A 79 kDa fragment of FlgE from C. jejuni has been crystallized.
Keywords: FlgE, hook proteins, bacterial flagella, motility, universal joints, nanomachines
Abstract
A 79 kDa fragment of the bacterial flagellar hook protein FlgE from Campylobacter jejuni was cloned, overexpressed, purified and crystallized. Two different crystal forms were obtained. Synchrotron X-ray diffraction data showed that the first crystal form, which diffracted to 4.9 Å resolution, belonged to the tetragonal crystal system, with space group I4122 and unit-cell parameters a = b = 186.2, c = 386.6 Å, α = β = γ = 90°. The second crystal form diffracted to 2.5 Å resolution and belonged to the monoclinic crystal system, with space group P21 and unit-cell parameters a = 75.7, b = 173.8, c = 150.8 Å, α = γ = 90, β = 106.5°. SeMet protein was also overexpressed, purified and crystallized, and a 2.6 Å resolution MAD data set was collected.
1. Introduction
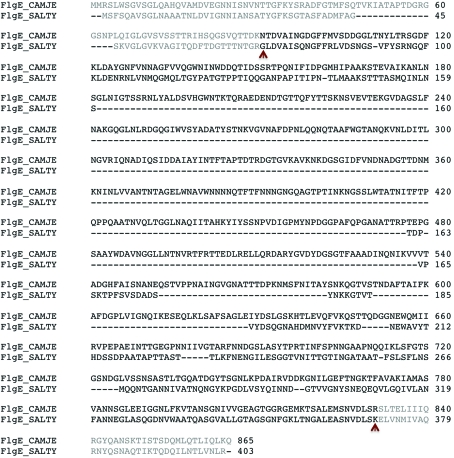
To swim in their living environment, bacteria have developed a nanomachine called the flagellum, which is found in both Gram-positive and Gram-negative bacteria (Berg & Anderson, 1973 ▶). The flagellum is made up of about 30 different proteins. It can be divided into three major parts: the filament, the hook and the basal body (Macnab, 2003 ▶; Berg, 2003 ▶). The basal body is a complex structure made up of many different proteins. It comprises the motor and the rotor, both of which are embedded in the inner membrane, and the bushing, which is located in the outer membrane. The hook and the filament are each made by the assembly of multiple copies of a single protein: FlgE and FliC, respectively (Namba & Vonderviszt, 1997 ▶). The filament is made by the assembly of a few thousand FliC molecules. The hook is shorter than the filament and consists of about 100 molecules of FlgE. The hook and the filament are connected by two junction proteins called hook-associated proteins 1 and 3 (HAP1 and HAP3; Homma et al., 1990 ▶). FlgE, HAP1, HAP3 and FliC are also known as the flagellar axial proteins (DePamphilis & Adler, 1971 ▶). The filament of Salmonella enterica serovar Typhimurium (S. typhimurium) is one of the most studied and has been used as a model for structural and functional studies of bacterial flagella in general. Although the proteins that make up the axial substructure of the flagellum have a wide range of molecular weights, depending on the bacterium, it has long been accepted that for all bacteria both the filament and the hook can be represented as tubular structures made by the assembly of 11 protofilaments. However, a recent study of the filament of Campylobacter jejuni proposed that its filament is made of seven protofilaments (Galkin et al., 2008 ▶). Based on the way that the bacterial flagellum is built, this model implies that the hook of C. jejuni is either also made of seven protofilaments or that it has a completely different junction structure compared with the S. typhimurium hook–filament junction. To obtain a better understanding of the flagellum of C. jejuni and to observe the structural differences from the hook of S. typhimurium, we proceeded to crystallize a 79 kDa fragment of FlgE2, gene Cj1729c, from C. jejuni strain NCTC11168. This fragment, consisting of amino-acid residues 91–931, was named FlgE79cj. C. jejuni possesses two flgE genes, Cj0043 and Cj1729c, which produce two distinct FlgE proteins: FlgE (58 kDa) and FlgE2 (92 kDa), respectively. FlgE and FlgE2 have 27% sequence identity; they have 29 and 33% identity, respectively, to the S. typhimurium FlgE protein. Studies performed by various groups have shown that FlgE2, not FlgE, makes the functional hook and that the deletion of the Cj0043 gene has no effect on motility (Hendrixson & DiRita, 2003 ▶). We wished to determine where the extra sequence of FlgE2 would fit when compared with the structure of S. typhimurium FlgE and to understand how the FlgE2 protein might fit into a seven-protofilament hook. A 79 kDa fragment of FlgE2 that starts at Asn91 and ends at Arg831 was selected based on sequence alignment with FlgE from S. typhimurium (Fig. 1 ▶); a 31 kDa fragment of FlgE from S. typhimurium has previously been crystallized (Samatey, Matsunami, Imada, Nagashima & Namba, 2004 ▶) and its atomic structure has been solved by X-ray crystallography (Samatey, Matsunami, Imada, Nagashima, Shaikh et al., 2004 ▶). Here, we describe the expression, purification and crystallization of native and selenomethionine (SeMet) derivative FlgE79cj protein and the complete high-resolution X-ray diffraction data of these two crystals.
Figure 1.
Sequence alignment of FlgE2 from C. jejuni (FlgE_CAMJE) and FlgE from S. typhimurium (FlgE_SALTY). The brown arrows indicate the start and the end of the 79 kDa fragment of FlgE2 and the 31 kDa fragment of FlgE from S. typhimurium.
2. Experimental procedures
2.1. Cloning, expression and purification
The DNA sequence encoding FlgE79cj (amino-acid residues 91–831) was amplified by polymerase chain reaction (PCR) from C. jejuni strain NCTC11168 genomic DNA with the 5′ primer CAAGCATATGAACACTGACGTTGCTATAAATGGC, generating an NdeI site and a start codon for an additional methionine, and the 3′ primer GGTGCTCGAGTTAACGACTTAAATCCACATTTGACATTTC, generating a stop codon and a XhoI site. The PCR fragment was digested with NdeI and XhoI and ligated into the T7 expression vector pET22b (Novagen). The plasmid was transformed into Escherichia coli Rosetta (DE3) cells for expression. SeMet-substituted FlgE79cj was expressed in methionine-auxotrophic E. coli strain B834 (DE3)/pRARE cells (Novagen) using SeMet core medium (Wako Pure Chemical Industries).
A single colony of Rosetta (DE3) cells harbouring FlgE79cj was inoculated into 50 ml Luria broth medium containing 50 µg ml−1 ampicillin and 34 µg ml−1 chloramphenicol and allowed to grow overnight at 310 K at 275 rev min−1. The culture was used to inoculate 5 l Luria broth containing antibiotics in a 10 l fermentor. The cells were induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside at an OD600 of 0.5 and cultivation continued for 3 h at 310 K. The culture broth was harvested by centrifugation at 8000g for 20 min. The cell pellet was suspended in 50 mM NaCl, 20 mM Tris–HCl pH 7.5 and disrupted by sonication. The solution was centrifuged at 10 000g for 15 min to remove the cell debris. The supernatant was centrifuged at 100 000g for 1 h and the supernatant was loaded onto a HiTrap Q FF column (GE Healthcare) equilibrated with 20 mM Tris–HCl pH 7.5. Elution was performed with the same buffer solution with a linear gradient of NaCl concentration from 0 to 0.5 M. The main peak fraction was dialyzed against 20 mM Tris–HCl pH 7.5 and loaded onto a HiLoad Q Sepharose ‘High Performance’ anion-exchange column (GE Healthcare) equilibrated with 20 mM Tris–HCl pH 7.5. Elution was performed with a linear gradient of NaCl concentration from 0 to 0.5 M. To enhance the purification, the elution fractions containing the protein were gathered together and loaded onto a Superdex 200 gel-filtration column (GE Healthcare) equilibrated with 100 mM NaCl, 10 mM Tris–HCl pH 7.5 and eluted with the same solution. The fractions containing the protein were gathered together and dialyzed overnight against 5 mM Tris–HCl pH 7.5. The protein was concentrated to a final concentration of 30 mg ml−1 using a Centriprep centrifugal filter device (Millipore). This protein stock solution was kept at 277 K.
2.2. Crystallization and data collection
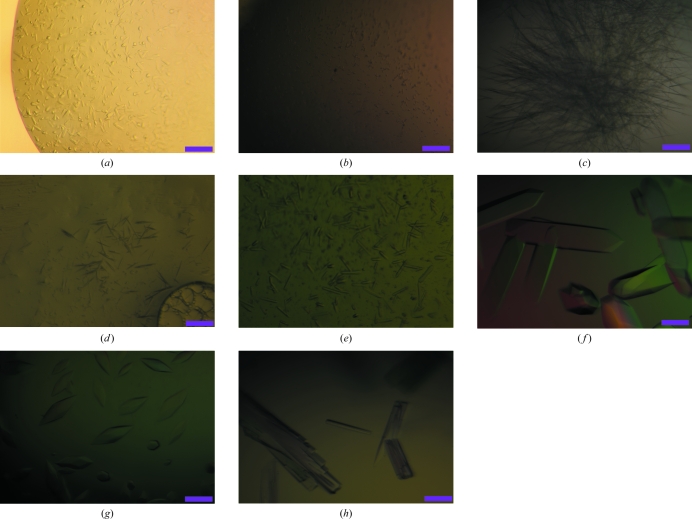
Initial screening of crystallization conditions was carried out using the sitting-drop vapour-diffusion method. The crystallization drops were initially prepared with an automated nanolitre liquid-handling system (Mosquito, TTP LabTech) in 96-well plates. Three different protein concentrations, 5, 10 and 15 mg ml−1, were used and crystallization plates were prepared by mixing 150 nl protein solution with 150 nl reservoir solution and were equilibrated against 120 ml reservoir solution. Crystallization plates were equilibrated at 293, 288 and 283 K. Screening kits from Hampton Research and Emerald BioSystems were used in the initial crystallization trials. Many conditions from the initial screen gave interesting results, mainly at 293 and 288 K, which were used as starting points for further crystallization refinements. The reservoir solutions in most of these conditions contained sulfate salts or polyethylene glycol (PEG) as precipitant agents (Figs. 2 ▶ a and 2 ▶ b). Fine-tuning of the crystallization of FlgE79cj was performed manually using the hanging-drop vapour-diffusion method with a reservoir volume of 1 ml and a drop volume of 10 µl. The PEG precipitant was omitted because changes in the molecular weight and concentration of PEG did not provide the expected improvements in the crystal shape and size (Figs. 2 ▶ c and 2 ▶ d).
Figure 2.
Crystals of FlgE79cj from C. jejuni obtained in (a) 1.5 M ammonium sulfate, 0.1 M Tris–HCl pH 8.5; (b) 20% PEG 8K, 0.1 M NaCl, 0.1 M CAPS pH 10.5; (c) 10% PEG 10K, 4% dioxane, 0.1 M citrate pH 5.5; (d) 30% PEG 400, 0.1 M CHES pH 9.5; (e) 1.5 M ammonium sulfate, 50 mM NaCl, 10% 2-propanol, 0.1 M CHES pH 9.5; (f) 0.8 M K2HPO4, 5% 2-propanol, 0.1 M acetate pH 4.5; (g) 1.4 M MgSO4, 50 mM NaCl, 6 mM CdSO4, Tris–HCl pH 8.5; (h) 1.2 M K2HPO4, 50 mM NaCl, 5 mM Tl2SO4, 12% DMSO, 50 mM MES pH 6.0. The scale bar on the bottom right represents 0.1 mm.
After extensively screening many sulfate salts as precipitants, we obtained crystals at 288 K in 1.5 M ammonium sulfate, 10% 2-propanol, 50 mM NaCl, 100 mM CHES pH 9.5 (Fig. 2 ▶ e). Unfortunately, these crystals dissolved when we tried to pick them up. We decided to abandon crystallization conditions that contained ammonium sulfate and to focus on magnesium sulfate (MgSO4) and dipotassium phosphate (K2HPO4). Using these two precipitant agents, we obtained crystals after three weeks in a wide range of buffers with pH varying from 4.5 to 9.5 and with a protein concentration of 15 mg ml−1. The best crystals were obtained at 293 K in 0.8 M K2HPO4, 0.1 M acetate pH 4.5. Prior to X-ray diffraction experiments, these crystals were cryoprotected by a brief soak in a solution corresponding to the reservoir solution supplemented with glycerol to a final concentration of 20%. The crystals were mounted on a cryoloop and data were obtained at 100 K. The crystals belonged to space group I4122, with unit-cell parameters a = b = 186.2, c = 386.6 Å, α = β = γ = 90°. Unfortunately, these crystals did not diffract beyond 5.0 Å resolution on beamline BL44XU of the SPring-8 synchrotron. The addition of NaCl and changing the buffer did not provide any improvement. To improve the diffraction limit of the crystals, we decided to investigate the use of additives with the initial crystallization conditions. Dipotassium phosphate in combination with 5% 2-propanol gave better looking crystals (Fig. 2 ▶ f), but there was no improvement in the diffraction limit. Increasing the concentration of 2-propanol did not provide any improvement. 2-Propanol combined with magnesium sulfate did not give improved results. However, we found that by using cadmium sulfate with magnesium sulfate we obtained crystals that seemed to have a different shape (Fig. 2 ▶ g). We switched to dimethyl sulfoxide (DMSO) as an additive. The addition of less than 10% of DMSO to the dipotassium phosphate conditions produced crystals that were unchanged in size, shape and diffraction limit. When the concentration of DMSO in the reservoir was between 10 and 14% (5 and 7% in the drop) relatively thin long stack crystals were obtained (Fig. 2 ▶ h). The crystals obtained in conditions containing more than 12% DMSO were mounted on a cryoloop and flash-cooled in liquid nitrogen without adding cryoprotectant. X-ray diffraction data were collected at 100 K. These crystals diffracted to about 2.1 Å resolution and belonged to space group P21, with unit-cell parameters a = 75.7, b = 173.8, c = 150.8 Å, α = γ = 90, β = 106.5°. SeMet-containing crystals of FlgE79cj were obtained under similar conditions. The data-collection statistics are summarized in Table 1 ▶.
Table 1. Diffraction data statistics for FlgE79cj crystals.
Values in parentheses are for the highest resolution shell.
| Native | SeMet peak | SeMet edge | |
|---|---|---|---|
| X-ray source | BL41XU, SPring-8 | BL44XU, SPring-8 | |
| CCD detector | Rayonix MX225HE | ||
| Space group | P21 | P21 | P21 |
| Unit-cell parameters | |||
| a (Å) | 75.26 | 75.45 | 75.45 |
| b (Å) | 173.21 | 173.76 | 173.76 |
| c (Å) | 146.94 | 147.69 | 147.69 |
| β (°) | 102.67 | 102.98 | 102.98 |
| Wavelength (Å) | 1.0 | 0.9791 | 0.9794 |
| No. of images | 180 | 180 | 180 |
| Resolution (Å) | 32.09–2.50 (2.64–2.50) | 37.69–2.60 (2.74–2.60) | 37.71–2.60 (2.74–2.60) |
| Completeness (%) | 97.3 (94.6) | 99.9 (99.9) | 100 (100) |
| Total reflections | 440763 (58820) | 434336 (61878) | 435907 (62299) |
| Unique reflections | 123178 (17416) | 113491 (16522) | 113633 (16564) |
| Multiplicity | 3.6 (3.4) | 3.8 (3.7) | 3.8 (3.8) |
| Rmerge† (%) | 8.6 (39.9) | 13.0 (45.6) | 13.7 (52.1) |
| Mean I/σ(I) | 11.0 (3.1) | 8.0 (3.0) | 7.4 (2.7) |
R
merge = 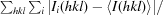
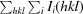 , where Ii(hkl) is the intensity of the ith measurement of reflection hkl and 〈I(hkl)〉 is the mean value of Ii(hkl) for all i measurements.
, where Ii(hkl) is the intensity of the ith measurement of reflection hkl and 〈I(hkl)〉 is the mean value of Ii(hkl) for all i measurements.
3. Results and discussion
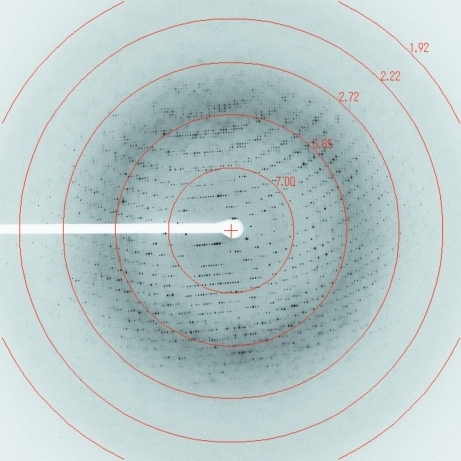
Diffraction data sets were collected at SPring-8 on beamline BL44XU for the SeMet-containing crystal and on beamline BL41XU for the native crystal (Fig. 3 ▶). The data sets were processed with MOSFLM (Leslie, 2006 ▶) and scaled with SCALA (Evans, 2006 ▶) from the CCP4 software suite (Winn et al., 2011 ▶). Data reduction and systematic absences of reflections indicated that the FlgE79cj crystals obtained in DMSO belonged to space group P21. The Matthews coefficient (V M; Matthews, 1968 ▶) was calculated to be 3.0 Å3 Da−1 for four molecules per asymmetric unit and 2.4 Å3 Da−1 for five molecules per asymmetric unit. These values correspond to solvent contents of 58% and 48%, respectively. Using the PHENIX software package (Adams et al., 2010 ▶), the program phenix.autosol found 42 SeMet sites. A few of the SeMet sites found had a very low occupancy. FlgE79cj contains ten Met residues (including the N-terminal methionine). The initial model-building calculations on the basis of four and five molecules per asymmetric unit were performed using the program phenix.autobuild. These preliminary calculations showed that the asymmetric unit contained four molecules. The structure of FlgE79cj is now in the final stages of refinement.
Figure 3.
Diffraction pattern from a native crystal of FlgE79cj.
The difficulties in the crystallization of FlgE79cj were a consequence of the fact that the initial crystallization trials gave many hits. It was difficult to refine all of these initially interesting conditions. The crystal used to collect the native data was obtained using a solution consisting of 1.2 M K2HPO4, 50 mM NaCl, 5 mM Tl2SO4, 12% DMSO, 50 mM MES pH 6.0. Although XAFS measurements did not confirm the presence of thallium in the crystal, crystals grown with thallium tended to diffract better than those grown without. The SeMet-containing crystal used to collect the MAD data grew using a [;solution consisting of 1.4 M K2HPO4, 50 mM NaCl, 14% DMSO, 0.1 M citrate pH 5.5 with a protein concentration of 15 mg ml−1.
Many of the initial crystals, which belonged to space group I4122, were large enough to be checked at 100 K on our laboratory X-ray source (Rigaku FR-E+); these crystals diffracted poorly despite a long exposure time. The need to check the crystals on a synchrotron beamline slowed down the project and prevented extensive screening of both pH and salt additives when using DMSO. As often happens in protein crystallization, the nice-looking crystals are not always the best diffracting.
Acknowledgments
We thank Dr O. Gundoglu for providing us with C. jejuni genomic DNA. We thank the staff members of beamline BL44XU for their great help in assessing the quality of our crystals and the staff members of beamline BL41XU for helping us to collect the native data. This work was supported by direct funding provided by OIST.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Berg, H. C. (2003). Annu. Rev. Biochem. 72, 19–54. [DOI] [PubMed]
- Berg, H. C. & Anderson, R. A. (1973). Nature (London), 245, 380–382. [DOI] [PubMed]
- DePamphilis, M. L. & Adler, J. (1971). J. Bacteriol. 105, 384–395. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Galkin, V. E., Yu, X., Bielnicki, J., Heuser, J., Ewing, C. P., Guerry, P. & Egelman, E. H. (2008). Science, 320, 382–385. [DOI] [PubMed]
- Hendrixson, D. R. & DiRita, V. J. (2003). Mol. Microbiol. 50, 687–702. [DOI] [PubMed]
- Homma, M., DeRosier, D. J. & Macnab, R. M. (1990). J. Mol. Biol. 213, 819–832. [DOI] [PubMed]
- Leslie, A. G. W. (2006). Acta Cryst. D62, 48–57. [DOI] [PubMed]
- Macnab, R. M. (2003). Annu. Rev. Microbiol. 57, 77–100. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Namba, K. & Vonderviszt, F. (1997). Q. Rev. Biophys. 30, 1–65. [DOI] [PubMed]
- Samatey, F. A., Matsunami, H., Imada, K., Nagashima, S. & Namba, K. (2004). Acta Cryst. D60, 2078–2080. [DOI] [PubMed]
- Samatey, F. A., Matsunami, H., Imada, K., Nagashima, S., Shaikh, T. R., Thomas, D. R., Chen, J. Z., DeRosier, D. J., Kitao, A. & Namba, K. (2004). Nature (London), 431, 1062–1068. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.