Dihydrodipicolinate synthase from the common grapevine V. vinifera has been cloned, expressed, purified and crystallized in the presence of the substrate pyruvate by in-drop hexahistidine-tag cleavage. A diffraction data set has been collected to a resolution of 2.2 Å.
Keywords: diaminopimelate, dihydrodipicolinate, grapes, herbicide, lysine, metabolism, quaternary structure, thrombin, wine
Abstract
Dihydrodipicolinate synthase (DHDPS) catalyses the first committed step of the lysine-biosynthesis pathway in bacteria, plants and some fungi. This study describes the cloning, expression, purification and crystallization of DHDPS from the grapevine Vitis vinifera (Vv-DHDPS). Following in-drop cleavage of the hexahistidine tag, cocrystals of Vv-DHDPS with the substrate pyruvate were grown in 0.1 M Bis-Tris propane pH 8.2, 0.2 M sodium bromide, 20%(w/v) PEG 3350. X-ray diffraction data in space group P1 at a resolution of 2.2 Å are presented. Preliminary diffraction data analysis indicated the presence of eight molecules per asymmetric unit (V M = 2.55 Å3 Da−1, 52% solvent content). The pending crystal structure of Vv-DHDPS will provide insight into the molecular evolution in quaternary structure of DHDPS enzymes.
1. Introduction
Lysine biosynthesis is unique to bacteria, plants and some fungi (Hutton et al., 2007 ▶; Dogovski et al., 2009 ▶). Study of this pathway has attracted much interest as lysine is an essential bacterial metabolite (Kobayashi et al., 2003 ▶) and offers potential for the development of novel antimicrobials (Hutton et al., 2007 ▶; Dogovski et al., 2009 ▶). Accordingly, we have been engaged in the study of the structure, function, regulation and molecular evolution of enzymes of lysine biosynthesis from a variety of bacteria (Dobson, Griffin et al., 2005 ▶, 2008 ▶; Perugini et al., 2005 ▶; Burgess, Dobson, Bailey et al., 2008 ▶; Burgess, Dobson, Dogovski et al., 2008 ▶; Devenish et al., 2008 ▶; Griffin et al., 2008 ▶, 2010 ▶; Kefala et al., 2008 ▶; Pearce et al., 2008 ▶; Atkinson et al., 2009 ▶; Domigan et al., 2009 ▶; Voss et al., 2009 ▶, 2010 ▶; Dommaraju et al., 2010 ▶, 2011 ▶; Hor et al., 2010 ▶; Sibarani et al., 2010 ▶; Wubben et al., 2010 ▶; Evans et al., 2011 ▶). In particular, knowledge gained from these studies is being applied to the rational design of inhibitors (Turner et al., 2005 ▶; Boughton, Dobson et al., 2008 ▶; Boughton, Griffin et al., 2008 ▶; Mitsakos et al., 2008 ▶). In contrast, studies of lysine biosynthesis in plants is of interest to agricultural science, given that lysine is one of the most limiting nutrients in plants (Bright & Shewry, 1983 ▶; Kumpaisal et al., 1987 ▶; Perl et al., 1992 ▶; Shaul & Galili, 1992 ▶; Chatterjee et al., 1994 ▶; Silk & Matthews, 1997 ▶). Of particular focus to the study of lysine biosynthesis in plants and bacteria is the enzyme dihydrodipicolinate synthase (DHDPS), which catalyses the first committed step of the pathway. Specifically, DHDPS catalyses the condensation of pyruvate and (S)-aspartate semialdehyde [(S)-ASA] to form (4S)-4-hydroxy-2,3,4,5-tetrahydro-(2S)-dipicolinic acid (HTPA) (Blickling, Renner et al., 1997 ▶; Hutton et al., 2007 ▶; Dogovski et al., 2009 ▶). The reaction proceeds via a ping-pong kinetic mechanism in which pyruvate binds as a Schiff base to an active-site lysine residue (Lys161 in E. coli DHDPS; Blickling, Renner et al., 1997 ▶; Dobson, Gerrard et al., 2004 ▶).
The structures of DHDPS enzymes from a number of bacterial species have been determined, including Bacillus anthracis (Blagova et al., 2006 ▶; Voss et al., 2010 ▶), Corynebacterium glutamicum (Rice et al., 2008 ▶), Escherichia coli (Mirwaldt et al., 1995 ▶; Dobson, Griffin et al., 2005 ▶), Hahella chejuensis (Kang et al., 2010 ▶), Methanococcus jannaschii (Padmanabhan et al., 2009 ▶), Mycobacterium tuberculosis (Kefala et al., 2008 ▶), Neisseria meningitides (Devenish et al., 2009 ▶), Pseudomonas aeruginosa (Kaur et al., 2011 ▶), Staphylococcus aureus (Burgess et al., 2008 ▶; Girish et al., 2008 ▶) and Thermotoga maritima (Pearce et al., 2006 ▶). The structure of DHDPS from Nicotiana sylvestris (Blickling, Beisel et al., 1997 ▶) is the only structure of the enzyme determined from a plant to date.
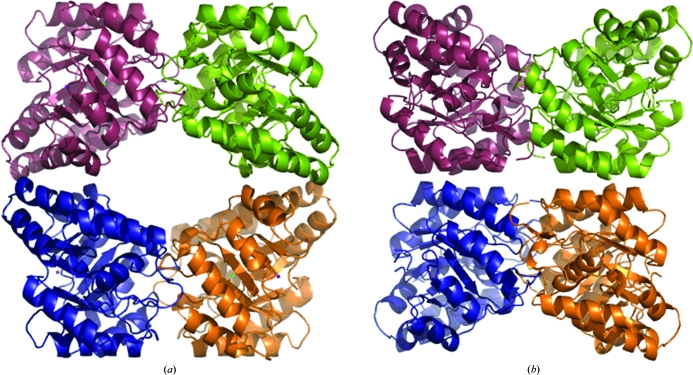
The bacterial form of the enzyme is usually a homotetramer of four identical (α/β)8-barrel monomers, with the active site situated near the centre of the barrel. The typical bacterial DHDPS tetramer can be described as a ‘head-to-head’ dimer of dimers, as depicted in Fig. 1 ▶(a). Both the active and allosteric sites are located at the so-called ‘tight-dimer interface’, with the allosteric cleft where lysine binds at the top and bottom of the tetramer (Dobson, Griffin et al., 2005 ▶; Fig. 1 ▶ a). Similarly, the structure of DHDPS from the plant N. sylvestris (Blickling, Beisel et al., 1997 ▶) also consists of a homotetramer. However, the plant DHDPS homotetramer forms an alternative dimer-of-dimers architecture that can be described as a ‘back-to-back’ conformation (Fig. 1 ▶ b). As in the bacterial structure, the active and allosteric sites are located at the tight-dimer interface, but given the ‘back-to-back’ architecture the allosteric clefts accommodating lysine face each other in the interior of the structure (Fig. 1 ▶ b).
Figure 1.
Dihydrodipicolinate synthase (DHDPS) from (a) E. coli (PDB entry 1dhp; Mirwaldt et al., 1995 ▶) and (b) N. sylvestris (Blickling, Beisel et al., 1997 ▶).
In this study, we sought to clone, express, purify and crystallize DHDPS from the common grapevine Vitis vinifera (Vv-DHDPS) in order to determine the macromolecular structure of this enzyme and to ascertain whether the ‘back-to-back’ architecture of N. sylvestris DHDPS (Fig. 1 ▶ b) is typical of plant DHDPS structures.
2. Material and methods
2.1. Identification of the gene encoding V. vinefera DHDPS
The putative Vv-DHDPS sequence was identified by performing a BLASTP (NCBI) search against the V. vinifera PN40024 genome (Jaillon et al., 2007 ▶) using E. coli K12 substrain W3110 (NCBI reference AP_003064.1) DHDPS as the query sequence. Inspection of the primary amino-acid sequence of the putative V. vinifera DHDPS enzyme (NCBI entry LOC 100262665) showed that it contains all the key conserved residues known to be essential for catalysis (Dobson et al., 2009 ▶; Dobson, Devenish et al., 2005 ▶; Dobson, Valegård et al., 2004 ▶; Dobson, Griffin et al., 2008 ▶). This sequence was used to design a codon-optimized gene intended for overexpression in E. coli. The gene was synthesized commercially by GeneArt and supplied in a cloning vector designated pMAdapA.
2.2. Construction of a Vv-DHDPS expression vector
A Vv-DHDPS expression vector with an amino-terminal hexahistidine tag and downstream thrombin cleavage site was constructed to facilitate efficient purification of recombinant protein by affinity chromatography. The gene encoding Vv-DHDPS was amplified by PCR using pMAdapA template DNA and the primer pair 5′-CATATGGCGGTGATTCCGAGCTTTCA-3′ (forward) and 5′-GGATCCTTAATAGCGGCCCAC-3′ (reverse), which contain NdeI and BamHI restriction sites (bold). The amplified product was then cloned into pCR-BluntII-TOPO (Invitrogen) and the presence of the gene encoding Vv-DHDPS and adaptor restriction sites was confirmed by dideoxynucleotide sequencing. The vector was digested with NdeI and BamHI and the Vv-DHDPS gene was subcloned into the corresponding sites in pET28a to create pET28a-dapA.
2.3. Expression and purification of Vv-DHDPS
E. coli BL21 (DE3) cells were transformed with pET28a-dapA and cultivated in flasks containing 1 l Luria–Bertani broth (25 µg ml−1 kanamycin) at 298 K with continuous shaking (180 rev min−1) until the culture reached an OD600nm of 0.6. The flasks were then transferred to a 289 K incubator for 1 h before treatment with 1.0 mM isopropyl β-d-1-thiogalactopyranoside to induce the expression of recombinant Vv-DHDPS. The cultures were incubated overnight at 289 K with continuous shaking (180 rev min−1) and the cells were pelleted by centrifugation at 16 000g for 20 min.
The cell pellet was resuspended in buffer I (20 mM Tris, 500 mM NaCl, 20 mM imidazole pH 8.0) and the cells were lysed by sonication using an MSE Soniprep 150 sonicator with an 18 mm diameter probe at a power output of 10–14 µm. Cellular debris was pelleted by centrifugation (30 min, 48 000g) and the soluble fraction was applied onto a 5 ml HisTrap HP column (GE Healthcare) which was pre-equilibrated with three column volumes of buffer I. The column was washed in buffer I until a steady baseline absorbance (A 280nm) was observed. Vv-DHDPS was eluted via a two-step gradient. Firstly, a gradient of 100–70% buffer I:0–30% buffer II (20 mM Tris, 500 mM NaCl, 500 mM imidazole pH 8.0) was applied over three column volumes for elution of contaminant proteins. Once a steady baseline absorbance (A 280nm) was observed, a gradient of 70–50% buffer I:30–50% buffer II was applied over three column volumes for elution of recombinant Vv-DHDPS. Vv-DHDPS eluted at an imidazole concentration of approximately 250 mM.
The eluate was dialysed overnight against buffer III (20 mM Tris, 150 mM NaCl pH 8.0 with or without 20 mM pyruvate) before storage at 193 K. Prior to crystallization, the protein was thawed overnight at 277 K followed by size-exclusion liquid chromatography using an XK 16/20 column packed with Superdex 200 resin (bed volume of 30 ml; GE Healthcare). If required, the protein solution was concentrated to approximately 10 mg ml−1 using a 10 kDa cutoff Centricon (Millipore) prior to gel-filtration chromatography. All purification steps were carried out at 277 K.
2.4. Analytical size-exclusion liquid chromatography
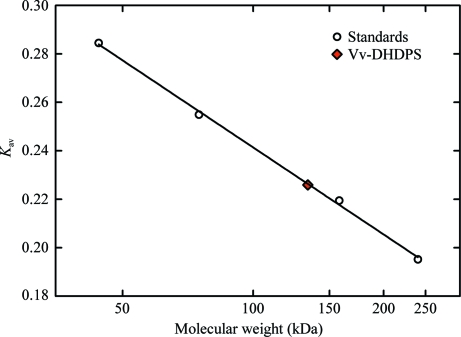
Analytical size-exclusion liquid chromatography was employed to estimate the native molecular weight of recombinant Vv-DHDPS. Samples of Vv-DHDPS (100 µl, 4 mg ml−1) and globular protein standards (ovalbumin, conalbumin, aldolase and catalase; 100 µl, 4 mg ml−1) were loaded onto a 10 × 300 mm Superose 12 column (GE Healthcare; fractionation range 1000–300 000 Da) and eluted with buffer III at 0.5 ml min−1 at 277 K. The absorbance at 280 nm was monitored as a function of elution volume. The void volume (V 0) and total volume (V t) were determined from the elution of blue dextran (2000 kDa) and riboflavin (376 Da), respectively. The partition coefficients (K av) of each sample were calculated using the formula K av = (V e − V 0)/(V t − V 0) and plotted as a function of log10(molecular weight) to construct a calibration curve to estimate the molecular weight of recombinant Vv-DHDPS (Fig. 2 ▶).
Figure 2.
Analytical gel-filtration liquid chromatography of Vv-DHDPS. The partition coefficient (K av) is plotted as a function of log10(molecular weight). Experiments were performed on a 10 × 300 mm Superose 12 column using ovalbumin (44 kDa), conalbumin (75 kDa), aldolase (158 kDa) and catalase (240 kDa) as globular protein standards (open circles) and employing blue dextran (2000 kDa) and riboflavin (376 Da) to determine the void and total volume of the column. Vv-DHDPS (red diamond) elutes at approximately 131 kDa, suggesting that the enzyme adopts either a trimeric or tetrameric quaternary structure (the M r of Vv-DHDPS is 37 876).
2.5. Protein crystallization
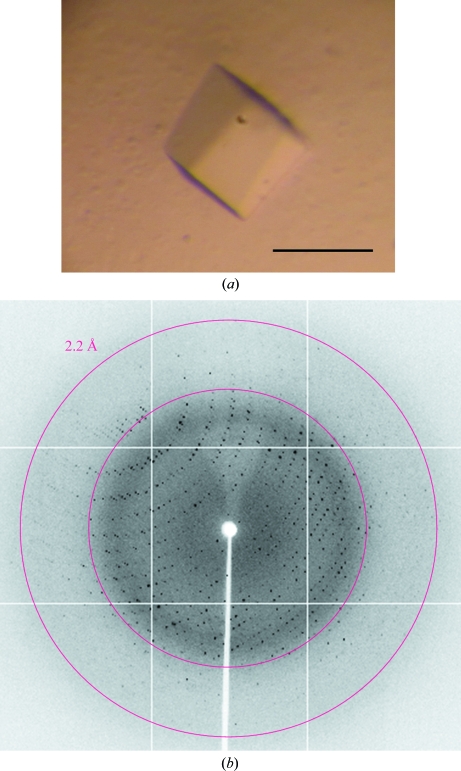
Crystallization studies were initially conducted using a 10 mg ml−1 preparation of Vv-DHDPS in 20 mM Tris, 150 mM NaCl pH 8.0. Protein crystallization trials were performed at the CSIRO node of the Bio21 Collaborative Crystallization Centre (C3; http://www.csiro.au/c3/) using the PACT Suite and the JCSG+ Suite crystal screens (Qiagen; Newman et al., 2005 ▶) at 281 and 293 K as previously described (Burgess et al., 2008 ▶; Dobson, Atkinson et al., 2008 ▶; Atkinson et al., 2009 ▶; Voss et al., 2009 ▶; Dommaraju et al., 2010 ▶; Hor et al., 2010 ▶; Sibarani et al., 2010 ▶; Wubben et al., 2010 ▶). Screens were performed using the sitting-drop vapour-diffusion method with droplets consisting of 150 nl protein solution and 150 nl reservoir solution. Crystals were not observed under these conditions, so the screen was repeated in the presence of 20 mM pyruvate, which is the first substrate to bind DHDPS (Dogovski et al., 2009 ▶) and has been shown to significantly stabilize other DHDPS enzymes (Blickling, Renner et al., 1997 ▶; Burgess et al., 2008 ▶; Kefala et al., 2008 ▶; Voss et al., 2010 ▶). Several PACT screen conditions produced crystals with needle-like morphology at 293 K. Optimization of the initial hits was carried out using the hanging-drop vapour-diffusion method. 2 µl protein solution and 2 µl precipitant solution were equilibrated against 1000 µl reservoir solution in 24-well Linbro plates at 293 K. It was noted that protein precipitation occurred in all conditions and crystal growth only resulted when the protein solution concentration was reduced to 2.5 mg ml−1. However, a needle morphology persisted, so an alternative approach which used in-drop protease cleavage (Wernimont & Edwards, 2009 ▶) of the hexahistidine tag was tested. 5 U (where 1 U cleaves 100 µg protein) of thrombin (GE Healthcare) and 100 mM CaCl2 were added to 115 µg Vv-DHDPS prior to hanging-drop crystallization as described above. A variety of crystal morphologies were observed, with the best diffracting crystal (Fig. 3 ▶ a) growing from a reservoir solution consisting of 0.1 M Bis-Tris propane pH 8.2, 0.2 M sodium bromide, 20%(w/v) PEG 3350.
Figure 3.
(a) Crystal of recombinant Vv-DHDPS with pyruvate. The bar indicates 100 µm. (b) X-ray diffraction image from the crystal of Vv-DHDPS with pyruvate in (a).
2.6. Data collection and processing
Diffraction data collection was carried out using a single crystal (Fig. 3 ▶ a) on the MX2 Micro Crystallography beamline (Evans & Pettifer, 2001 ▶; McPhillips et al., 2002 ▶) at the Australian Synchrotron, Clayton, Australia. This beamline uses a 03ID1 (3 m in-vacuum undulator) source and an ADSC Q315r CCD detector. A crystal which had been flash-cooled was mounted on the beamline in a cold nitrogen stream at 110 K. With the detector positioned 280 mm from the crystal, data were collected in 0.5° steps for one 360° pass, with 80% attenuation and an exposure time of 2 s.
Indexing and integration of the data was performed using the program iMOSFLM (Battye et al., 2011 ▶). POINTLESS from the CCP4 program suite (Winn et al., 2011 ▶) was run to verify the space group, and scaling and data reduction were performed using the program SCALA, also from CCP4. All relevant data-collection and processing parameters are given in Table 1 ▶. Images will be made available via the TARDIS server (Androulakis et al., 2008 ▶) once the structure has been solved and published.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution bin.
| Wavelength (Å) | 0.9536 |
| No. of images | 720 |
| Step range (°) | 0.5 |
| Space group | P1 |
| Unit-cell parameters (Å, °) | a = 70.6, b = 78.9, c = 135.4, α = 93.19, β = 95.02, γ = 100.61 |
| Resolution (Å) | 59–2.20 (2.26–2.20) |
| Observed reflections | 470484 (69507) |
| Unique reflections | 123307 (18027) |
| Completeness (%) | 97.4 (97.3) |
| Rmerge† | 0.108 (0.454) |
| Rr.i.m.‡ | 0.126 (0.527) |
| Rp.i.m.§ | 0.064 (0.267) |
| Mean I/σ(I) | 10.4 (3.1) |
| Multiplicity | 3.8 (3.9) |
| Wilson B value (Å2) | 22.33 |
| Molecules per asymmetric unit | 8 |
| Matthews coefficient VM (Å3 Da−1) | 2.55 |
| Solvent content (%) | 52 |
R
merge = 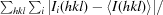
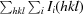 .
.
R
r.i.m. = 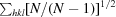
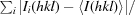
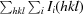 .
.
R
p.i.m. = 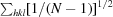
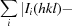
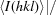
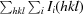 , where Ii(hkl) is the ith intensity measurement of reflection hkl, 〈I(hkl)〉 its average and N is the redundancy of a given reflection.
, where Ii(hkl) is the ith intensity measurement of reflection hkl, 〈I(hkl)〉 its average and N is the redundancy of a given reflection.
3. Results and discussion
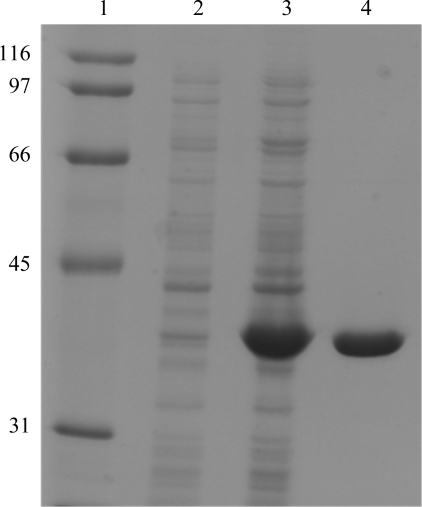
Approximately 5 mg pure protein was obtained from 1 l bacterial cell culture following the two-step purification procedure. The purity of the sample was at least 95% as estimated by SDS–PAGE (Fig. 4 ▶) and it was shown by size-exclusion liquid chromatography to exist as either a trimer or tetramer in aqueous solution (Fig. 2 ▶). As described in §2.5, initial attempts at crystallization using a Vv-DHDPS concentration of 10 mg ml−1 resulted in protein precipitation. Accordingly, the initial protein concentration was reduced fourfold and crystal growth was observed. In-drop cleavage of the hexahistidine tag using thrombin changed the morphology of the crystals from needle-like to a more three-dimensional habit. The crystallization conditions were then optimized to produce crystals of a flattened cubic shape (Fig. 3 ▶ a). In preparation for diffraction data collection, a single crystal was cryoprotected using 20%(v/v) glycerol in reservoir solution, mounted in a nylon loop and flash-cooled in liquid nitrogen. Diffraction data were collected from the crystal to a resolution of 2.2 Å (Fig. 3 ▶ b). The relevant data-collection and processing parameters are given in Table 1 ▶. The crystal belonged to space group P1, with unit-cell parameters a = 70.55, b = 78.90, c = 135.35 Å, α = 93.19, β = 95.02, γ = 100.61°. The P1 space group was verified using the program POINTLESS (Evans, 2006 ▶). Based on the deduced molecular weight of 35 850 Da, calculation of the Matthews coefficient suggested the presence of eight subunits per asymmetric unit, with an estimated solvent content of 52% (V M = 2.55 Å3 Da−1).
Figure 4.
Purification of recombinant Vv-DHDPS. SDS–PAGE gel showing the purification of Vv-DHDPS. Lane 1, molecular-weight markers (labelled in kDa); lane 2, non-induced whole cell lysate; lane 3, IPTG-treated whole cell lysate; lane 4: post IMAC chromatography.
Scaling and merging of the crystallographic data resulted in an overall R merge of 0.108 with an R merge of 0.454 in the highest resolution shell. Complete data-collection statistics are given in Table 1 ▶. Molecular replacement using the program Phaser (McCoy et al., 2007 ▶), employing E. coli DHDPS (monomer) as a search model (PDB entry 1yxc; Dobson, Griffin et al., 2005 ▶; 34% identity), showed an unambiguous solution with eight monomers in the asymmetric unit with a translation-function Z score of 582.1 and a final log-likelihood gain of 21 445. To verify that there was no higher order symmetry, we submitted the solution to the Zanuda server (http://www.york.ac.uk/chemistry/research/groups/ysbl). Further model building is currently under way. The pending structure will provide insight into the function and molecular evolution of plant DHDPS enzymes, thus complementing and developing the previous studies of N. sylvestris DHDPS (Blickling, Beisel et al., 1997 ▶; Fig. 1 ▶ b).
Acknowledgments
We would firstly like to acknowledge the support and assistance of the friendly staff at the CSIRO Collaborative Crystallization Centre at CSIRO Material Science and Engineering, Parkville, Melbourne, Australia and the MX beamline scientists at the Australian Synchrotron, Victoria, Australia. The views expressed herein are those of the authors and are not necessarily those of the owner or operator of the Australian Synchrotron. We would also like to thank all members of the Perugini laboratory for helpful discussions during the preparation of this manuscript. Finally, we acknowledge the Australian Research Council for providing a Future Fellowship for MAP and The University of Melbourne for providing project funding (FRGSS 2011 project grant). RCJD acknowledges the C. R. Roper Bequest for fellowship support and the New Zealand Royal Society Marsden Fund for funding support (UOC1013).
References
- Androulakis, S. et al. (2008). Acta Cryst. D64, 810–814.
- Atkinson, S. C., Dobson, R. C. J., Newman, J. M., Gorman, M. A., Dogovski, C., Parker, M. W. & Perugini, M. A. (2009). Acta Cryst. F65, 253–255. [DOI] [PMC free article] [PubMed]
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Blagova, E., Levdikov, V., Milioti, N., Fogg, M. J., Kalliomaa, A. K., Brannigan, J. A., Wilson, K. S. & Wilkinson, A. J. (2006). Proteins, 62, 297–301. [DOI] [PubMed]
- Blickling, S., Beisel, H. G., Bozic, D., Knäblein, J., Laber, B. & Huber, R. (1997). J. Mol. Biol. 274, 608–621. [DOI] [PubMed]
- Blickling, S., Renner, C., Laber, B., Pohlenz, H. D., Holak, T. A. & Huber, R. (1997). Biochemistry, 36, 24–33. [DOI] [PubMed]
- Boughton, B. A., Dobson, R. C. J., Gerrard, J. A. & Hutton, C. A. (2008). Bioorg. Med. Chem. Lett. 18, 460–463. [DOI] [PubMed]
- Boughton, B. A., Griffin, M. D. W., O’Donnell, P. A., Dobson, R. C. J., Perugini, M. A., Gerrard, J. A. & Hutton, C. A. (2008). Bioorg. Med. Chem. 16, 9975–9983. [DOI] [PubMed]
- Bright, S. W. J. & Shewry, P. R. (1983). CRC Crit. Rev. Plant Sci. 1, 49–93.
- Burgess, B. R., Dobson, R. C. J., Bailey, M. F., Atkinson, S. C., Griffin, M. D. W., Jameson, G. B., Parker, M. W., Gerrard, J. A. & Perugini, M. A. (2008). J. Biol. Chem. 283, 27598–27603. [DOI] [PubMed]
- Burgess, B. R., Dobson, R. C. J., Dogovski, C., Jameson, G. B., Parker, M. W. & Perugini, M. A. (2008). Acta Cryst. F64, 659–661. [DOI] [PMC free article] [PubMed]
- Chatterjee, S. P., Singh, B. K. & Gilvarg, C. (1994). Plant Mol. Biol. 26, 285–290. [DOI] [PubMed]
- Devenish, S. R. A., Gerrard, J. A., Jameson, G. B. & Dobson, R. C. J. (2008). Acta Cryst. F64, 1092–1095. [DOI] [PMC free article] [PubMed]
- Devenish, S. R. A., Huisman, F. H., Parker, E. J., Hadfield, A. T. & Gerrard, J. A. (2009). Biochim. Biophys. Acta, 1794, 1168–1174. [DOI] [PubMed]
- Dobson, R. C. J., Atkinson, S. C., Gorman, M. A., Newman, J. M., Parker, M. W. & Perugini, M. A. (2008). Acta Cryst. F64, 206–208. [DOI] [PMC free article] [PubMed]
- Dobson, R. C. J., Devenish, S. R. A., Turner, L. A., Clifford, V. R., Pearce, F. G., Jameson, G. B. & Gerrard, J. A. (2005). Biochemistry, 44, 13007–13013. [DOI] [PubMed]
- Dobson, R. C. J., Gerrard, J. A. & Pearce, F. G. (2004). Biochem. J. 377, 757–762. [DOI] [PMC free article] [PubMed]
- Dobson, R. C. J., Griffin, M. D. W., Devenish, S. R. A., Pearce, F. G., Hutton, C. A., Gerrard, J. A., Jameson, G. B. & Perugini, M. A. (2008). Protein Sci. 17, 2080–2090. [DOI] [PMC free article] [PubMed]
- Dobson, R. C. J., Griffin, M. D. W., Jameson, G. B. & Gerrard, J. A. (2005). Acta Cryst. D61, 1116–1124. [DOI] [PubMed]
- Dobson, R. C. J., Perugini, M. A., Jameson, G. B. & Gerrard, J. A. (2009). Biochimie, 91, 1036–1044. [DOI] [PubMed]
- Dobson, R. C. J., Valegård, K. & Gerrard, J. A. (2004). J. Mol. Biol. 338, 329–339. [DOI] [PubMed]
- Dogovski, C., Atkinson, S. C., Dommaraju, S. R., Hor, L., Hutton, C. A., Gerrard, J. A. & Perugini, M. A. (2009). Encyclopedia of Life Support Systems (EOLSS), edited by H. Doelle, Vol. II, pp. 116–136. Oxford: Eolss Publishers.
- Domigan, L. J., Scally, S. W., Fogg, M. J., Hutton, C. A., Perugini, M. A., Dobson, R. C. J., Muscroft-Taylor, A. C., Gerrard, J. A. & Devenish, S. R. A. (2009). Biochim. Biophys. Acta, 1794, 1510–1516. [DOI] [PubMed]
- Dommaraju, S., Dogovski, C., Czabotar, P. E., Hor, L., Smith, B. J. & Perugini, M. A. (2011). Arch. Biochem. Biophys. 512, 167–174. [DOI] [PubMed]
- Dommaraju, S., Gorman, M. A., Dogovski, C., Pearce, F. G., Gerrard, J. A., Dobson, R. C. J., Parker, M. W. & Perugini, M. A. (2010). Acta Cryst. F66, 57–60. [DOI] [PMC free article] [PubMed]
- Evans, G. & Pettifer, R. F. (2001). J. Appl. Cryst. 34, 82–86.
- Evans, G., Schuldt, L., Griffin, M. D. W., Devenish, S. R. A., Pearce, F. G., Perugini, M. A., Dobson, R. C. J., Jameson, G. B., Weiss, M. S. & Gerrard, J. A. (2011). Arch. Biochem. Biophys. 512, 154–159. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Girish, T. S., Sharma, E. & Gopal, B. (2008). FEBS Lett. 582, 2923–2930. [DOI] [PubMed]
- Griffin, M. D. W., Dobson, R. C. J., Gerrard, J. A. & Perugini, M. A. (2010). Arch. Biochem. Biophys. 494, 58–63. [DOI] [PubMed]
- Griffin, M. D. W., Dobson, R. C. J., Pearce, F. G., Antonio, L., Whitten, A. E., Liew, C. K., Mackay, J. P., Trewhella, J., Jameson, G. B., Perugini, M. A. & Gerrard, J. A. (2008). J. Mol. Biol. 380, 691–703. [DOI] [PubMed]
- Hor, L., Dobson, R. C. J., Dogovski, C., Hutton, C. A. & Perugini, M. A. (2010). Acta Cryst. F66, 37–40. [DOI] [PMC free article] [PubMed]
- Hutton, C. A., Perugini, M. A. & Gerrard, J. A. (2007). Mol. Biosyst. 3, 458–465. [DOI] [PubMed]
- Jaillon, O. et al. (2007). Nature (London), 449, 463–467.
- Kang, B. S., Kim, Y.-G., Ahn, J.-W. & Kim, K.-J. (2010). Int. J. Biol. Macromol. 46, 512–516. [DOI] [PubMed]
- Kaur, N., Gautam, A., Kumar, S., Singh, A., Singh, N., Sharma, S., Sharma, R., Tewari, R. & Singh, T. P. (2011). Int. J. Biol. Macromol. 48, 779–787. [DOI] [PubMed]
- Kefala, G., Evans, G. L., Griffin, M. D. W., Devenish, S. R. A., Pearce, F. G., Perugini, M. A., Gerrard, J. A., Weiss, M. S. & Dobson, R. C. (2008). Biochem. J. 411, 351–360. [DOI] [PubMed]
- Kobayashi, K. et al. (2003). Proc. Natl Acad. Sci. USA, 100, 4678–4683.
- Kumpaisal, R., Hashimoto, T. & Yamada, Y. (1987). Plant Physiol. 85, 145–151. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- McPhillips, T. M., McPhillips, S. E., Chiu, H.-J., Cohen, A. E., Deacon, A. M., Ellis, P. J., Garman, E., Gonzalez, A., Sauter, N. K., Phizackerley, R. P., Soltis, S. M. & Kuhn, P. (2002). J. Synchrotron Rad. 9, 401–406. [DOI] [PubMed]
- Mirwaldt, C., Korndörfer, I. & Huber, R. (1995). J. Mol. Biol. 246, 227–239. [DOI] [PubMed]
- Mitsakos, V., Dobson, R. C. J., Pearce, F. G., Devenish, S. R. A., Evans, G. L., Burgess, B. R., Perugini, M. A., Gerrard, J. A. & Hutton, C. A. (2008). Bioorg. Med. Chem. Lett. 18, 842–844. [DOI] [PubMed]
- Newman, J., Egan, D., Walter, T. S., Meged, R., Berry, I., Ben Jelloul, M., Sussman, J. L., Stuart, D. I. & Perrakis, A. (2005). Acta Cryst. D61, 1426–1431. [DOI] [PubMed]
- Padmanabhan, B., Strange, R. W., Antonyuk, S. V., Ellis, M. J., Hasnain, S. S., Iino, H., Agari, Y., Bessho, Y. & Yokoyama, S. (2009). Acta Cryst. F65, 1222–1226. [DOI] [PMC free article] [PubMed]
- Pearce, F. G., Dobson, R. C. J., Weber, A., Lane, L. A., McCammon, M. G., Squire, M. A., Perugini, M. A., Jameson, G. B., Robinson, C. V. & Gerrard, J. A. (2008). Biochemistry, 47, 12108–12117. [DOI] [PubMed]
- Pearce, F. G., Perugini, M. A., McKerchar, H. J. & Gerrard, J. A. (2006). Biochem. J. 400, 359–366. [DOI] [PMC free article] [PubMed]
- Perl, A., Shaul, O. & Galili, G. (1992). Plant Mol. Biol. 19, 815–823. [DOI] [PubMed]
- Perugini, M. A., Griffin, M. D. W., Smith, B. J., Webb, L. E., Davis, A. J., Handman, E. & Gerrard, J. A. (2005). Eur. Biophys. J. 34, 469–476. [DOI] [PubMed]
- Rice, E. A., Bannon, G. A., Glenn, K. C., Jeong, S. S., Sturman, E. J. & Rydel, T. J. (2008). Arch. Biochem. Biophys. 480, 111–121. [DOI] [PubMed]
- Shaul, O. & Galili, G. (1992). Plant J. 2, 203–209.
- Sibarani, N. E., Gorman, M. A., Dogovski, C., Parker, M. W. & Perugini, M. A. (2010). Acta Cryst. F66, 32–36. [DOI] [PMC free article] [PubMed]
- Silk, G. W. & Matthews, B. F. (1997). Plant Mol. Biol. 33, 931–933. [DOI] [PubMed]
- Turner, J. J., Healy, J. P., Dobson, R. C. J., Gerrard, J. A. & Hutton, C. A. (2005). Bioorg. Med. Chem. Lett. 15, 995–998. [DOI] [PubMed]
- Voss, J. E., Scally, S. W., Taylor, N. L., Atkinson, S. C., Griffin, M. D. W., Hutton, C. A., Parker, M. W., Alderton, M. R., Gerrard, J. A., Dobson, R. C. J., Dogovski, C. & Perugini, M. A. (2010). J. Biol. Chem. 285, 5188–5195. [DOI] [PMC free article] [PubMed]
- Voss, J. E., Scally, S. W., Taylor, N. L., Dogovski, C., Alderton, M. R., Hutton, C. A., Gerrard, J. A., Parker, M. W., Dobson, R. C. J. & Perugini, M. A. (2009). Acta Cryst. F65, 188–191. [DOI] [PMC free article] [PubMed]
- Wernimont, A. & Edwards, A. (2009). PLoS One, 4, e5094. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Wubben, J. M., Dogovski, C., Dobson, R. C. J., Codd, R., Gerrard, J. A., Parker, M. W. & Perugini, M. A. (2010). Acta Cryst. F66, 1511–1516. [DOI] [PMC free article] [PubMed]