β-Ketoacyl-acyl carrier protein synthase I (XoFabB) from X. oryzae pv. oryzae (Xoo) plays a crucial role in fatty-acid synthesis and has been considered as a target for the development of antibacterial agents against Xoo. XoFabB was expressed, purified and crystallized to determine its atomic resolution structure.
Keywords: Xanthomonas oryzae pv. oryzae, bacterial blight, XoFabB, β-ketoacyl-acyl carrier protein synthase I
Abstract
The proteins in the fatty-acid synthesis pathway in bacteria have significant potential as targets for the development of antibacterial agents. An essential elongation step in fatty-acid synthesis is performed by β-ketoacyl-acyl carrier protein synthase I (FabB). The organism Xanthomonas oryzae pv. oryzae (Xoo) causes a destructive bacterial blight disease of rice. The XoFabB protein from Xoo was expressed, purified and crystallized for the three-dimensional structure determination that is essential for the development of specific inhibitors of the enzyme. An XoFabB crystal diffracted to 3.0 Å resolution and belonged to the tetragonal space group P41, with unit-cell parameters a = b = 82.2, c = 233.2 Å. Assuming that the crystallographic structure contains four molecules in the asymmetric unit, the corresponding V M would be 2.18 Å3 Da−1 and the solvent content would be 43.5%. The initial structure was determined by the MOLREP program with an R factor of 44.0% and does contain four monomers in the asymmetric unit.
1. Introduction
Bacterial blight, which is caused by Xanthomonas oryzae pv. oryzae (Xoo), is a destructive disease of rice which has caused huge damage in most rice-growing countries (Ezuka & Kaku, 2000 ▶). However, no effective antibacterial agents against Xoo are available to date. In 2005 the whole genome sequence of Xoo was determined (Lee et al., 2005 ▶) and approximately 100 different essential genes of Xoo were selected as targets for the development of antibacterial agents against Xoo (Payne et al., 2004 ▶, 2007 ▶). The gene coding for β-ketoacyl-acyl carrier protein synthase I was one such target.
Fatty-acid synthesis (FAS) in bacteria has been clarified as type II, in which each catalytic step is performed by an individual enzyme (Rock & Jackowski, 2002 ▶). In contrast, the type I FAS found in eukaryotes (in particular in mammals) consists of a large single polypeptide chain for all catalytic steps (Smith et al., 2003 ▶). In FAS type II pathways, the condensation of acyl groups onto the elongating fatty-acid chain is catalyzed by β-ketoacyl-acyl carrier protein synthases (KASs). Three enzymes related to KAS activity have been identified and called KAS I, KAS II and KAS III; these enzymes are also known as FabB, FabF and FabH, respectively (Heath & Rock, 2002 ▶; Garwin et al., 1980 ▶). FabB can catalyze the condensation reaction of both saturated and unsaturated acyl-acyl carrier proteins (ACPs), whereas FabF can only carry out the condensation reaction of saturated acyl ACPs (Cronan & Rock, 1987 ▶). Since FabB plays a vital role in bacterial survival and has no counterpart in eukaryotes, it has been considered as a potential target for drug development. The crystal structure of FabB from Escherichia coli was solved a decade ago (Olsen et al., 1999 ▶) and Price and coworkers reported that two natural compounds, cerulenin and thiolactomycin, could inhibit E. coli FabB (Price et al., 2001 ▶). Although they inhibit the growth of several bacteria successfully, the limited selectivity of cerulenin and problems with the synthesis and stability of thiolactomycin have decreased their usage as antibacterial drugs (Heath et al., 2001 ▶; Price et al., 2001 ▶). Recently, using a structure-based screening method, it has been suggested that aminothiazole is a promising compound that inhibits E. coli FabB (Pappenberger et al., 2007 ▶).
In this study, we cloned the XoFabB gene from Xoo and the gene product (402 residues, 44 kDa) was expressed, purified and crystallized. We believe that the three-dimensional crystal structure of XoFabB will provide valuable information in the development of antibacterial agents against Xoo.
2. Materials and methods
2.1. Cloning
The XofabB gene coding for β-ketoacyl-acyl carrier protein synthase I from Xoo (ATCC10331) was amplified by PCR using the genomic DNA of Xoo as a template. The oligonucleotides used were 5′-GGG CAT ATG CGT CGC GTC GTC ATC ACC GGA AT-3′ for the forward primer and 5′-GG GGA TCC TCA GAC CCG GCC GAA CAC CAG G-3′ for the reverse primer (NdeI and BamHI sites are shown in bold). The PCR fragments were digested with the restriction enzymes and then inserted into a pET11a vector (Novagen, Republic of Korea). The plasmid pET11a was modified to include the sequence MGHHHHHHSSENLYFQGH containing six histidine residues at the N-terminus of the cloned target protein to facilitate protein purification. The XofabB gene in the recombinant vector pET-XofabB was verified to be free of mutations by DNA sequencing (Macrogen, Republic of Korea).
2.2. Overexpression and purification
The recombinant vector pET-XofabB was transformed into E. coli BL21 (DE3) cells grown in Luria–Bertani medium supplemented with ampicillin (50 µg ml−1). The transformant was cultured in a shaking incubator at 310 K. When the OD600 of the culture reached 0.6, 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to induce expression of the XoFabB protein. The culture was further incubated for 6 h at 303 K and the cells were harvested by centrifugation at 277 K for 30 min at 800g. The cell pellet was resuspended in ice-cold lysis buffer (25 mM Tris–HCl pH 7.5, 300 mM NaCl, 15 mM imidazole, 3 mM β-mercaptoethanol) and lysed by sonication on ice (Sonomasher, S&T Science, Republic of Korea). The lysate was centrifuged for 30 min at 25 000g (Vision VS24-SMTi V508A rotor) at 277 K to pellet the cellular debris. The supernatant containing soluble XoFabB was loaded onto a column containing Ni–NTA resin (Novagen) according to the the manufacturer’s protocol. The unbound material which flowed through was discarded and nonspecifically bound proteins were eluted with lysis buffer containing 30 mM imidazole. XoFabB was eluted using lysis buffer containing 250 mM imidazole. The fractions containing the target protein were collected and dialyzed for 12 h at 277 K in buffer A (25 mM Tris–HCl pH 7.5, 3 mM β-mercaptoethanol). The protein was further purified by anion-exchange chromatography using a Uno-Q column (Amersham, Republic of Korea) with buffer B (25 mM Tris–HCl pH 7.5, 1 M NaCl, 3 mM β-mercaptoethanol) as the high-salt buffer in a step gradient. XoFabB protein was eluted at an NaCl concentration of between 380 and 420 mM. The homogeneity of the purified protein was confirmed via SDS–PAGE: only one band of XoFabB was visible on 10% SDS–PAGE, at a position corresponding to 44 kDa (Fig. 1 ▶). The purified XoFabB was concentrated to 7 mg ml−1 in buffer A for crystallization purposes using a spin concentrator with a molecular-weight cutoff of 30 000 Da.
Figure 1.
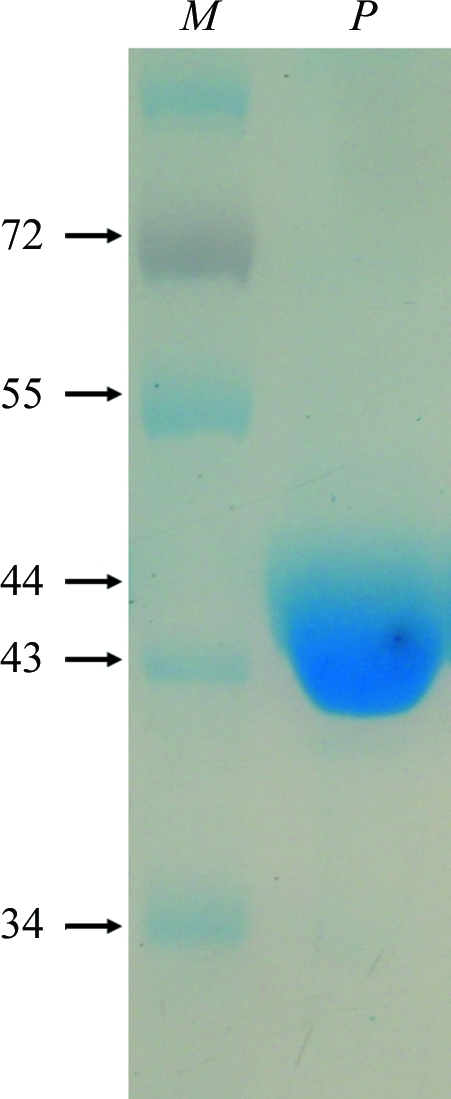
Purified XoFabB shown on 10% SDS–PAGE. Lane M, PageRuler Prestained Protein Ladder (Fermentas, Republic of Korea; labelled in kDa), lane P, protein.
2.3. Crystallization and X-ray data collection
Crystallization conditions were initially screened using the sitting-drop vapour-diffusion method with a Hydra II eDrop automated pipetting system (Matrix) at 287 K. Drops consisted of 0.5 µl XoFabB protein and 0.5 µl reservoir solution and were equilibrated against 70 µl reservoir solution at 287 K. The initial crystallization conditions tested were from Index and Grid Screen Ammonium Sulfate kits (Hampton Research, USA) and the Wizard kit (Emerald BioSystems, USA). After 2 d, needle-like crystals were observed in one condition from Grid Screen Ammonium Sulfate (condition No. 14; 0.1 M citric acid pH 5.0, 2.4 M ammonium sulfate; Fig. 2 ▶). Since the sitting drops did not produce good crystals, we tried using hanging drops and also tried the seeding method with various concentrations of the protein. The final well diffracting crystals were obtained from hanging drops consisting of 2 µl 5.5 mg ml−1 XoFabB and 2 µl reservoir solution (0.1 M citric acid pH 5.5, 2.2 M ammonium sulfate). An optimized crystal (0.3 × 0.05 × 0.03 mm; Fig. 3 ▶) was mounted in a loop and transferred to a cryoprotectant solution prior to cooling in liquid nitrogen. Several cryoprotectant solutions were created by mixing the reservoir solution with various concentrations of glycerol and ethylene glycol; reservoir solution with an additional 20%(v/v) ethylene glycol was finally selected as the most suitable cryosolution. The crystal was analyzed on beamline BL26B1 of SPring-8 in Japan using a Jupiter210 (Rigaku/MSC) CCD detector. Diffraction data were collected to 3.0 Å resolution and were integrated and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶). The crystal belonged to a tetragonal primitive space group (P41, P42 or P43), with unit-cell parameters a = b = 82.2, c = 233.2 Å. Matthews coefficient analysis (Matthews, 1968 ▶) indicated that there might be four molecules present in the asymmetric unit (V M = 2.18 Å3 Da−1), giving a solvent content of 43.5%. The final statistics of data collection and processing details are summarized in Table 1 ▶. Monomer A of the FabB crystal structure from E. coli (Olsen et al., 1999 ▶) was used as a search model for molecular replacement (MR) using the MOLREP (Vagin & Teplyakov, 2010 ▶) and Phaser (McCoy et al., 2007 ▶) programs. The resulting model was given by MOLREP, with an initial R factor of 44.0%.
Figure 2.
Initial crystals of XoFabB protein obtained by the sitting-drop vapour-diffusion method using condition No. 14 of the Grid Screen Ammonium Sulfate kit (0.1 M citric acid pH 5.0, 2.4 M ammonium sulfate) at 287 K.
Figure 3.
Optimized XoFabB crystals (0.3 × 0.05 × 0.03 mm) were obtained using a reservoir solution consisting of 0.1 M citric acid pH 5.5, 2.2 M ammonium sulfate at 287 K.
Table 1. Data-collection statistics.
Values in parentheses are for the last shell.
| Beamline | Spring-8 BL26B1 |
| Wavelength (Å) | 1.0 |
| Resolution range (Å) | 50.0–3.0 (3.05–3.00) |
| Space group | P41 |
| Unit-cell parameters (Å) | a = b = 82.2, c = 233.2 |
| Total No. of reflections | 1169323 |
| No. of unique reflections | 30477 |
| Completeness (%) | 98.2 (95.9) |
| Molecules per asymmetric unit | 4 |
| Solvent content (%) | 43.5 |
| Average I/σ(I) | 10.6 (2.4) |
| Rmerge† (%) | 11.5 (38.2) |
| Rp.i.m.‡ (%) | 9.5 (35.5) |
| Multiplicity | 4.2 (3.1) |
R
merge = 
 .
.
R
p.i.m. = 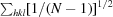

 , where Ii(hkl) is the intensity of reflection hkl,
, where Ii(hkl) is the intensity of reflection hkl,  is the sum over all reflections,
is the sum over all reflections,  is the sum over i measurements of reflection hkl and 〈I(hkl)〉 is the weighted average intensity of all observations i of reflection hkl.
is the sum over i measurements of reflection hkl and 〈I(hkl)〉 is the weighted average intensity of all observations i of reflection hkl.
3. Results and discussion
We expressed the β-ketoacyl-acyl carrier protein synthase I from Xoo (XoFabB) in E. coli with a 6×His-TEV tag and purified and crystallized it. Initial needle-shaped crystals were observed using condition No. 14 from Grid Screen Ammonium Sulfate in sitting drops at 287 K. After optimization of this crystallization condition, large single crystals were obtained and a complete diffraction data set was collected to 3.0 Å resolution. The initial space group derived from the auto-indexing program was P4 (P41, P42 or P43) or P422. Analysis of self-rotation functions calculated by MOLREP confirmed that the XoFabB crystal possessed one fourfold and two twofold crystallographic axes and an additional twofold noncrystallographic axis (Fig. S11). The MOLREP and Phaser programs were used for MR with the E. coli β-ketoacyl-ACP synthase I structure (PDB entry 1dd8; 62% sequence identity; Olsen et al., 1999 ▶) as a template. Although all possible space groups were used in MR, MOLREP only provided a solution in space group P41. The resulting model has four molecules in the asymmetric unit, with an initial R factor of 44.0%. The resulting electron-density maps are of high quality and no clashes were found between symmetry-related molecules (Fig. S31). Currently, structure refinement is being carried out. We believe that the XoFabB structure will provide insights into its catalytic mechanism and will provide valuable information for the development of antibacterial agents against Xoo.
Supplementary Material
Supplementary material file. DOI: 10.1107/S1744309111040590/nj5100sup1.pdf
Acknowledgments
We are grateful to the staff members at beamline BL26B1 of SPring-8 in Japan and beamline 17A of the Photon Factory, High Energy Accelerator Research Organization, Japan for their assistance. This work was supported by a grant from the Next-Generation BioGreen 21 Program (PJ008174), Rural Development Administration, Republic of Korea, by the National Research Foundation of Korea Grant funded by the Korean Government (NRF-2011-619-E0002), and by a WCU (World Class University, R33-2008-000-1071) program through the Korea Science and Engineering Foundation funded by the Ministry of Education, Science and Technology, South Korea.
Footnotes
Supplementary material has been deposited in the IUCr electronic archive (Reference: NJ5100).
References
- Cronan, J. E. & Rock, C. O. (1987). Escherichia Coli and Salmonella Typhimurium: Cellular and Molecular Biology, edited by F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter & H. E. Umbarger, Vol. 1, pp. 474–477. Washington DC: American Society for Microbiology.
- Ezuka, A. & Kaku, H. (2000). Bull. Natl Inst. Agrobiol. Resour. (Jpn), 15, 53–54.
- Garwin, J. L., Klages, A. L. & Cronan, J. E. Jr (1980). J. Biol. Chem. 255, 3263–3265. [PubMed]
- Heath, R. J. & Rock, C. O. (2002). Nat. Prod. Rep. 19, 581–596. [DOI] [PubMed]
- Heath, R. J., White, S. W. & Rock, C. O. (2001). Prog. Lipid Res. 40, 467–497. [DOI] [PubMed]
- Lee, B. M. et al. (2005). Nucleic Acids Res. 33, 577–586. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Olsen, J. G., Kadziola, A., von Wettstein-Knowles, P., Siggaard-Andersen, M., Lindquist, Y. & Larsen, S. (1999). FEBS Lett. 460, 46–52. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pappenberger, G., Schulz-Gasch, T., Kusznir, E., Müller, F. & Hennig, M. (2007). Acta Cryst. D63, 1208–1216. [DOI] [PMC free article] [PubMed]
- Payne, D. J., Gwynn, M. N., Holmes, D. J. & Pompliano, D. L. (2007). Nature Rev. Drug Discov. 6, 29–40. [DOI] [PubMed]
- Payne, D. J., Gwynn, M. N., Holmes, D. J. & Rosenberg, M. (2004). Methods Mol. Biol. 266, 231–259. [DOI] [PubMed]
- Price, A. C., Choi, K.-H., Heath, R. J., Li, Z., White, S. W. & Rock, C. O. (2001). J. Biol. Chem. 276, 6551–6559. [DOI] [PubMed]
- Rock, C. O. & Jackowski, S. (2002). Biochem. Biophys. Res. Commun. 292, 1155–1166. [DOI] [PubMed]
- Smith, S., Witkowski, A. & Joshi, A. K. (2003). Prog. Lipid Res. 42, 289–317. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material file. DOI: 10.1107/S1744309111040590/nj5100sup1.pdf




