Expression, purification and crystallization of Srr-1-K4BD, a human keratin 4-binding domain of serine-rich repeat protein 1 from S. agalactiae, was carried out. Native crystals of Srr-1-K4BD diffracted to 3.8 Å resolution using synchrotron radiation.
Keywords: serine-rich repeat protein 1, Streptococcus agalactiae, keratin 4-binding domain
Abstract
Serine-rich repeat protein 1 (Srr-1) is a surface protein from Streptococcus agalactiae. A 17 kDa region of this protein has been identified to bind to human keratin 4 (K4) and is termed the Srr-1 K4-binding domain (Srr-1-K4BD). Recombinant Srr-1-K4BD was overexpressed in Escherichia coli BL21 (DE3) cells. Native and selenomethionine-substituted proteins were prepared using Luria–Bertani (LB) and M9 minimal media, respectively. A two-step purification protocol was carried out to obtain a final homogenous sample of Srr-1-K4BD. Crystals of native Srr-1-K4BD were obtained using PEG 3350 as a precipitant. The crystals diffracted to 3.8 Å resolution using synchrotron radiation and belonged to space group P21, with unit-cell parameters a = 47.56, b = 59.48, c = 94.71 Å, β = 93.95°.
1. Introduction
Streptococcus agalactiae, also called group B streptococcus (GBS), is a Gram-positive streptococcal bacterium and a causative agent of invasive neonatal sepsis and meningitis (Glaser et al., 2002 ▶). Immunocompromised adults who acquire invasive GBS infections may develop infections of the bloodstream (sepsis), lung (pneumonia) or, rarely, the fluid and lining tissues surrounding the brain (meningitis). GBS is observed in 15–45% of all healthy women as a commensal bacterium in their intestine, vagina and rectum (Gilbert, 2004 ▶).
Both commensal and pathogenic lifestyles necessitate the attachment of bacteria to host tissues (Doran & Nizet, 2004 ▶; Jenkinson & Lamont, 1997 ▶; Patti et al., 1994 ▶). Surface proteins are known to mediate this key step of bacteria–host interaction in many microbial infections (Navarre & Schneewind, 1999 ▶). S. agalactiae expresses many surface-exposed proteins that are known to adhere to the extracellular matrix (ECM) molecules of the host. For example, the proteins FbsA and FbsB bind to fibrinogen (Schubert et al., 2002 ▶), Lmb binds to laminin (Spellerberg et al., 1999 ▶) and ScpB (C5a peptidase) attaches to fibronectin (Beckmann et al., 2002 ▶; Cheng et al., 2002 ▶).
Serine-rich repeat protein 1, termed Srr-1 (1310 amino acids), is one such surface protein encoded by the gbs1529 gene of S. agalactiae (Seifert et al., 2006 ▶). Srr-1 (UniProt accession Q8E473) belongs to the streptococcal and staphylococcal serine-rich repeat (Srr) protein family. The Srr family of proteins contain a short serine-rich repeat region in the N-terminal region, a nonrepeat adhesive region, a region containing numerous serine-rich repeats, and a cell-wall-anchoring region comprising an LPXTG motif, a hydrophobic domain and a short C-terminal charged tail (Fig. 1 ▶; Navarre & Schneewind, 1999 ▶; Samen et al., 2007 ▶). A previous study identified a 17 kDa region (residues 485–642) of Srr-1 as a human keratin 4 binding domain (Samen et al., 2007 ▶) termed Srr-1-K4BD. In that study, five different constructs, namely Srr-1-N (residues 7–642), Srr-1-N1 (7–327), Srr-1-N2 (322–485), Srr-1-N3 (485–642) and Srr-1-N2N3 (322–642), were produced, and using binding assays it was demonstrated that Srr-1-N3 (Srr-1-K4BD) is sufficient and necessary for binding to human keratin 4.
Figure 1.
Structural organization of S. agalactiae Srr-1. Srr-1 contains 1310 amino acids. The human keratin 4-binding domain (K4BD) is shown. The serine-rich repeat regions are indicated by grey boxes. At the C-terminal end the LPXTG motif required for sortase-mediated anchoring of the protein to the cell-wall peptidoglycan and the membrane-spanning region (dotted box) are indicated.
Srr-1 is present in many GBS strains and a previous study showed that Srr-1 promotes penetration of the blood–brain barrier (BBB), causing invasion of the central nervous system (CNS) and leading to the development of GBS meningitis (van Sorge et al., 2009 ▶). Despite universal screening and preventive measures, early-onset GBS disease still prevails and remains a significant public health issue. Maternal immunization against GBS is an excellent way to prevent not only neonatal disease but also stillbirth and maternal disease. Vaccines against GBS are expected to be the most effective and sustainable long-term preventive strategy (Gilbert, 2004 ▶). Since the adhesin Srr-1 paves the way for the critical step in microbial infection, which is the attachment of the bacterium to the host molecule, understanding the structure–function relationship of Srr-1-K4BD will aid in the development of new therapeutics for GBS infection.
Here, we report the expression, purification, crystallization and preliminary X-ray analysis of recombinant Srr-1-K4BD.
2. Materials and methods
2.1. Expression and purification of Srr-1-K4BD
The srr-1 gene encoding the region comprised of residues 485–642 (Srr-1-K4BD) was PCR-amplified using the primers CATGCCAT GGTCATGAAGCTTGATGATGAAAGAC and CCGCGGATCCTCTGATAAAAGTTTAATTTCGGC and cloned into the pET-28a(+) expression vector (Novagen) using the restriction enzymes NcoI and BamHI (sites shown in bold) as reported previously (Samen et al., 2007 ▶). The pET-28a::Srr-1-K4BD construct was transformed into Escherichia coli BL21 (DE3) cells and protein expression was carried out in Luria–Bertani (LB) medium. A 10 ml overnight culture was prepared and transferred to 1 l LB medium supplemented with 50 µg ml−1 kanamycin and allowed to grow at 310 K until an optimum optical density (OD600 of 0.6–0.8) was reached. Following this, protein expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). After 4 h induction, the cells were spun down at 2468g for 20 min at 277 K. The harvested cells were resuspended in 15 ml buffer A (20 mM Tris–HCl pH 7.0, 300 mM NaCl, 5% glycerol, 5 mM β-mercaptoethanol) with 1 mM phenylmethylsulfonyl fluoride (PMSF). The cell suspension was lysed by ultrasonication on ice. The lysate was spun down at 9838g for 40 min. The supernatant and pellet were checked on a 15% SDS–PAGE gel and the protein was found in the soluble fraction (Fig. 2 ▶).
Figure 2.
Expression and purification analysis of recombinant Srr-1-K4BD on a 15% SDS–PAGE gel. Lane 1, molecular-weight marker (labelled in kDa); lane 2, cell lysate before IPTG induction; lane 3, supernatant after lysis; lane 4, pellet after lysis; lane 5, unbound material from Ni–NTA column; lane 6, buffer wash with 10 ml buffer A; lane 7, pooled fractions of protein eluted from Ni–NTA column with 100–250 mM imidazole; lane 8, final purified sample after size-exclusion chromatography.
Initial purification was carried out by the immobilized metal-affinity chromatography (IMAC) technique using an Ni–NTA column. The bound protein was eluted using a linear gradient of 0–1 M imidazole in buffer A. The peak fractions corresponding to Srr-1-K4BD were collected and checked on a 15% SDS–PAGE gel (Fig. 2 ▶). The fractions containing the protein were pooled and concentrated to 2 ml using a Vivaspin 20 concentrator (Sartorius Stedim Biotech). The yield of the protein was around 9 mg per litre of culture.
The concentrated sample was further purified using size-exclusion chromatography by loading it onto a Superdex 75 column (130 ml; GE Healthcare) pre-equilibrated with a buffer consisting of 20 mM Tris–HCl pH 7.0, 100 mM NaCl, 5% glycerol, 5 mM β-mercaptoethanol. The fractions corresponding to the peak were subjected to 15% SDS–PAGE and pure fractions were pooled and concentrated to 200 µl. A final yield of 5.6 mg protein was obtained from 1 l culture and was concentrated to 28 mg ml−1 for crystallization trials.
Selenomethionine incorporation of Srr-1-K4BD was carried out using the methionine-auxotroph strain E. coli B834 (DE3) (Novagen). A 10 ml overnight culture was grown in LB medium at 310 K. The cells were harvested at 2468g for 20 min and the cell pellet was used as an inoculum for 1 l 1× M9 minimal medium supplemented with kanamycin (50 µg ml−1) and other additives (Sambrook et al., 1989 ▶; Guerrero et al., 2001 ▶). l-Selenomethionine (40 µg ml−1) was added 1 h after inoculation. The culture was grown for 10 h before IPTG induction. A cell-harvesting and purification protocol similar to that used for native Srr-1-K4BD was carried out except that the buffers were degassed in order to avoid oxidation of selenomethionine.
2.2. Crystallization
Several trials were conducted to crystallize the Srr-1-K4BD protein. Initial conditions were obtained using commercial screens (Hampton Research) and home-made screens of ammonium sulfate versus pH (4–9) and polyethylene glycol (PEG; 600, 800, 1500 and 3500) versus pH (4–9). Crystallization was performed using the hanging-drop vapour-diffusion method at 293 K. 1 µl protein sample was mixed with an equal volume of reservoir solution and equilibrated against 1 ml of the latter at 293 K. Small two-dimensional plate-like crystals were obtained in 2 d in conditions with 32–35%(w/v) PEG 3350 as a precipitant and imidazole (pH 6.3 and 6.5) as a buffer. To obtain diffraction-quality crystals, extensive trials such as reducing the volume of the reservoir solution (from 1000 to 600 µl), changing the amounts of protein solution and reservoir solution in the drop (1 + 1 µl, 2 + 2 µl, 2 + 3 µl and 3 + 2 µl), addition of reducing agents and salts to the reservoir solution and microseeding (Bergfors, 2009 ▶) were carried out.
A microseed stock solution was prepared by adding 10 µl mother liquor to a crystallization drop which contained very thin plate-like crystals. This was pipetted into an 1.5 ml Eppendorf tube kept on ice. 50–100 µl mother liquor was added and spun down to wash the crystals. The supernatant was discarded and 50–100 µl mother liquor was again added and spun down. This procedure was repeated three times to wash the crystals. 50 µl mother liquor was then added to the washed crystals and a small glass bead was placed in the Eppendorf tube. The tube was vortexed for a few minutes to crush the crystals. This 50 µl solution was used as a seed stock. From this, dilutions of 1:50, 1:100 etc. were made which were used to streak the crystallization drops. Finally, a microseeded drop (1:100 dilution, streaked using a whisker) containing 1 µl protein solution and 1 µl reservoir solution [35%(w/v) PEG 3350, 0.1 M imidazole pH 6.5, 40 mM MgCl2] equilibrated against 1 ml reservoir solution gave stable native crystals of Srr-1-K4BD (0.3 × 0.15 × 0.05 mm) in one week (Fig. 3 ▶).
Figure 3.
Thin plate-like crystals of Srr-1-K4BD. The scale bar represents 0.1 mm.
2.3. Data collection and processing
Initial diffraction experiments were carried out using an in-house MAR 345 image-plate detector and Bruker MICROSTAR copper rotating-anode generator operating at 60 mA and 45 kV. Crystals were taken directly from the crystallization drop and flash-cooled in a liquid-nitrogen stream. The PEG 3350 in the crystallization drop acted as a cryoprotectant. The crystals diffracted to 5.0 Å resolution with an exposure time of 5 min per frame. Subsequently, diffraction experiments were carried out at the Elettra synchrotron-radiation laboratory in Italy. A native data set was collected on the XRD1 beamline from one crystal at 100 K. An oscillation range of 1° was used to collect 271 frames with an exposure of 55 s per frame at a crystal-to-detector distance of 150 mm. The diffraction data were processed using the program iMOSFLM (Battye et al., 2011 ▶). Data-collection statistics are given in Table 1 ▶.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 0.9789 |
| Space group | P21 |
| Unit-cell parameters | |
| a (Å) | 47.56 |
| b (Å) | 59.48 |
| c (Å) | 94.71 |
| β (°) | 93.95 |
| Resolution (Å) | 28.44–3.80 (4.01–3.80) |
| Total No. of reflections | 23741 |
| No. of unique reflections | 5320 |
| Completeness (%) | 99.5 (100) |
| Rmerge† (%) | 11.8 (39.7) |
| 〈I/σ(I)〉 | 8.3 (3.6) |
| Average mosaicity (°) | 1.5 |
| Multiplicity | 4.1 |
R
merge = 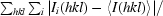
 , where 〈I(hkl)〉 is the mean of the intensity measurements Ii(hkl) and the summation extends over all reflections.
, where 〈I(hkl)〉 is the mean of the intensity measurements Ii(hkl) and the summation extends over all reflections.
3. Results and discussion
Assuming a molecular weight of 17 kDa for the protein and three molecules in the asymmetric unit, the resultant Matthews coefficient (V M; Matthews, 1968 ▶) was calculated to be 2.62 Å3 Da−1 with a solvent content of 53.06%. Alternatively, if there were two molecules in the asymmetric unit the Matthews coefficient was calculated to be 3.93 Å3 Da−1 with a solvent content of 68.71%. To determine the location of the noncrystallographic twofold or threefold symmetry, a self-rotation function study was performed with the program POLARRFN (Winn et al., 2011 ▶) using data between 20 and 4 Å resolution. The results of this study were inconclusive and therefore the noncrystallographic symmetry will be determined during subsequent structure-solution steps.
Srr-1-K4BD does not have a homologous structure in the PDB and therefore determination of its structure by molecular replacement is not possible. Srr-1-K4BD contains three methionine residues which can be modified to selenomethionine (SeMet) in order to resolve the phase problem using the multi-wavelength anomalous dispersion (MAD) or single-wavelength anomalous dispersion (SAD) techniques. SeMet-substituted protein was prepared using a methionine-auxotroph strain in minimal medium (M9 medium) as described in §2. Microcrystals of SeMet-substituted Srr-1-K4BD were obtained under similar conditions to those used for the native protein. However, the crystals were too small (the longest dimension was about 0.05 mm) to mount in a loop for diffraction analysis. Further crystallization trials are under way in order to obtain diffraction-quality crystals of SeMet-incorporated Srr-1-K4BD. A three-wavelength MAD data set or a SAD data set will be collected to solve the structure of Srr-1-K4BD.
Acknowledgments
KP thanks the Department of Biotechnology (DBT), Government of India for financial support in the form of a grant. KP thanks the Department of Science and Technology (DST), India and the Ministry of Foreign Affairs/Sincrotrone Trieste, Italy for financial support to visit the Elettra synchrotron facility for data collection. RS thanks the DBT and the University Grants Commission (UGC), Government of India for providing a fellowship.
References
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Beckmann, C., Waggoner, J. D., Harris, T. O., Tamura, G. S. & Rubens, C. E. (2002). Infect. Immun. 70, 2869–2876. [DOI] [PMC free article] [PubMed]
- Bergfors, T. (2009). Protein Crystallization, 2nd ed. La Jolla: International University Line.
- Cheng, Q., Stafslien, D., Purushothaman, S. S. & Cleary, P. (2002). Infect. Immun. 70, 2408–2413. [DOI] [PMC free article] [PubMed]
- Doran, K. S. & Nizet, V. (2004). Mol. Microbiol. 54, 23–31. [DOI] [PubMed]
- Gilbert, R. (2004). Int. J. Epidemiol. 33, 2–8. [DOI] [PubMed]
- Glaser, P., Rusniok, C., Buchrieser, C., Chevalier, F., Frangeul, L., Msadek, T., Zouine, M., Couvé, E., Lalioui, L., Poyart, C., Trieu-Cuot, P. & Kunst, F. (2002). Mol. Microbiol. 45, 1499–1513. [DOI] [PubMed]
- Guerrero, S. A., Hecht, H.-J., Hofmann, B., Biebl, H. & Singh, M. (2001). Appl. Microbiol. Biotechnol. 56, 718–723. [DOI] [PubMed]
- Jenkinson, H. F. & Lamont, R. J. (1997). Crit. Rev. Oral Biol. Med. 8, 175–200. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Navarre, W. W. & Schneewind, O. (1999). Microbiol. Mol. Biol. Rev. 63, 174–229. [DOI] [PMC free article] [PubMed]
- Patti, J. M., Allen, B. L., McGavin, M. J. & Höök, M. (1994). Annu. Rev. Microbiol. 48, 585–617. [DOI] [PubMed]
- Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual New York: Cold Spring Harbor Laboratory Press.
- Samen, U., Eikmanns, B. J., Reinscheid, D. J. & Borges, F. (2007). Infect. Immun. 75, 5405–5414. [DOI] [PMC free article] [PubMed]
- Schubert, A., Zakikhany, K., Schreiner, M., Frank, R., Spellerberg, B., Eikmanns, B. J. & Reinscheid, D. J. (2002). Mol. Microbiol. 46, 557–569. [DOI] [PubMed]
- Seifert, K. N., Adderson, E. E., Whiting, A. A., Bohnsack, J. F., Crowley, P. J. & Brady, L. J. (2006). Microbiology, 152, 1029–1040. [DOI] [PubMed]
- Sorge, N. M. van, Quach, D., Gurney, M. A., Sullam, P. M., Nizet, V. & Doran, K. S. (2009). J. Infect. Dis. 199, 1479–1487. [DOI] [PMC free article] [PubMed]
- Spellerberg, B., Rozdzinski, E., Martin, S., Weber-Heynemann, J., Schnitzler, N., Lütticken, R. & Podbielski, A. (1999). Infect. Immun. 67, 871–878. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.





