The crystal structure of the H107R variant of the extracellular domain of mouse NKR-P1A was determined using X-ray diffraction from a merohedrally twinned crystal.
Keywords: NKR-P1A, merohedral twinning, mutation
Abstract
The structure of the H107R variant of the extracellular domain of the mouse natural killer cell receptor NKR-P1A has been determined by X-ray diffraction at 2.3 Å resolution from a merohedrally twinned crystal. Unlike the structure of the wild-type receptor in space group I4122 with a single chain per asymmetric unit, the crystals of the variant belonged to space group I41 with a dimer in the asymmetric unit. Different degrees of merohedral twinning were detected in five data sets collected from different crystals. The mutation does not have a significant impact on the overall structure, but led to the binding of an additional phosphate ion at the interface of the molecules.
1. Introduction
Natural killer (NK) cells play an important role in the innate immune response against viruses, parasites, intracellular bacteria and tumour cells (Makrigiannis et al., 2001 ▶; Bezouška et al., 1994 ▶). NK cells eliminate their targets using two distinct mechanisms: cytokine secretion and cell-mediated cytotoxicity (Mesci et al., 2006 ▶). The activity towards the target cells is regulated by a large number of receptors on the cell surface (Vivier et al., 2008 ▶).
The activating and inhibitory receptors of NK cells fall into two distinct structural classes: those belonging to the immunoglobulin superfamily and those of the C-type lectin-like (CTL) type. The receptors of the NKR-P1 family belong to the CTL type (Kolenko et al., 2011 ▶). When an NK cell encounters a target cell, the NKR-P1 receptors are expected to engage in interactions with their protein partners expressed on the surface of the target. Although the protein ligands have been identified as cell-surface CTL proteins, the details of the interactions as well as the exact route of NKR-P1 signalling are not yet fully understood. Recent studies have supported the participation of NKR-P1 receptors in protein–protein interactions with C-type lectin-related (Clr) molecules, e.g. LLT1 is a ligand of human NKR-P1 (Aldemir et al., 2005 ▶; Rosen et al., 2005 ▶) and Clrb/Ocil binds mouse NKR-P1B and NKR-D (Iizuka et al., 2003 ▶; Carlyle et al., 2004 ▶; Kveberg et al., 2009 ▶). The function of the remaining receptors is predicted from the amino-acid composition of the intracellular part, i.e. whether it contains the immunoreceptor tyrosin-based inhibitory motif (ITIM) or activation motif (ITAM) (Mesci et al., 2006 ▶).
The recently determined crystal structure of the extracellular domain of the mouse NKR-P1A receptor (mNKR-P1A) was the first structure of a representative of the NKR-P1 family. mNKR-P1A is formed by a compact CTL core and an extended loop that participates in domain swapping. A potential role of the swapped loop in natural ligand binding has been suggested by in silico studies (Sovová et al., 2011 ▶). Even if a protein ligand (or ligands) for mNKR-P1A were suggested, firm proof of the interactions on the molecular level and of complex formation and consequent signalling-cascade events is still lacking.
In a series of cloning and protein-expression experiments, a mutant H107R was unintentionally generated which could be purified to sufficient purity and homogeneity and crystallized. In the process of structure solution the mutation of residue 107 was observed and was confirmed by sequencing. The mutation causes interesting behaviour of the receptor dimers, affects the symmetry of the dimer interface and gives rise to twinning.
2. Methods
2.1. Protein preparation and crystallization
The plasmid for the expression of mNKR-P1A was constructed as described previously (Kolenko et al., 2011 ▶). DNA-sequence analysis of the constructed plasmid revealed a novel point mutation in the gene for NKR-P1A leading to the substitution of His107 by Arg. Recombinant H107R variant was expressed and purified as described previously. Briefly, the plasmid was transformed into Escherichia coli BL21 (DE3) Gold (Stratagene). The protein was produced as inclusion bodies and refolded in vitro. The refolded protein was purified using ion-exchange chromatography on Q Sepharose FF (1.0 × 6.0 cm) and Superdex 75 HR 10/300 GL columns (GE Healthcare). The monodispersity of the monomeric protein solution was further assessed by dynamic light-scattering and SDS–PAGE analysis.
Prior to crystallization, the protein was concentrated to 10 mg ml−1 in a buffer solution consisting of 50 mM NaCl, 1 mM NaN3, 10 mM HEPES pH 7.5. The search for a novel crystallization condition was also performed using the PolyA and PolyB screens (Skálová et al., 2010 ▶). However, the only well diffracting crystals grew from 0.3 M ammonium phosphate, which was the original crystallization condition for wild-type mNKR-P1A. Tetragonal bipyramids appeared after 4 d at a temperature of 291 K. Prior to diffraction experiments, the crystals were flash-cooled in liquid nitrogen with 20% ethylene glycol as a cryoprotectant.
2.2. Data collection and structure determination
Four diffraction data sets were collected from different crystals on a Gemini Enhanced Ultra diffractometer at copper wavelength using an Atlas CCD detector (Agilent Technologies) at the Institute of Physics AS CR, v.v.i., Prague. The data set with the highest resolution was collected on beamline 14.1 of BESSY II at the Helmholtz-Zentrum Berlin. The synchrotron data set was collected at 100 K; all other data sets were collected at 120 K. Integration of the diffraction images was performed using the XDS program package (Kabsch, 2010 ▶) and scaling was performed using SCALA from the CCP4 program package (Winn et al., 2011 ▶). The crystals belonged to space group I41. Analysis of the diffraction data revealed variable merohedral twinning of all crystals (αtwin ≃ 0.09–0.26, twin law k, h, −l). The data-processing statistics are given in Table 1 ▶.
Table 1. Data-collection statistics.
Values in parentheses are for the outer resolution shell.
| X-ray source† | BESSY II | IP | IP | IP | IP |
| Wavelength () | 0.918 | 1.542 | 1.542 | 1.542 | 1.542 |
| Resolution () | 50.02.3 (2.422.30) | 40.82.6 (2.672.60) | 30.03.0 (3.163.00) | 40.53.1 (3.273.10) | 40.03.5 (3.623.50) |
| Space group | I41 | I41 | I41 | I41 | I41 |
| Unit-cell parameters () | |||||
| a = b | 64.7 | 64.3 | 64.0 | 64.4 | 64.1 |
| c | 156.1 | 158.1 | 157.0 | 156.1 | 157.5 |
| No. of observations | 107152 (15469) | 37107 (2658) | 20972 (2880) | 14941 (2168) | 12419 (1223) |
| No. of unique reflections | 14248 (2064) | 9801 (719) | 6301 (908) | 5417 (804) | 3933 (387) |
| Completeness (%) | 100.0 (100.0) | 99.4 (98.8) | 99.5 (99.6) | 99.5 (95.0) | 97.8 (99.2) |
| Multiplicity | 7.5 (7.5) | 3.8 (3.7) | 3.3 (3.2) | 2.8 (2.7) | 3.2 (3.2) |
| I/(I) | 16.0 (3.4) | 9.4 (2.5) | 10.0 (2.5) | 7.6 (2.4) | 9.2 (3.3) |
| Wilson B factor (2) | 49 | 76 | 71 | 80 | 46 |
| R merge | 0.072 (0.546) | 0.049 (0.696) | 0.107 (0.460) | 0.100 (0.406) | 0.120 (0.321) |
| twin | 0.14 | 0.09 | 0.26 | 0.19 | 0.21 |
BESSY II, beamline 14.1 at the BESSY II synchrotron-radiation source, Helmholtz-Zentrum Berlin; IP, Agilent Technologies Gemini Enhanced Ultra sealed tube at Institute of Physics AS CR, v.v.i., Prague.
The structure was solved by molecular replacement with the program MOLREP (Vagin & Teplyakov, 2010 ▶) using the structure of wild-type mNKR-P1A as a search model (PDB entry 3m9z; Kolenko et al., 2011 ▶). Structure refinement against experimental intensities with automated twin detection was carried out with REFMAC5 (Murshudov et al., 2011 ▶) using a built-in twin-refinement protocol. Manual corrections of the model were performed with the program Coot (Emsley & Cowtan, 2004 ▶).
Structure validation was carried out with MolProbity (Chen et al., 2010 ▶). All residues of the model were found in the allowed regions of the Ramachandran plot. The statistics of structure refinement are given in Table 2 ▶. The coordinates and structure factors have been deposited in the PDB (Berman et al., 2000 ▶) with access code 3t3a.
Table 2. Refinement statistics for the synchrotron data set.
| Resolution () | 50.02.3 |
| No. of residues in the asymmetric unit | 241 |
| Localized ions | 3 PO4 3 |
| No. of water molecules | 56 |
| No. of localized non-H atoms | 2016 |
| Mean ADP for protein atoms (2) | 45 |
| Mean ADP for solvent (2) | 48 |
| Mean ADP for all atoms (2) | 45 |
| R.m.s.d. bonds () | 0.017 |
| R.m.s.d. angles () | 1.620 |
| Ramachandran plot | |
| No. of residues in favoured region | 224 |
| No. of residues in allowed region | 9 |
| No. of residues in outlier region | 0 |
| R work | 0.188 |
| R free | 0.216 |
| No. of reflections for R free calculation | 708 (5%) |
| R all | 0.191 |
3. Results and discussion
3.1. Overall structure
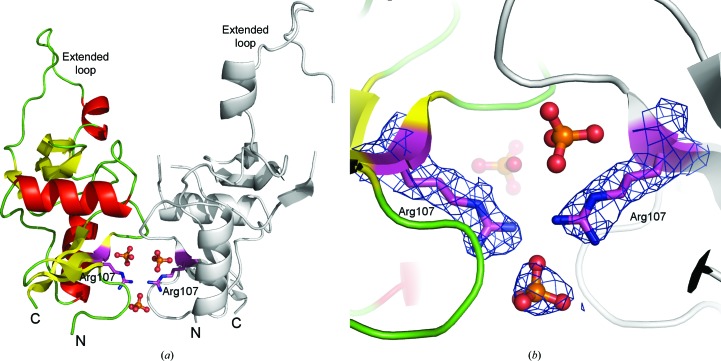
We produced and crystallized the H107R variant of the extracellular domain of the mouse NK-cell activation receptor NKR-P1A. The crystals belonged to space group I41 with merohedral twinning. In a search for a sample that would provide untwinned data, five data sets were collected. However, twinning with a variable twin factor was repeatedly observed. The structure was determined by molecular replacement using the extracellular domain of wild-type mNKR-P1A as a search model. The structure contained two protein chains consisting of residues Glu93–Leu214 with three phosphate ions attached at the interface of the molecules (see Fig. 1 ▶). The quality of the observed electron density did not allow the localization of all of the atoms of residues Tyr149, Pro161 and Asp162 in chain B. Two phosphate ions are bound close to the interface between the monomers in the asymmetric unit (similar to the structure of the wild type) and a third phosphate ion with partial occupancy was localized between Arg107 residues belonging to opposite chains. The latter participates in interactions with different side chains from the two monomers (see Fig. 1 ▶ b) and represents some asymmetry in this region.
Figure 1.
Overall structure of the H107R variant of mouse NKR-P1A and a detailed view of the interface between monomers. (a) Chain A is represented by coloured secondary-structure elements, chain B is shown in grey, phosphate ions are shown in ball-and-stick representation and side chains of Arg107 are represented by sticks. (b) A detailed view of the interface between the monomers and the binding of the additional phosphate ion. The 2F o − F c map around the phosphate and Arg107 is contoured at the 1σ level (blue).
The two protein chains are highly similar in structure (r.m.s.d. of 1.7 Å for Cα atoms, secondary-structure matching superposition algorithm) and each is formed by a compact core region similar to most C-type lectins (two main α-helices and two antiparallel β-sheets stabilized by three disulfide bridges) and a loop consisting of 30 residues (Tyr158–Asp187) which exhibits domain swapping with a symmetry-related molecule.
3.2. Interactions within the crystal lattice
Surprisingly, a more extensive interaction was found between two symmetry-related molecules within the crystal lattice than between the monomers in the asymmetric unit. This interaction corresponds to the domain-swapping effect of the extended loops Tyr158–Asp187, with a total interface area of about 1700 Å2 involving hydrogen bonds and hydrophobic interactions. The potential participation of the loop in interactions with Clr molecules has been suggested, but has not yet been structurally confirmed (Kolenko et al., 2011 ▶). The mutated residue Arg107 is not localized in this region, so the mutation at this residue could not affect the formation of the domain-swapped dimers observed in the wild-type structure.
The monomers in the asymmetric unit are related by a noncrystallographic twofold axis and form a noncovalent dimer with an interface area of 730 Å (Krissinel & Henrick, 2007 ▶). The difference in the size of the asymmetric unit of the wild-type structure and the mutant structure is exactly reflected by the presence of the additional twofold symmetry (monomer and dimer in the asymmetric unit; space groups I4122 and I41, respectively). The twinning observed in the case of the H107R variant also corresponds to the ‘missing’ twofold symmetry with respect to the wild-type crystals. The arrangement of the molecules in the asymmetric unit of the H107R variant has previously been confirmed to be a potential biological unit by cross-linking experiments (Kolenko et al., 2011 ▶). The mutated residue Arg107 is located at this interface and together with the partially occupied phosphate ion contributes to stabilization of the interface (see Fig. 1 ▶ b). The His107 residues in the wild-type structure are too distant from each other to participate in any interaction.
3.3. H107R mutant versus wild type
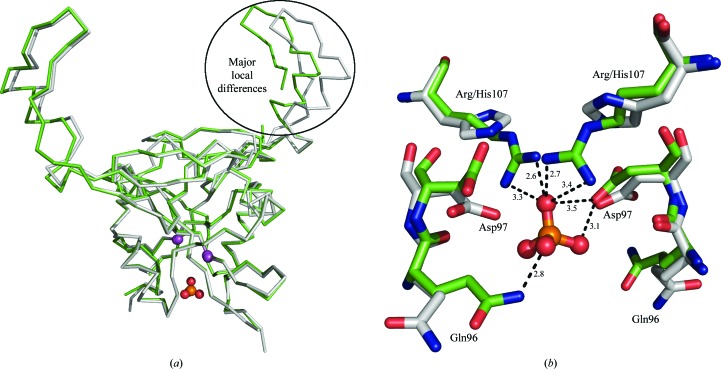
The overall structure of the monomers does not differ significantly from the wild-type structure (Fig. 2 ▶). The r.m.s.d. between Cα atoms of the wild type and chains A and B of the mutant are 0.6 and 1.8 Å, respectively. The more similar chain A of the mutant contains only one region, consisting of seven residues (174–180), in which the differences between Cα atoms exceed 1 Å. The most significant structural differences in the mutated chain B are observed in the region 148–150 (in which the differences exceed 3 Å) and in a large C-terminal region containing residues 163–205, in which the differences between equivalent Cα atoms exceed 2 Å for the majority of superimposed pairs. However, these parts of the protein are not involved in the formation of the interface containing residue 107. Nevertheless, the mutation and binding of phosphate ion at the interface probably plays a role in the twinning of the crystals of the H107R mutant.
Figure 2.
Structural alignment of dimers of H107R-variant (green) and wild-type (grey) mNKR-P1A. (a) Chains are represented by Cα traces, the Cα atoms of Arg/His107 are shown as magenta spheres and the additional phosphate ion is shown in ball-and-stick representation. The region of major local differences, the extended loop (chain B in the H107R variant), is highlighted by an oval. (b) A detailed view of the mutation-induced asymmetric binding of the phosphate ion compared with the wild type. Amino acids are represented by sticks and the phosphate ion is shown in ball-and-stick representation; the hydrogen-bonding network with distances in Å is only displayed for the discussed phosphate ion.
We analyzed the interface containing residues 107 using the PISA server (Krissinel & Henrick, 2007 ▶). In the cystal structure of the wild-type receptor, a buried surface area of about 650 Å2 containing 11 hydrogen bonds was observed. The interface of the H107R mutant variant contains 13 hydrogen bonds, with a buried surface of about 730 Å2. Contrary to our expectations, this analysis suggests that the H107R mutation caused a strengthening of the interface of interest.
Although many trials to improve the quality of the crystals were performed, the highest resolution achieved for the H107R variant (2.3 Å) is not sufficient for interpretation of the fine structural differences that lead to merohedral twinning in this case. Our interpretation of the observed phenomenon is as follows. The asymmetric binding of the phosphate ions causes a loss of the perfect symmetry in dimers of mNKR-P1A, which are then organized in two possible orientations in the crystal owing to a very high structure similarity between the monomers. The ratio between the orientations is random (varying twin factor in different crystals). The structure of the mutant in the absence of phosphate would be of particular interest. Unfortunately, despite an extensive search such conditions were not found. The present structure shows how small structural differences may lead to defects in crystal growth that affect structure analysis.
The occurrence of nonsymmetrical dimers in the asymmetric unit of the crystal unfortunately does not imply their presence in solution or their natural role in the NK cells, although to some extent it increases support for the natural occurrence of such dimers when previously confirmed by alternative methods (Kolenko et al., 2011 ▶).
Supplementary Material
PDB reference: H107R mNKR-P1A, 3t3a
Structure factors: contains datablock(s) r3t3asf. DOI: 10.1107/S1744309111046203/hv52033T3Asup1.hkl
Acknowledgments
The authors wish to acknowledge the support of the EC Integrated project SPINE2-Complexes (LSHG-CT-2006-031220), ELISA grant No. 226716 (synchrotron-access funding, projects 09.1.81077 and 09.2.90262), the Academy of Sciences (Praemium Academiae), the Czech Science Foundation (project Nos. 305/07/1073 and P302/11/0855), the Ministry of Education of the Czech Republic (MSM 21620808 and 1M0505) and Charles University Prague (263209/2011 and 403211/2011).
References
- Aldemir, H., Prod’homme, V., Dumaurier, M.-J., Retiere, C., Poupon, G., Cazareth, J., Bihl, F. & Braud, V. M. (2005). J. Immunol. 175, 7791–7795. [DOI] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed]
- Bezouška, K., Yuen, C.-T., O’Brien, J., Childs, R. A., Chai, W., Lawson, A. M., Drbal, K., Fišerová, A., Pospíšil, M. & Feizi, T. (1994). Nature (London), 372, 150–157. [DOI] [PubMed]
- Carlyle, J. R., Jamieson, A. M., Gasser, S., Clingan, C. S., Arase, H. & Raulet, D. H. (2004). Proc. Natl Acad. Sci. USA, 101, 3527–3532. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Iizuka, K., Naidenko, O. V., Plougastel, B. F., Fremont, D. H. & Yokoyama, W. M. (2003). Nature Immunol. 4, 801–807. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kolenko, P., Rozbeský, D., Vaněk, O., Kopecký, V., Hofbauerová, K., Novák, P., Pompach, P., Hašek, J., Skálová, T., Bezouška, K. & Dohnálek, J. (2011). J. Struct. Biol. 175, 434–441. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Kveberg, L., Dai, K. Z., Westgaard, I. H., Daws, M. R., Fossum, S., Naper, C. & Vaage, J. T. (2009). Eur. J. Immunol. 39, 541–551. [DOI] [PubMed]
- Makrigiannis, A. P., Pau, A. T., Saleh, A., Winkler-Pickett, R., Ortaldo, J. R. & Anderson, S. K. (2001). J. Immunol. 166, 5034–5043. [DOI] [PubMed]
- Mesci, A., Ljutic, B., Makrigiannis, A. P. & Carlyle, J. R. (2006). Immunol. Res. 35, 13–26. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Rosen, D. B., Bettadapura, J., Alsharifi, M., Mathew, P. A., Warren, H. S. & Lanier, L. L. (2005). J. Immunol. 176, 7796–7799. [DOI] [PubMed]
- Skálová, T., Dušková, J., Hašek, J., Kolenko, P., Štěpánková, A. & Dohnálek, J. (2010). J. Appl. Cryst. 43, 737–742.
- Sovová, Z., Kopecký, V., Pazderka, T., Hofbauerová, K., Rozbeský, D., Vaněk, O., Bezouška, K. & Ettrich, R. (2011). J. Mol. Model. 17, 1353–1370. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Vivier, E., Tomasello, E., Baratin, M., Walzer, T. & Ugolini, S. (2008). Nature Immunol. 9, 503–510. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: H107R mNKR-P1A, 3t3a
Structure factors: contains datablock(s) r3t3asf. DOI: 10.1107/S1744309111046203/hv52033T3Asup1.hkl