tag (cleavable by TEV protease). The C-terminal His×6 tag was left as a remainder of cloning. The cDNA for GST was the synthetic product from Schistosoma japonicum (GenBank ID CAX71419.1). The protein was overexpressed in Escherichia coli BL21 (DE3) Star at 310 K in LB medium in the presence of 25 µg ml−1 kanamycin. When the cell density reached an OD600 of 0.6, isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to the medium to a final concentration of 0.5 mM for induction. After 16 h of incubation with constant shaking, the cells were harvested by centrifugation at 4500g for 30 min at 277 K, washed with 50 mM sodium phosphate buffer pH 7.5, 300 mM NaCl, 10% glycerol and stored at 203 K until use.
The cell pellets (21 g from 3 l culture) were resuspended in buffer consisting of 50 mM sodium phosphate pH 6.5, 500 mM NaCl, 20 mM imidazole, 10 µg ml−1 DNase and disrupted by sonication. The lysate was centrifuged for 30 min at 40 000g and 277 K. The supernatant was collected and filtered through 0.22 µm pore-size filters (Sterivex; Millipore). The supernatant was loaded onto a 5 ml HisTrap HP column (GE Healthcare Biosciences) previously equilibrated with suspension buffer using an ÄKTApurifier system (GE Healthcare). Next, the column was washed twice with 4 and 8% elution buffer (50 mM sodium phosphate pH 6.5, 500 mM NaCl, 500 mM imidazole). The protein was eluted with a linear gradient from 8 to 100% elution buffer. The N-terminal His-GST tag was removed by TEV protease in cleavage buffer (10 mM Tris–HCl pH 7.4, 500 mM NaCl, 1 mM DTT). To separate the protein and the His-GST tag, the sample was loaded onto a 5 ml HisTrap HP column previously equilibrated with TEV protease cleavage buffer. The sample was further purified using a 5 ml HisTrap Heparin column (GE Healthcare Biosciences) equilibrated with 40 mM sodium phosphate pH 6.5 and 50 mM NaCl. The elution was carried out with buffer consisting of 40 mM sodium phosphate pH 6.5 and 500 mM NaCl. The purity of the protein was assessed by 15% SDS–PAGE and one band around 13 kDa corresponding to the molecular weight of hL30e was confirmed to be nearly homogeneous. The purified protein harbours an additional Gly as part of the TEV recognition site at the N-terminus and additional GAPHHHHHH residues at the C-terminus. 4.2 mg L30e was purified from 3 l culture. All purification steps were carried out at 277 K or on ice.
3. Crystallization
Prior to crystallization trials, the purified protein was concentrated to 14 mg ml−1 in buffer consisting of 10 mM Tris–HCl pH 8.0, 300 mM NaCl, 1 mM EDTA using a Millipore centrifugal filter device (Amicon Ultra-4, 10 kDa cutoff; Millipore). Crystallization screening was performed using Wizard I and II (Emerald BioStructures), JCSG Core I–IV Suites, MPD Suite and Classics Suite (Qiagen) by the sitting-drop vapour-diffusion method in 96-well plates (SWISSCI MRC 2 Well, Jena Bioscience). A 0.5 µl drop of the sample was mixed with an equal volume of reservoir solution and the mixture was equilibrated against 0.1 ml reservoir solution at 293 K. Crystals were grown from Classics Suite condition No. 47 (0.1 M sodium acetate pH 4.6, 2.0 M sodium formate; Fig. 1 ▶). The crystals from the screening were directly used for X-ray data collection.
Figure 1.
Crystal of human L30e.
4. Data collection and reduction
X-ray diffraction data were collected on beamline NW12 of the Photon Factory Advanced Ring (PF-AR; Tsukuba, Japan) using an ADSC Q210 CCD detector. Prior to diffraction data collection, the crystals were cryoprotected by transfer into a solution containing 25%(v/v) glycerol for a few seconds and flash-cooled. The data set was integrated, merged and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶); the crystal diffracted to 1.6 Å resolution. The hL30e crystal belonged to space group H32, with unit-cell parameters a = b = 56.3, c = 203.6 Å. Based on the value of the Matthews coefficient (V M; Matthews, 1968 ▶), it was estimated that there was one molecule in the asymmetric unit with V M = 2.21 Å3 Da−1 (V solv = 44.5%). Details of the data-collection and processing statistics are given in Table 1 ▶.
Table 1. Data statistics.
Values in parentheses are for the highest resolution shell.
| PDB entry | 3vi6 |
| Data collection | PF-AR NW12 |
| Wavelength (Å) | 1.000 |
| Resolution range (Å) | 50–1.59 (1.62–1.59) |
| Space group | H32 |
| Unit-cell parameters (Å) | a = b = 56.3, c = 203.6 |
| No. of observations | 181997 |
| No. of unique reflections | 16978 (814) |
| Completeness (%) | 99.6 (98.3) |
| Multiplicity | 10.7 (10.1) |
| Average I/σ(I) | 39.0 (8.2) |
| Rmerge† | 0.052 (0.310) |
| Refinement | |
| No. of reflections | 15242 |
| Protein atoms | 753 |
| Formate atoms | 3 |
| Water atoms | 83 |
| Resolution range (Å) | 50–1.59 (1.64–1.59) |
| Rwork‡ | 0.192 (0.217) |
| Rfree§ | 0.216 (0.257) |
| Mean B factor (Å2) | 19.0 |
| Overall anisotropic B factors (Å2) | |
| B11, B22, B33 | 0.79, 0.79, −1.18 |
| B12, B12, B23 | 0.39, 0.00, 0.00 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.157 |
R
merge = 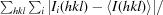
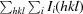 , where 〈I(hkl)〉 is the mean intensity of symmetry-equivalent reflections.
, where 〈I(hkl)〉 is the mean intensity of symmetry-equivalent reflections.
R
work = 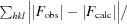
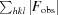 , where F
obs and F
calc are the observed and calculated structure-factor amplitudes, respectively.
, where F
obs and F
calc are the observed and calculated structure-factor amplitudes, respectively.
The R free value was calculated as for the R factor but using only a test set of reflections (5% of the total) that were not used in the refinement.
5. X-ray structure analysis
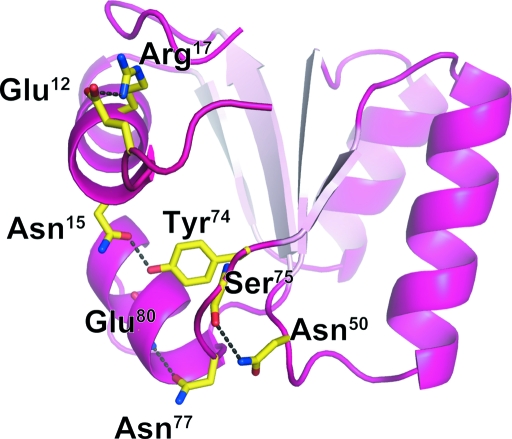
The structure was solved by the molecular-replacement method using the program MOLREP (Vagin & Teplyakov, 2010 ▶). The crystal structure of L30e from the yeast Saccharomyces cerevisiae in a fusion with MBP (PDB entry 1nmu, chain B; Chao et al., 2003 ▶) was used as a search model. The sequence identity between hL30e and yeast L30e is 53.9%. Structure refinement was carried out using REFMAC5 (Murshudov et al., 2011 ▶). After introducing an anisotropic B-factor model, the final model was refined to an R free factor of 19.0% and an R factor of 21.6% with root-mean-square deviations of 0.008 Å in bond lengths and 1.157° in bond angles for all reflections between 50 and 1.6 Å resolution. Table 1 ▶ presents a summary of the statistics of structure refinement. The stereochemical properties of the structure were assessed by MolProbity (Chen et al., 2010 ▶) and Coot (Emsley et al., 2010 ▶) and showed no residues in the disallowed or generously allowed regions of the Ramachandran plot. The final model comprises Lys9–Ile105 out of a total of 115 residues of hL30e; the N-terminal residues containing part of TEV-recognition site and the C-terminal residues and extra His×6 tag are missing. The structure of hL30e was very similar to that of yeast L30e, being folded into a three-layer α/β/α sandwich topology (Fig. 2 ▶), and superimposed with an r.m.s.d. of 1.07 Å using 97 Cα atoms. However, amino-acid residues that are not conserved between human and yeast (but are conserved in higher animals) contribute to stabilizing sharp turns via hydrogen bonds. The interactions between these residues are Glu12 (side chain)–Arg17 (side chain), Asn15 (side chain)–Tyr74 (side chain), Asn50 (side chain)–Ser75 (main-chain carbonyl) and Asn77 (side chain)–Glu80 (main-chain amide). These interactions may play roles in making these side chains less mobile in higher eukaryotes.
Figure 2.
Cartoon representation of human L30e. Specific intramolecular interactions that are not observed in yeast L30e are shown with amino-acid residues in stick representation and hydrogen bonds as black dashed lines.
Supplementary Material
PDB reference: ribosomal protein L30e, 3vi6
Acknowledgments
We are grateful to the beamline staff of PF-AR (Tsukuba, Japan) for technical help during data collection. This project was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
References
- Allmang, C., Carbon, P. & Krol, A. (2002). RNA, 8, 1308–1318. [DOI] [PMC free article] [PubMed]
- Ben-Shem, A., Jenner, L., Yusupova, G. & Yusupov, M. (2010). Science, 330, 1203–1209. [DOI] [PubMed]
- Chao, J. A., Prasad, G. S., White, S. A., Stout, C. D. & Williamson, J. R. (2003). J. Mol. Biol. 326, 999–1004. [DOI] [PubMed]
- Chao, J. A. & Williamson, J. R. (2004). Structure, 12, 1165–1176. [DOI] [PubMed]
- Chavatte, L., Brown, B. A. & Driscoll, D. M. (2005). Nature Struct. Mol. Biol. 12, 408–416. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Cléry, A., Bourguignon-Igel, V., Allmang, C., Krol, A. & Branlant, C. (2007). Nucleic Acids Res. 35, 1868–1884. [DOI] [PMC free article] [PubMed]
- Copeland, P. R. & Driscoll, D. M. (1999). J. Biol. Chem. 274, 25447–25454. [DOI] [PubMed]
- Copeland, P. R., Fletcher, J. E., Carlson, B. A., Hatfield, D. L. & Driscoll, D. M. (2000). EMBO J. 19, 306–314. [DOI] [PMC free article] [PubMed]
- Donovan, J., Caban, K., Ranaweera, R., Gonzalez-Flores, J. N. & Copeland, P. R. (2008). J. Biol. Chem. 283, 35129–35139. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Eng, F. J. & Warner, J. R. (1991). Cell, 65, 797–804. [DOI] [PubMed]
- Fagegaltier, D., Hubert, N., Yamada, K., Mizutani, T., Carbon, P. & Krol, A. (2000). EMBO J. 19, 4796–4805. [DOI] [PMC free article] [PubMed]
- Halic, M., Becker, T., Frank, J., Spahn, C. M. & Beckmann, R. (2005). Nature Struct. Mol. Biol. 12, 467–468. [DOI] [PubMed]
- Klein, D. J., Schmeing, T. M., Moore, P. B. & Steitz, T. A. (2001). EMBO J. 20, 4214–4221. [DOI] [PMC free article] [PubMed]
- Latrèche, L., Jean-Jean, O., Driscoll, D. M. & Chavatte, L. (2009). Nucleic Acids Res. 37, 5868–5880. [DOI] [PMC free article] [PubMed]
- Li, B., Vilardell, J. & Warner, J. R. (1996). Proc. Natl Acad. Sci. USA, 93, 1596–1600. [DOI] [PMC free article] [PubMed]
- Mao, H., White, S. A. & Williamson, J. R. (1999). Nature Struct. Biol. 6, 1139–1147. [DOI] [PubMed]
- Mao, H. & Williamson, J. R. (1999). J. Mol. Biol. 292, 345–359. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Takeuchi, A., Schmitt, D., Chapple, C., Babaylova, E., Karpova, G., Guigo, R., Krol, A. & Allmang, C. (2009). Nucleic Acids Res. 37, 2126–2141. [DOI] [PMC free article] [PubMed]
- Tujebajeva, R. M., Copeland, P. R., Xu, X.-M., Carlson, B. A., Harney, J. W., Driscoll, D. M., Hatfield, D. L. & Berry, M. J. (2000). EMBO Rep. 1, 158–163. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Vilardell, J. & Warner, J. R. (1994). Genes Dev. 8, 211–220. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: ribosomal protein L30e, 3vi6