The Saccharomyces cerevisiae NAD+-dependent deacetylase HST1156–503 was expressed and crystallized. Crystals grown by the hanging-drop vapour-diffusion method diffracted to 2.90 Å resolution.
Keywords: HST1, NAD-dependent deacetylases, SIR2
Abstract
The Saccharomyces cerevisiae NAD+-dependent deacetylase HST1 belongs to the class III HDAC family; it acts as a transcriptional corepressor for the specific middle sporulation and de novo NAD+-biosynthesis genes and also takes part in the SET3C and SUM1–RFM1–HST1 complexes. Structural information on HST1 and its related complexes would be helpful in order to understand the structural basis of its deacetylation mechanism and the assembly of these complexes. Here, HST1156–503 was expressed and crystallized. Crystals grown by the hanging-drop vapour-diffusion method diffracted to 2.90 Å resolution and belonged to space group P21, with unit-cell parameters a = 40.2, b = 101.7, c = 43.9 Å, β = 103.9°. Both Matthews coefficient analysis and the self-rotation function suggested the presence of four molecules per asymmetric unit in the crystal, with a solvent content of 49.76% (V M = 2.45 Å3 Da−1).
1. Introduction
The acetylation of histones affects gene expression via its influence on chromatin conformation. The counteracting enzyme families histone deacetylases (HDACs) and histone acetyltransferases (HATs) control transcription by selectively deacetylating or acetylating the ∊-amino groups of lysine located near the N-terminal extensions of core histones (Marks et al., 2003 ▶). Chromatin deacetylation is correlated with gene silencing, whereas acetylation is correlated with transcriptional activity. HDACs are also involved in the reversible acetylation of nonhistone proteins (Gallinari et al., 2007 ▶).
Eukaryotic HDACs have been classified into three classes according to phylogenetic analyses and sequence homology. Class I HDACs are homologues of yeast RPD3 and localize to the nucleus. Class II HDACs are homologues of yeast Hda1 and are found in both the nucleus and cytoplasm. The class I and class II proteins are evolutionarily related and share a common zinc-dependent enzymatic mechanism (de Ruijter et al., 2003 ▶). Class III HDACs are homologues of yeast SIR2, which is evolutionarily unrelated to the class I or II proteins, and are found in both the nucleus and cytoplasm (Blander & Guarente, 2004 ▶). Class III proteins are strictly dependent on NAD+ as a cofactor and transfer the acetyl group from acetylated lysine residues to the ADP-ribose moiety of NAD+, generating deacetylated histone tails, nicotinamide and the novel metabolite O-acetyl-ADP ribose (Tong & Denu, 2010 ▶).
In Saccharomyces cerevisiae there are five NAD+-dependent deacetylases, SIR2 and HST1–HST4, which vary in their cellular localization and possibly in their substrate specificity. SIR2 is the founding member of the entire family; it functions as a regional transcriptional silencer and is involved in modifying chromatin structure (Mead et al., 2007 ▶). HST1, the most closely related paralogue to SIR2, deacetylates histones H3 and H4 and acts as a transcriptional corepressor for the specific middle sporulation and de novo NAD+-biosynthesis genes (Li et al., 2010 ▶). HST2, a more distantly related paralogue of SIR2, is predominantly a cytoplasmic protein, but also has a cell-cycle-specific nuclear localization that is required for rDNA and centromeric silencing (Durand-Dubief et al., 2007 ▶). HST3 and HST4 are the most distantly related to SIR2 but are closely related to each other; they deacetylate Lys56 on histone H3 and have been implicated in cell-cycle regulation, heterochromatin silencing and genome integrity (Starai et al., 2003 ▶).
Despite an overall sequence conservation of 63% between SIR2 and HST1 (76% similarity), they have nonoverlapping functions: SIR2 functions as a transcriptional silencer of relatively large regions of the genome, while HST1 functions as a transcriptional repressor acting locally on a specific set of promoters (Mead et al., 2007 ▶). Structure determination of HST1 is necessary in order to reveal the structural basis of the different functions of these two NAD+-dependent deacetylases.
In S. cerevisiae, HST1 alone has NAD+-dependent deacetylation activity in vivo (Sutton et al., 2001 ▶). It also belongs to the seven-subunit SET3C complex (consisting of SET3, SNT1, YIL112W, SIF2, CPR1, HOS2 and HST1) and is involved in the NAD+-dependent deacetylase activity of the whole complex (Pijnappel et al., 2001 ▶). SET3C represses genes in the early/middle stages of the yeast sporulation program, including the key meiotic regulators IME2 and NDT80. HST1 is also present in the SUM1–RFM1–HST1 complex that represses meiotic genes during vegetative growth via histone H4 Lys5 deacetylation by HST1. Structural information on HST1 and its related complexes would be helpful in understanding the complex-assembly processes and operation mechanisms of both the SET3C and the SUM1 complexes. Here, we describe the crystallization and preliminary X-ray analysis of the region of HST1 (Asp156–Gln503) corresponding to the core catalytic domain of SIR2.
2. Materials and methods
2.1. Cloning and expression
Primers of sense strand 5′-CGCGGATCCGATCCCTTAGAGAAAAAGCATG-3′ and antisense strand 5′-CCCAAGCTTTTACTGTTGTTTCTTTCGTGGCTG-3′ (Invitrogen) were used to amplify the target gene by polymerase chain reaction from the S. cerevisiae genomic DNA. The PCR fragment was inserted into expression vector pETgst [Novagen; modified to add a hexahistidine tag (MGHHHHHH) before the GST at the N-terminus and to insert the TEV cleavage sequence ENLYFQSL after the GST] to create recombinant HST1156–503. After sequencing, the plasmid was transformed into Escherichia coli BL21 (DE3) competent cells (Novagen). The transformant was grown in 1.6 l Luria–Bertani (LB) medium containing 50 µg ml−1 kanamycin at 289 K. When an OD600 of 0.6–0.8 was reached, 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added for induction. After 16 h induction at 289 K, the cells were harvested by centrifugation at 6000g for 10 min.
2.2. Purification
The harvested cells were suspended in buffer A (50 mM Tris–HCl pH 8.0, 300 mM NaCl) and lysed by sonication on ice. The cell lysate was centrifuged. The clear supernatant was passed through an Ni–NTA column (Qiagen) previously equilibrated with buffer A. Unbound proteins were washed away with buffer A containing 50 mM imidazole. The bound protein was eluted with buffer A containing 200 mM imidazole. The protein was dialyzed against buffer B (50 mM Tris–HCl pH 8.0, 60 mM NaCl) and the His-GST tags were removed by TEV protease digestion (1 OD280 TEV protease per 100 OD280 substrate) at room temperature (293 K) for 6 h. The digested fraction was loaded onto an Ni–NTA column (Qiagen) previously equilibrated with buffer B. The target protein was recovered in the flowthrough. After ultrafiltration to 2 ml using a Millipore 10 kDa centrifugal device, the target protein was purified using a Superdex 200 (GE Healthcare) gel-filtration chromatography column previously equilibrated with buffer B. Fractions containing the recombinant protein were determined by SDS–PAGE. The identity of the protein was further confirmed by mass-spectrometric analysis (Thermo Fisher).
2.3. Crystallization
The recombinant HST1156–503 protein fractions were concentrated to 20 mg ml−1 (calculated from the OD280 using a molar absorption coefficient of 32 890 M −1 cm−1; Eppendorf BioPhotometer Plus) by centrifugal ultrafiltration (Millipore; 10 kDa cutoff) prior to crystallization trials. Crystallization screens with native protein were performed with a Mosquito liquid-handling robot (TTP LabTech) using the vapour-diffusion method in 96-well crystallization plates at 289 K. Drops were prepared by mixing 0.25 µl protein solution containing 20 mg ml−1 protein with 0.25 µl reservoir solution and were equilibrated against 100 µl reservoir solution under 480 different conditions based on the Crystal Screen, Crystal Screen 2, Index, SaltRx, Grid Screen and ProPlex kits from Hampton Research and Molecular Dimensions. One week later, the best crystals were observed using condition No. 23 of the Molecular Dimensions ProPlex kit (0.1 M HEPES pH 7.0, 15% polyethylene glycol 4000). Subsequent screening was performed by varying the pH and the polyethylene glycol 4000 concentration. Drops were prepared by mixing 1 µl protein solution containing 20 mg ml−1 protein with 1 µl reservoir solution and were equilibrated against 200 µl reservoir solution in 24-well crystallization plates at 289 K. The optimal crystals appeared in drops containing 15%(w/v) polyethylene glycol 4000 and 0.1 M HEPES pH 7.0.
A crystal mounted in a loop was soaked briefly in a cryoprotectant solution consisting of the corresponding reservoir solution supplemented with 25%(v/v) glycerol, in which the glycerol replaced water, and was then flash-cooled in liquid nitrogen. X-ray diffraction data were collected on beamline 17U1 of the Shanghai Synchrotron Radiation Facility (SSRF) using a Jupiter CCD detector. All frames were collected at 100 K using a 1° oscillation angle with an exposure time of 1.2 s per frame. The crystal-to-detector distance was set to 350 mm. The complete diffraction data set was subsequently processed using HKL-2000 (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
Recombinant HST1156–503 (the region corresponding to the core catalytic domain of human SIRT2 histone deacetylase) was expressed and purified, and grew as plate-like crystals under an optimized precipitant condition consisting of 0.1 M HEPES pH 6.8 and 15% polyethylene glycol 4000 (Fig. 1 ▶). A total of 180 diffraction images were recorded from a single crystal. The HST1156–503 crystal diffracted to a maximum resolution of 2.90 Å and belonged to the monoclinic space group P21, with unit-cell parameters a = 40.2, b = 101.7, c = 43.9 Å, β = 103.9°. The number of molecules in the asymmetric unit was assumed to be four based on the Matthews coefficient (2.45 Å3 Da−1), with a solvent content of 49.76%. The detailed data-processing statistics are shown in Table 1 ▶.
Figure 1.

Crystals of HST1 protein growing under the condition 0.1 M HEPES pH 6.8, 15% polyethylene glycol 4000.
Table 1. Data-collection statistics for HST1.
Values in parentheses are for the highest resolution shell.
| Space group | P21 |
| Wavelength (Å) | 0.9796 |
| Unit-cell parameters (Å, °) | a = 40.2, b = 101.7, c = 43.9, β = 103.9 |
| Resolution limits (Å) | 50.00–2.90 (3.00–2.90) |
| No. of observed reflections | 26952 |
| No. of unique reflections | 7495 |
| Completeness (%) | 98.1 (91.9) |
| Rmerge† (%) | 9.8 (33.0) |
| Mean I/σ(I) | 11.8 (2.3) |
| VM (Å3 Da−1) | 2.45 |
| No. of subunits per asymmetric unit | 4 |
| Solvent content (%) | 49.76 |
R
merge = 
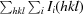 , where I
i(hkl) is the intensity of an individual reflection and 〈I(hkl)〉 is the average intensity of that reflection.
, where I
i(hkl) is the intensity of an individual reflection and 〈I(hkl)〉 is the average intensity of that reflection.
Structure determination of S. cerevisiae HST1156–503 was attempted by the molecular-replacement method using the S. cerevisiae SIR2 deacetylase structure (PDB entry 2hjh; B. E. Hall, J. R. Buchberger, S. A. Gerber, A. L. B. Ambrosio, S. P. Gygi, D. Filman, D. Moazed & T. Ellenberger, unpublished work) as a search model. However, to date this has not been successful.
Acknowledgments
We are grateful to the staff members at SSRF for the collection of diffraction data. Financial support for this project was provided by the Fundamental Research Funds for the Central Universities, the Chinese National Natural Science Foundation (grant Nos. 30900224, 31130018 and 10979039), the Chinese Ministry of Science and Technology (grant Nos. 2006AA02A318 and 2009CB825500), the Science and Technological Fund of Anhui Province for Outstanding Youth (grant No. 10040606Y11), the Anhui Provincial Natural Science Foundation (grant No. 090413081) and the Education Department of Anhui Province (grant No. 2009SQRZ007ZD).
References
- Blander, G. & Guarente, L. (2004). Annu. Rev. Biochem. 73, 417–435. [DOI] [PubMed]
- Durand-Dubief, M., Sinha, I., Fagerström-Billai, F., Bonilla, C., Wright, A., Grunstein, M. & Ekwall, K. (2007). EMBO J. 26, 2477–2488. [DOI] [PMC free article] [PubMed]
- Gallinari, P., Di Marco, S., Jones, P., Pallaoro, M. & Steinkühler, C. (2007). Cell Res. 17, 195–211. [DOI] [PubMed]
- Li, M., Petteys, B. J., McClure, J. M., Valsakumar, V., Bekiranov, S., Frank, E. L. & Smith, J. S. (2010). Mol. Cell. Biol. 30, 3329–3341. [DOI] [PMC free article] [PubMed]
- Marks, P. A., Miller, T. & Richon, V. M. (2003). Curr. Opin. Pharmacol. 3, 344–351. [DOI] [PubMed]
- Mead, J., McCord, R., Youngster, L., Sharma, M., Gartenberg, M. R. & Vershon, A. K. (2007). Mol. Cell. Biol. 27, 2466–2475. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pijnappel, W. W., Schaft, D., Roguev, A., Shevchenko, A., Tekotte, H., Wilm, M., Rigaut, G., Séraphin, B., Aasland, R. & Stewart, A. F. (2001). Genes Dev. 15, 2991–3004. [DOI] [PMC free article] [PubMed]
- Ruijter, A. J. de, van Gennip, A. H., Caron, H. N., Kemp, S. & van Kuilenburg, A. B. (2003). Biochem. J. 370, 737–749. [DOI] [PMC free article] [PubMed]
- Starai, V. J., Takahashi, H., Boeke, J. D. & Escalante-Semerena, J. C. (2003). Genetics, 163, 545–555. [DOI] [PMC free article] [PubMed]
- Sutton, A., Heller, R. C., Landry, J., Choy, J. S., Sirko, A. & Sternglanz, R. (2001). Mol. Cell. Biol. 21, 3514–3522. [DOI] [PMC free article] [PubMed]
- Tong, L. & Denu, J. M. (2010). Biochim. Biophys. Acta, 1804, 1617–1625. [DOI] [PMC free article] [PubMed]


