Dodecyl-β-d-selenomaltoside in a leukotriene C4 synthase crystal exhibited sufficient anomalous diffraction for multiwavelength anomalous diffraction phasing.
Keywords: membrane proteins, MAD phasing, seleno-detergents, leukotriene C4 synthase
Abstract
Dodecyl-β-d-selenomaltoside (SeDDM) is a seleno-detergent with a β-glycosidic seleno-ether in place of the ether moiety in dodecyl-β-d-maltoside. Seleno-detergents are candidates for heavy-atom agents in experimental phasing of membrane proteins in protein crystallography. Crystals of a nuclear membrane-embedded enzyme, leukotriene C4 synthase (LTC4S), in complex with SeDDM were prepared and a multiwavelength anomalous diffraction (MAD) experiment was performed. The SeDDM in the LTC4S crystal exhibited sufficient anomalous diffraction for determination of the structure using MAD phasing.
1. Introduction
The discovery of heterologous expression has driven a rapid increase in the structure determination of membrane proteins. The number of membrane-protein structures identified using heterologous expression since 2007 is twice that identified from natural sources (Bill et al., 2011 ▶). Recombinant techniques allow crystals of novel membrane proteins that cannot be obtained from natural sources to be prepared, which enables data collection for experimental phasing. This situation is similar to that of water-soluble proteins in the late 1980s. The dramatic increase in the structure determination of water-soluble proteins was achieved by MAD phasing methods using energy-tunable synchrotron-radiation X-rays and selenomethionine (SeMet) labelling of recombinant proteins to give heavy-atom derivatives, in conjunction with sophisticated computer hardware and software (Joachimiak, 2009 ▶)
SeMet is widely applicable for the preparation of heavy-atom derivatives owing to the diverse array of hosts for SeMet protein expression, which include Escherichia coli, yeast, insect cells and mammalian cells (Walden, 2010 ▶). Eukaryotic hosts favourably produce functionally folded membrane proteins from higher organisms; however, the production of sufficient quantities of SeMet-labelled membrane proteins for crystallography remains a challenge owing to the time-consuming and expensive large-scale culture required, as well as the toxicity of SeMet. SeMet phasing needs a Met residue(s) for substitution, not including the N-terminal Met residue in the protein; therefore, it may be necessary to mutate certain residue(s) to methionine in the well ordered region (Ago et al., 2007 ▶). There are still difficulties associated with the recombinant expression and SeMet labelling of membrane proteins. Derivatizations of membrane-protein crystals are achieved by extensive screening of heavy-atom agents.
Heavy-atom soaking and cocrystallization remain important methods for phasing membrane proteins that have novel folds (Morth et al., 2006 ▶). To derivatize proteins, heavy metals or halogens have been used to prepare isomorphous derivative(s) of the covalent/noncovalent bonds at certain hydrophilic residues via soaking or cocrystallization methods. However, derivatized crystals frequently diffract less well than native crystals and the incorporation of heavy atom(s) occasionally causes non-isomorphism with the native crystal. These problems are particularly serious for labile membrane-protein crystals because native crystals of membrane proteins often diffract poorly.
Dodecyl-β-d-selenomaltoside (SeDDM; Fig. 1 ▶ a) is selenosubstituted dodecyl-β-d-maltoside (DDM), which is a standard detergent for membrane-protein crystals. The substitution of oxygen by selenium maintains similar chemical properties, including geometry, because selenium and oxygen are in the same group of the periodic table (group 16). Therefore, this seleno-detergent may be a suitable heavy-atom derivative for membrane proteins, since DDM has been used for solubilization and crystallization (Sonoda et al., 2011 ▶).
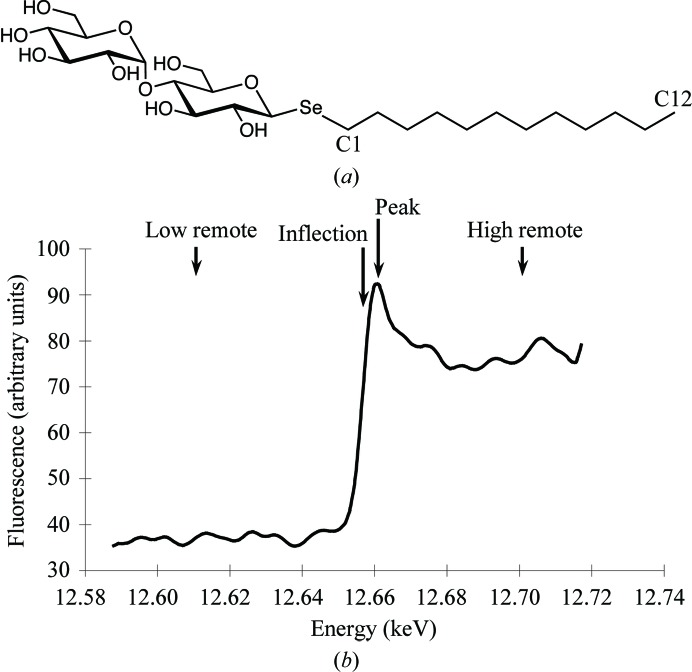
Figure 1.
(a) The chemical structure of dodecyl-β-d-selenomaltoside (SeDDM). (b) Fluorescence scan of the Se K edge of the LTC4S crystal complex indicating the data-collection energies.
Detergent molecules are occasionally ordered in crystals; for example, 60 membrane-protein structures have been reported in complex with alkyl-β-d-maltosides or alkyl-β-d-glucosides (Appendix A ). This represents one fifth of the 299 unique membrane proteins among a total of 847 Protein Data Bank (PDB) entries (http://blanco.biomol.uci.edu/mpstruc/). This includes 12 membrane proteins complexed with DDM, 11 proteins complexed with nonyl-β-d-glucoside, 29 proteins complexed with octyl-β-d-glucoside and eight proteins complexed with other alkyl-β-d-maltosides (Appendix A ). These structures suggest that seleno-detergents are potentially applicable for experimental phasing of membrane proteins. Leukotriene C4 synthase (LTC4S) should be a good candidate to validate SeDDM phasing since we have previously solved the crystal structure of this membrane protein via X-ray crystallography with DDM (Ago et al., 2007 ▶; Saino et al., 2011 ▶). There are several defined DDM molecules per 17 kDa monomer of LTC4S and the biologically functional unit of LTC4S is a homotrimer.
In addition to its use in experimental phasing, the anomalous signal of the Se atom can act as a reference to more accurately define the position of SeDDM. The reference should be useful especially when the detergent molecule binds to functionally important sites of the membrane protein such as the lipid-binding site. In the crystal structure of LTC4S the DDM molecule occupies the active-site cleft at the interface of adjacent monomers (Ago et al., 2007 ▶; Saino et al., 2011 ▶). LTC4S catalyzes the conjugation of glutathione (GSH) and leukotriene A4 (LTA4), which is an unsaturated fatty acid that is involved in eicosanoid biosynthesis. The DDM binding site is the putative binding site of the substrate LTA4, since the alkyl chain of DDM resides in the hydrophobic cleft connected to the bound GSH site. Based on this DDM-binding mode, the LTA4-binding model was proposed using the DDM molecule as a substrate mimic (Ago et al., 2007 ▶; Martinez Molina et al., 2007 ▶). However, the position of the alkyl tail and glycosidic O atom of the DDM molecule remained ambiguous in the previous structure because the electron densities of oxygen and carbon could not be accurately discriminated even in the high-resolution structure (Saino et al., 2011 ▶).
In this study, we formed complex crystals of SeDDM with LTC4S to show the applicability of SeDDM as a heavy-atom agent for MAD phasing. We successfully executed structural determination via MAD phasing of SeDDM. The anomalous electronic densities of the Se atoms provided a more defined structure of the SeDDM molecules that included the precise orientation of the alkyl chain in the proposed LTA4-binding cleft of LTC4S.
2. Materials and methods
2.1. Purification and crystallization
Human LTC4S was overexpressed by Schizosaccharomyces pombe with a His6 tag at the C-terminus and was purified using DDM as described previously (Ago et al., 2007 ▶; Saino et al., 2011 ▶). In brief, LTC4S was solubilized using a DDM/deoxycholic acid mixture and was purified using S-hexylglutathione affinity resin, Ni–agarose resin and size-exclusion chromatography. A PD-10 desalting column was equilibrated with 25 ml 0.04%(w/v) SeDDM (Affymetrix), 20 mM MES–NaOH pH 6.5, 5 mM GSH; 2.5 ml purified protein solution was then applied and eluted with 3.5 ml buffer for detergent exchange. The SeDDM-exchanged protein was concentrated to 6 mg ml−1 and stored at 193 K. Crystals of LTC4S with SeDDM were grown at 293 K using the sitting-drop vapour-diffusion method with equal amounts of protein solution and reservoir solution (0.1 M MES–NaOH pH 6.5, 1.6 M ammonium sulfate, 0.4 M MgCl2). The crystals were transferred into a harvesting solution that did not contain SeDDM (0.1 M MES–NaOH pH 6.5, 2.4 M ammonium sulfate, 50 mM GSH). The crystals were then dipped into a cryosolution supplemented with 15%(v/v) ethylene glycol and cooled in liquid nitrogen.
2.2. Data collection, processing and phasing
MAD data were collected at wavelengths of 0.97909 Å (12.663 keV, peak), 0.97938 Å (12.659 keV, inflection), 0.97600 Å (12.703 keV, high-energy remote) and 0.98300 Å (12.613 keV, low-energy remote), as estimated from a fluorescence scan of the Se K edge, at 100 K using the BL26B2 beamline at the SPring-8 facility (Fig. 1 ▶ b). A total of 720 images were collected from one crystal, with 180 images for each wavelength and an oscillation of 1°. The data-collection statistics are given in Table 1 ▶. The images were processed using MOSFLM (Leslie, 1992 ▶; Winn et al., 2011 ▶) and scaled using SCALA from CCP4 (Winn et al., 2011 ▶). SOLVE in PHENIX (Adams et al., 2010 ▶; Terwilliger, 2000 ▶) was used to determine the selenium substructure and calculate the initial phase; the resultant electron density was improved using RESOLVE in PHENIX (Adams et al., 2010 ▶; Terwilliger, 2000 ▶). Structural refinement was carried out using REFMAC5 (Murshudov et al., 2011 ▶; Winn et al., 2011 ▶); CNS (Brünger et al., 1998 ▶) was used for simulated annealing and Coot (Emsley et al., 2010 ▶) was used for model building. The mean phase error of the initial phase angles against the model phases of the refined structure was calculated using CPHASEMATCH (Winn et al., 2011 ▶).
Table 1. Data-collection, phasing and refinement statistics.
Values in parentheses are for the highest shell.
| Low remote | High remote | Peak | Inflection | |
|---|---|---|---|---|
| Unit-cell parameters () | a = b = c = 168.6 | a = b = c = 168.8 | a = b = c = 168.5 | a = b = c = 168.7 |
| Space group | F23 | |||
| Wavelength () | 0.98300 | 0.97600 | 0.97909 | 0.97938 |
| Resolution () | 26.73.20 (3.373.20) | 26.73.20 (3.373.20) | 26.63.20 (3.373.20) | 26.73.20 (3.373.20) |
| Total No. of images | 180 | 180 | 180 | 180 |
| R merge | 0.13 (0.37) | 0.13 (0.40) | 0.13 (0.40) | 0.13 (0.37) |
| Completeness (%) | 89.9 (69.3) | 90.0 (68.9) | 90.0 (69.5) | 90.0 (68.8) |
| Multiplicity | 21.9 (21.1) | 21.9 (22.2) | 21.9 (22.1) | 21.9 (22.2) |
| I/(I) | 5.6 (2.1) | 5.3 (1.9) | 5.4 (1.9) | 5.3 (2.1) |
| Phasing | ||||
| No. of Se sites | 3 | |||
| FOM | 0.24 | |||
| Refined f/f | 5.53/3.81 | 8.04/4.22 | 9.61/2.55 | |
| Density modification | ||||
| Solvent content (%) | 68 | |||
| FOM | 0.75 | |||
| Refinement | ||||
| Resolution range () | 25.43.20 | |||
| R/R free | 0.203/0.210 | |||
| B factor (2) | ||||
| Protein | 30.5 | |||
| Ligands | 31.1 | |||
| Detergents | 62.3 | |||
| Se site 1 | 68.5 | |||
| Se site 2 | 94.5 | |||
| Se site 3 | 59.0 | |||
| R.m.s. deviations | ||||
| Bond lengths () | 0.002 | |||
| Bond angles () | 0.511 | |||
| Chiral volumes (3) | 0.032 | |||
| Ramachandran plot | ||||
| Favoured (%) | 95.2 | |||
| Allowed (%) | 4.8 | |||
| Disallowed (%) | 0 | |||
3. Results and discussion
3.1. Selenium MAD phasing
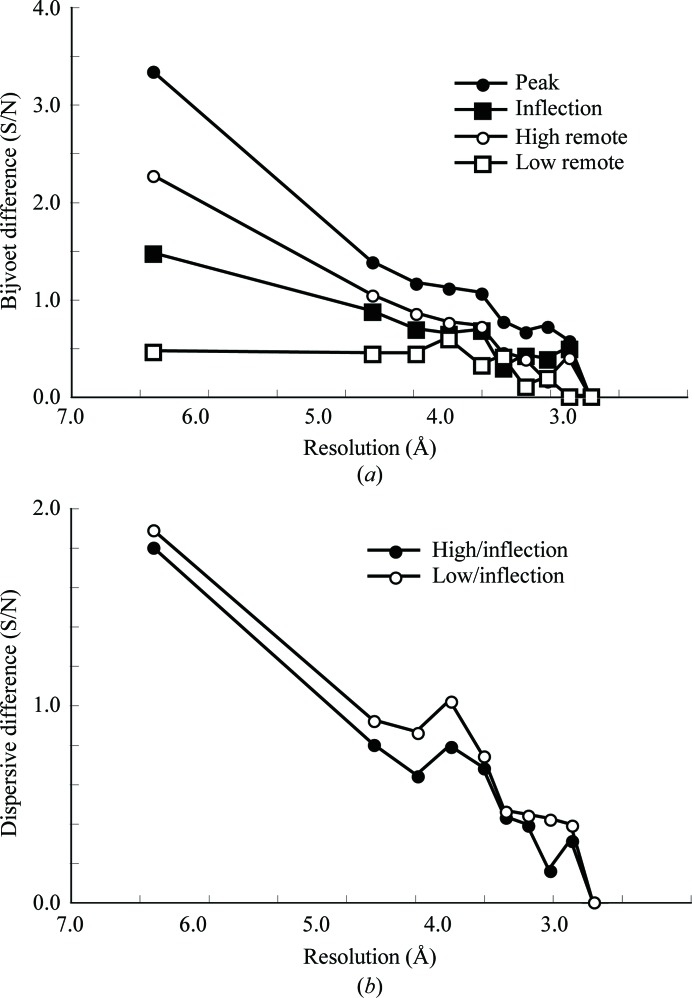
The cocrystallized SeDDM in the LTC4S crystal gave a significant anomalous signal (Fig. 2 ▶). The signal-to-noise ratio was over 1.0 up to 4.0 Å resolution for the peak data set (Adams et al., 2010 ▶; Terwilliger, 2000 ▶). The inflection and high-energy remote data also exhibited higher signal-to-noise ratios than the low-energy remote data set up to 4.0 Å resolution, which suggested that the anomalous contribution of the Se atoms in the diffraction data was statistically significant. The dispersive signals estimated with high-remote/inflection and low-remote/inflection data were comparable and the peak, inflection and high-energy remote data sets were used for substructure determination of the Se atoms. An anomalous difference Patterson map (Fig. 3 ▶) showed significant peaks corresponding to the selenium sites, as described below.
Figure 2.
(a) Bijvoet difference signal-to-noise ratio (S/N) for the four MAD data sets. S/N ratios were calculated using SOLVE in PHENIX (Adams et al., 2010 ▶; Terwilliger, 2000 ▶). (b) Dispersive difference (S/N) of the MAD data sets.
Figure 3.
Anomalous difference Patterson map calculated from the peak data set at a resolution of 3.2 Å. The asymmetric area of the Harker section (u = 0) is drawn at contour levels from 1.5σ to 10σ in 0.5σ steps. The numbered crosses indicate symmetry-related self-vectors of the three selenium sites assigned using self-vectors calculated from the solution given by SOLVE in PHENIX (Adams et al., 2010 ▶; Terwilliger, 2000 ▶).
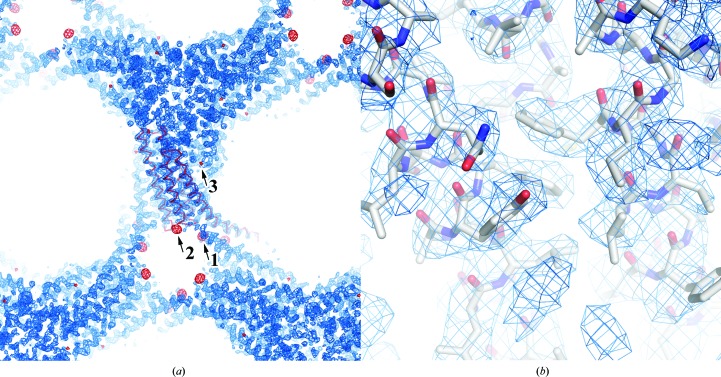
There were three selenium sites: two were found using SOLVE in PHENIX (Adams et al., 2010 ▶; Terwilliger, 2000 ▶), while the other was manually picked from the anomalous difference Fourier map (Fig. 4 ▶). The first two sites had high occupancy in absolute scaling: 0.64 (B factor of 85.0 Å2) at site 1 and 0.54 (B factor of 86.9 Å2) at site 2, as determined by heavy-atom searching. The initial phases were calculated using these two heavy-atom positions and this was followed by phase improvement to 3.2 Å resolution. The corrected overall figure of merit was 0.75 and the solvent content was 68%. An anomalous difference Fourier map was calculated using the experimental phases of the two sites and the given corresponding peaks of 25σ and 15σ, respectively. Besides these two peaks, an additional peak corresponding to a minor site (6.3σ) was found. With the additional site incorporated, the occupancy and B-factor values refined to 0.41 and 63.0 Å2, respectively, for site 1, 0.28 and 61.1 Å2 for site 2 and 0.07 and 35.0 Å2 for site 3. The refined values of f′ and f′′ were −8.04 and 4.22, respectively, for the peak data, −9.16 and 2.25 for the inflection data and −5.53 and 3.81 for the high-remote data. In the anomalous difference Patterson peaks, self-cross-vector peaks were present with their symmetry-related positions; site 1 was the highest peak (Fig. 3 ▶). The phases were recalculated to include the additional site and applied to RESOLVE in PHENIX (Adams et al., 2010 ▶; Terwilliger, 2000 ▶). The mean phase errors of the final model were 46.4° and 43.8° for the two-site and the three-site calculations, respectively (Adams et al., 2010 ▶; Terwilliger, 2000 ▶). The latter phase angles are closer to those of the refined phases; this indicates that site 3 contributes substantially to phasing even though its occupancy is not high. The resultant election-density map was sufficient to unambiguously build an atomic model (Fig. 4 ▶).
Figure 4.
The electron-density map calculated from the experimental phases after density modification. (a) The blue mesh is the electron-density map contoured at 1.5σ. The red mesh is the anomalous difference Fourier map calculated from the experimental phases and the anomalous differences of the peak data set contoured at 5σ. The red ribbon shows the Cα trace of an asymmetric unit. (b) Magnified view of the electron-density map with the corresponding structure of the refined LTC4S model.
De novo model building was carried out manually to avoid model bias from the previous DDM-complex structure of LTC4S (Ago et al., 2007 ▶; Saino et al., 2011 ▶). The structure of LTC4S in complex with SeDDM was refined using the low-remote data set to R = 0.203 and R free = 0.210 at 3.2 Å resolution using REFMAC5 (Murshudov et al., 2011 ▶; Winn et al., 2011 ▶; Table 1 ▶). The root-mean-square deviation between the refined structure and the previous high-resolution structure (PDB entry 3pcv; Saino et al., 2011 ▶) was 0.62 Å for all atoms.
3.2. SeDDM binding sites
There were three SeDDM binding sites with selenium peaks in the anomalous difference Fourier map (Fig. 5 ▶ a). Site 1 was between helices IV and V and helices IV* and V* in a twofold symmetry-related molecule. Site 2 was along the transmembrane helices I and III, and site 3 was in the active site of LTC4S. The alkyl chain of SeDDM in each site was bound in the same mode as that of the corresponding DDM in the previous structures (PDB entries 3pcv, 3leo, 2pno and 2uuh; Saino et al., 2011 ▶, Rinaldo-Matthis et al., 2010 ▶; Ago et al., 2007 ▶; Martinez Molina et al., 2007 ▶). These results indicate that SeDDM molecules bound competitively with DDM molecules.
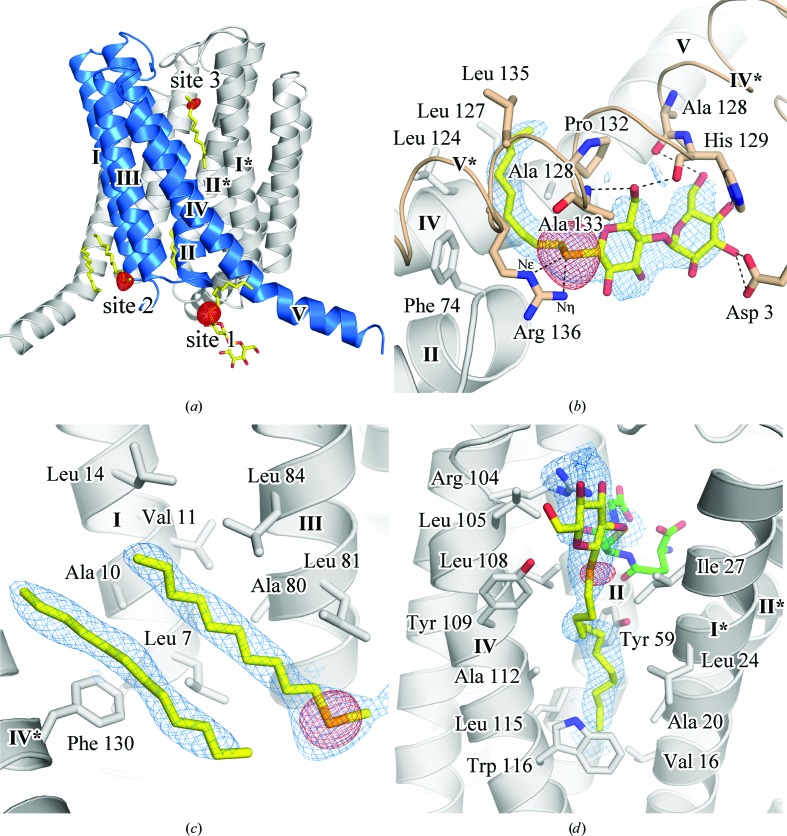
Figure 5.
(a) SeDDM molecules in an LTC4S trimer with anomalous difference Fourier map. The red mesh is the anomalous difference Fourier map calculated from the refined phases and the anomalous differences of the peak data set contoured at 5σ of an asymmetric unit. SeDDM is represented by a stick model with yellow C atoms. The blue cartoon model indicates a monomer in the LTC4S trimer and the roman numerals show the sequential order of the helices. Magnified views of the SeDDM binding sites are shown in (b), (c) and (d), with the electron densities of the SeDDM molecules represented by a blue mesh contoured at 1.0σ. (b) SeDDM and its surrounding residues in site 1. The dashed lines indicate hydrogen bonds between SeDDM and amino-acid residues. The wheat-coloured models labelled IV* and V* indicate helices IV and V of a twofold-symmetry-related molecule in the crystal packing. (c) SeDDM of site 2. The neighbouring alkyl chain and its election density are also displayed. (d) SeDDM in site 3. The bound glutathione is represented by green C atoms.
At site 1 the Se atom was located close to the guanidino group of Arg136 at distances of 3.3 and 3.5 Å to Nη and N∊, respectively (Fig. 5 ▶ b). The maltoside of SeDDM was surrounded by polar groups, i.e. Asp3 of helix I*, the carbonyl O atom of Ala128, the Nδ and carbonyl O atoms of His129 of helix IV* and the backbone amide of Ala133 of helix V* (Fig. 5 ▶ b). Electron density for the alkyl chains was observed for the C1–C9 C atoms; the alkyl chains were close to (i.e. within 4 Å of) hydrophobic residues, including Phe74, Leu124, Leu127, Ala128, Pro132 and Leu135.
The strong peak (25σ) in the anomalous difference Fourier map indicates that the selenium is stably located at this site. In addition, the electron densities corresponding to the maltose moiety were clearer with SeDDM than with DDM (PDB entries 2pno, 2uui, 2uuh, 3leo and 3pcv; Ago et al., 2007 ▶; Martinez Molina et al., 2007 ▶; Rinaldo-Matthis et al., 2010 ▶; Saino et al., 2011 ▶). These results suggest that polar interactions with selenium and maltoside, such as hydrogen bonding, provide tighter binding of SeDDM than DDM.
The SeDDM in site 2 (Fig. 5 ▶ c) was surrounded by hydrophobic residues, i.e. Leu7, Ala10, Val11 and Leu14 in helix I and Ala80, Leu81 and Leu84 in helix III. These residues were within 4.2 Å of the alkyl chain of SeDDM. Electron densities for the Se atom and alkyl chains were clearly observed, whereas that of the maltose moiety was not definitive. According to the orientation of the alkyl chain and the Se atom, the maltose moiety beyond the seleno-ether must extend into the solvent. Electron density corresponding to the putative alkyl chain together with the SeDDM molecule of site 2 was present, but no anomalous peak was observed (Fig. 5 ▶ c).
Site 3 is the putative LTA4-binding site in the active site of LTC4S. The anomalous peak of the Se atom overlapped with the end of the long electron density of the C1–C12 alkyl chain, which indicates the position of the seleno-ether (Fig. 5 ▶ d). The alkyl chain was inserted into the valley between helices composed of hydrophobic residues, including Leu105, Leu108, Tyr109, Ala112, Leu115 and Trp116 of helix IV, Tyr59 of helix II and Val16, Ala20, Leu24 and Ile27 of helix I* in the adjacent monomer. The electron density of maltoside was located next to the thiol group of the GSH.
The electron density of the alkyl chain of SeDDM was consistent with the previous DDM structure and the LTA4-binding model (Ago et al., 2007 ▶). The LTA4-binding model was constructed based around the alkyl chain of DDM. The aliphatic chain of LTA4 was embedded along the alkyl chain (C12) of DDM at the bottom of the cavity covered by the indole ring of Trp116 (Ago et al., 2007 ▶; Martinez Molina et al., 2007 ▶; Rinaldo-Matthis et al., 2010 ▶; Saino et al., 2011 ▶). This binding model implies that SeDDM and DDM affect the activity of LTC4S: experimentally, both SeDDM and DDM showed inhibitory activity against LTC4S catalysis in a preliminary enzyme assay (data not shown).
4. Conclusion
The SeDDM in the LTC4S crystal provided sufficient anomalous signal for selenium MAD phasing. The alkyl chains of the SeDDM molecules were surrounded by hydrophobic residues in all three sites, indicating that hydrophobic interactions are involved in the binding of SeDDM. This work suggests that SeDDM is applicable for phase determination. DDM molecules were found in the complex structures of 11 membrane proteins (Appendix A ); their alkyl chains also form hydrophobic interactions in their binding sites. The molecular weight per detergent ratio (kDa per detergent molecule; Appendix A ) shows that several membrane proteins bind a larger number of detergent molecules than LTC4S.
The alkyl chains of SeDDM formed hydrophobic interactions similar to those of DDM in the previously reported LTC4S structures. The SeDDM molecules bound three sites in competition with DDM molecules; therefore, it is possible that the DDM-binding sites were not fully substituted by SeDDM. Detergent exchange by a more thorough method, such as washing with SeDDM on an affinity column, may be necessary if the anomalous signal is insufficient.
In addition to experimental phasing, the anomalous scattering of the Se atom in the seleno-detergent allows the binding mode of these detergent molecules to be defined more accurately than with common detergents. The positions of the Se atoms and linked alkyl chains were confirmed by using the anomalous peaks as positional references. The position of the SeDDM alkyl chain in the putative LTA4-binding site was clearly defined. This result supports the previous LTA4-binding model based on the binding of DDM in the active site (Ago et al., 2007 ▶).
Supplementary Material
PDB reference: LTC4S with SeDDM, 3b29
Structure factors: contains datablock(s) r3b29sf. DOI: 10.1107/S1744309111042345/wd5163sup1.hkl
Acknowledgments
We appreciate the support of the RIKEN Structural Genomics Beamline staff. This work was supported in part by a grant-in-aid for Scientific Research in the Global Center of Excellence program (A-12) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to HA).
Appendix A. 60 membrane-protein structures with modelled alkyl-β-d-glucoside or alkyl-β-d-maltoside in the PDB
There are 299 unique membrane-protein structures in 847 PDB entries for membrane proteins (http://blanco.biomol.uci.edu/mpstruc/). The total molecular weight of the protein and number of detergents in the asymmetric unit were investigated for 141 of the detergent-containing structures (Table 2 ▶).
Table 2. Membrane-protein structures with modelled alkyl--D-glucoside and alkyl--D-maltoside in the PDB.
kDa/detergent indicates the molecular weight of protein per detergent molecule in the asymmetric unit.
| Detergent (PDB residue code) | Unique† | Name | PDB code | Structure weight (kDa) | No. of detergent molecules | kDa/detergent (kDa) |
|---|---|---|---|---|---|---|
| Octyl--D-glucoside (BOG) | 29‡ | Ram prostaglandin H2 synthase-1 (COX-1; Ovis aries) | 1pth | 133 | 2 | 66 |
| 1cqe | 133 | 5 | 27 | |||
| 1eqh | 133 | 3 | 44 | |||
| 1ht5 | 127 | 3 | 42 | |||
| 1ht8 | 127 | 3 | 42 | |||
| 1eqg | 133 | 3 | 44 | |||
| 1q4g | 127 | 8 | 16 | |||
| 2ayl | 127 | 7 | 18 | |||
| 3n8w | 127 | 3 | 42 | |||
| 3n8x | 127 | 2 | 64 | |||
| 3n8y | 127 | 4 | 32 | |||
| 3n8z | 127 | 5 | 25 | |||
| 3n8v | 127 | 5 | 25 | |||
| Glycerol-3-phosphate dehydrogenase (GlpD, native; Escherichia coli) | 2r4j | 114 | 6 | 19 | ||
| 2r45 | 114 | 6 | 19 | |||
| 2r46 | 114 | 5 | 23 | |||
| 2r4e | 114 | 6 | 19 | |||
| 2qcu | 114 | 6 | 19 | |||
| Dihydroorotate dehydrogenase in complex with atovaquone (Rattus rattus) | 1uum | 81 | 2 | 40 | ||
| Omp32 anion-selective porin (Delftia acidovorans) | 2fgq | 35 | 1 | 35 | ||
| OmpF matrix porin in complex with colicin peptide OBS1 (E. coli) | 3o0e | 232 | 1 | 232 | ||
| OmpG monomeric porin in open state (E. coli) | 2iww | 66 | 12 | 5 | ||
| VceC outer membrane protein (Vibrio cholerae) | 1yc9 | 48 | 1 | 48 | ||
| OmpT outer membrane protease (E. coli) | 1i78 | 67 | 4 | 17 | ||
| OmpLA (PldA) outer membrane phospholipase A monomer | 1qd5 | 32 | 5 | 6 | ||
| 1ild | 31 | 5 | 6 | |||
| 1im0 | 31 | 4 | 8 | |||
| 1ilz | 31 | 5 | 6 | |||
| Cytolysin pore-forming toxin protomer (V. cholerae) | 1xez | 80 | 1 | 80 | ||
| Sensory rhodopsin II (SRII; Natronomonas pharaonis) | 1jgj | 23 | 1 | 23 | ||
| 1h2s | 30 | 1 | 30 | |||
| Rhodopsin in meta II state (bovine rod outer segment) | 3pqr | 40 | 2 | 20 | ||
| 3pxo | 39 | 2 | 20 | |||
| 3dqb | 40 | 3 | 13 | |||
| Rhodopsin (squid) | 2z73 | 100 | 2 | 50 | ||
| M2 proton channel (influenza A) | 3bkd | 22 | 6 | 4 | ||
| SLAC1 anion channel, TehA homologue (wild type; Haemophilus influenzae A) | 3m73 | 35 | 4 | 9 | ||
| 3m74 | 35 | 4 | 9 | |||
| 3m75 | 35 | 4 | 9 | |||
| 3m76 | 35 | 4 | 9 | |||
| 3m71 | 35 | 4 | 9 | |||
| AQP4 aquaporin water channel (human) | 3gd8 | 24 | 1 | 24 | ||
| AqpM aquaporin water channel (Methanothermobacter marburgensis) | 2evu | 25 | 2 | 13 | ||
| AqpZ aquaporin water channel (E. coli) | 2o9g | 24 | 2 | 12 | ||
| 3nka | 48 | 4 | 12 | |||
| 3nkc | 48 | 3 | 16 | |||
| 3nk5 | 48 | 4 | 12 | |||
| GlpF glycerol facilitator channel (E. coli) | 1fx8 | 30 | 3 | 10 | ||
| 1ldi | 30 | 2 | 15 | |||
| 1lda | 30 | 2 | 15 | |||
| 1ldf | 30 | 2 | 15 | |||
| PfAQP aquaglyceroporin (Plasmodium falciparum) | 3c02 | 28 | 1 | 28 | ||
| Aqy1 yeast aquaporin (pH 3.5; Pichia pastoris) | 2w1p | 30 | 4 | 7 | ||
| 2w2e | 30 | 6 | 5 | |||
| FocA formate transporter without formate (V. cholerae) | 3klz | 153 | 14 | 11 | ||
| 3kly | 153 | 16 | 10 | |||
| AmtB ammonia channel (mutant; E. coli) | 1u7g | 40 | 1 | 40 | ||
| 2ns1 | 56 | 8 | 7 | |||
| Rh protein, possible ammonia or CO2 channel (Nitrosomonas europaea) | 3b9z | 41 | 2 | 20 | ||
| 3b9y | 41 | 2 | 20 | |||
| 3b9w | 43 | 1 | 43 | |||
| Human Rh C glycoprotein ammonia transporter (Homo sapiens) | 3hd6 | 54 | 1 | 54 | ||
| LeuTAa leucine transporter (Aquifex aeolicus) | 2a65 | 58 | 5 | 12 | ||
| 2q6h | 58 | 5 | 12 | |||
| 2qb4 | 58 | 5 | 12 | |||
| 2qei | 58 | 5 | 12 | |||
| 3f4j | 58 | 4 | 15 | |||
| 3f48 | 58 | 6 | 10 | |||
| 3f3e | 58 | 7 | 8 | |||
| 3f3d | 58 | 5 | 12 | |||
| 3f4i | 58 | 5 | 12 | |||
| 3f3c | 58 | 5 | 12 | |||
| 3f3a | 58 | 7 | 8 | |||
| 2qju | 57 | 4 | 14 | |||
| 3gjc | 115 | 8 | 14 | |||
| 3gjd | 58 | 6 | 10 | |||
| 3mpq | 57 | 3 | 19 | |||
| 3mpn | 57 | 6 | 9 | |||
| Oestrone sulfatase (human placenta) | 1p49 | 63 | 2 | 32 | ||
| Cytochrome bc 1 (Gallus gallus) | 2bcc | 229 | 1 | 229 | ||
| 1bcc | 229 | 1 | 229 | |||
| Light-harvesting complex (Rhodopseudomonas acidophila) | 1nkz | 31 | 6 | 5 | ||
| Nonyl--D-glucoside (BNG) | 11‡ | Archaerhodopsin-2 (aR-2; Halorubrum sp. aus-2) | 1vgo | 55 | 12 | 5 |
| Rhodopsin (bovine rod outer segment; Bos taurus) | 1hzx | 78 | 7 | 11 | ||
| 1l9h | 78 | 7 | 11 | |||
| Kir3.1 prokaryotic Kir chimera (Mus musculus/Burkholderia xenovorans) | 2qks | 72 | 1 | 72 | ||
| AQP0 aquaporin water channel (bovine lens) | 1ymg | 28 | 2 | 14 | ||
| AQP1 aquaporin red blood cell water channel (B. taurus) | 1j4n | 29 | 3 | 10 | ||
| GlpG rhomboid-family intramembrane protease (E. coli) | 2ic8 | 21 | 12 | 2 | ||
| 3b44 | 20 | 17 | 1 | |||
| 3b45 | 20 | 17 | 1 | |||
| 2o7l | 20 | 1 | 20 | |||
| 2xow | 20 | 16 | 1 | |||
| 2xov | 20 | 19 | 1 | |||
| 2xtu | 20 | 18 | 1 | |||
| FucP fucose transporter in outward-facing conformation (E. coli) | 3o7p | 48 | 1 | 48 | ||
| 3o7q | 48 | 1 | 48 | |||
| UraA uracil/H+ symporter (E. coli) | 3qe7 | 45 | 1 | 45 | ||
| AdiC arginine:agmatine antiporter (E. coli) | 3l1l | 47 | 1 | 47 | ||
| 3rlb | 42 | 11 | 4 | |||
| Cytochrome ba 3 (Thermus thermophilus) | 1ehk | 85 | 3 | 28 | ||
| Light-harvesting complex LHC-II, spinach photosystem II (Spinacia oleracea) | 1rwt | 250 | 10 | 25 | ||
| Nonyl--D-maltoside (ZDM) | 1 | ChbC EIIC phosphorylation-coupled saccharide transporter (Bacillus cereus) | 3qnq | 192 | 4 | 48 |
| Decyl--D-maltoside (DMU) | 3 | CorA Mg2+ transporter (Thermotoga maritima) | 2bbh | 32 | 4 | 8 |
| Cytochrome c oxidase, aa 3 (bovine heart mitochondria) | 1v55 | 410 | 2 | 205 | ||
| 1v54 | 410 | 2 | 205 | |||
| Cytochrome c oxidase, two-subunit catalytic core (Rhodobacter sphaeroides) | 2gsm | 185 | 10 | 19 | ||
| Dodecyl--D-maltoside (LMT) | 12‡ | Sulfide:quinone oxidoreductase in complex with decylubiquinone (A.aeolicus) | 3hyv | 285 | 6 | 48 |
| 3hyx | 285 | 6 | 48 | |||
| 3hyw | 285 | 6 | 48 | |||
| Prokaryotic pentameric ligand-gated ion channel (GLIC; Gloeobacter violaceus) | 3eam | 181 | 6 | 30 | ||
| 3p4w | 182 | 6 | 30 | |||
| 3p50 | 182 | 6 | 30 | |||
| GluCl anion-selective receptor (Fabivermectin complex; Caenorhabditis elegans) | 3rif | 431 | 3 | 144 | ||
| 3ri5 | 431 | 3 | 144 | |||
| 3ria | 431 | 3 | 144 | |||
| 3rhw | 431 | 3 | 144 | |||
| GlpG rhomboid-family intramembrane protease (E. coli) | 2irv | 41 | 1 | 41 | ||
| MexB bacterial multidrug efflux transporter (Pseudomonas aeruginosa) | 2v50 | 682 | 8 | 85 | ||
| Leukotriene C4 synthase in complex with glutathione (human) | 2pno | 209 | 57 | 4 | ||
| 2uui | 17 | 2 | 9 | |||
| 2uuh | 17 | 1 | 17 | |||
| NrfH cytochrome c quinol dehydrogenase (Desulfovibrio vulgaris) | 2j7a | 795 | 6 | 132 | ||
| Fumarate reductase complex (Wolinella succinogenes) | 1qlb | 260 | 2 | 130 | ||
| 2bs2 | 261 | 2 | 131 | |||
| Cytochrome c oxidase, aa 3 (Paracoccus denitrificans) | 3ehb | 123 | 12 | 10 | ||
| 3hb3 | 123 | 14 | 9 | |||
| Photosystem II (Thermosynechococcus elongatus) | 1s5l | 305 | 2 | 152 | ||
| 2axt | 304 | 6 | 51 | |||
| 3bz2 | 306 | 7 | 44 | |||
| 3bz1 | 306 | 7 | 44 | |||
| Photosystem II (T. vulcanus) | 3arc | 295 | 12 | 25 | ||
| Undecyl--D-maltoside (UMQ) | 4 | Rotor of V-type Na+-ATPase (Enterococcus hirae) | 2bl2 | 160 | 22 | 7 |
| Cytochrome bc 1 (Saccharomyces cerevisiae) | 1kb9 | 244 | 1 | 244 | ||
| 1p84 | 244 | 1 | 244 | |||
| 3cxh | 520 | 2 | 260 | |||
| 3cx5 | 520 | 2 | 260 | |||
| Cytochrome b 6 f complex (Mastigocladus laminosus) | 2e75 | 108 | 4 | 27 | ||
| 2e76 | 108 | 4 | 27 | |||
| 2e74 | 108 | 4 | 27 | |||
| Cytochrome b 6 f complex (Nostoc sp. PCC 7120) | 2zt9 | 106 | 3 | 35 |
The unique number of membrane proteins counts the same proteins from different species differently.
There are two membrane-protein structures in complex with two kinds of alkyl--D-glycosides each, i.e. BOG/BNG and BNG/LMT.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Ago, H., Kanaoka, Y., Irikura, D., Lam, B. K., Shimamura, T., Austen, K. F. & Miyano, M. (2007). Nature (London), 448, 609–612. [DOI] [PubMed]
- Bill, R. M., Henderson, P. J., Iwata, S., Kunji, E. R., Michel, H., Neutze, R., Newstead, S., Poolman, B., Tate, C. G. & Vogel, H. (2011). Nature Biotechnol. 29, 335–340. [DOI] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Joachimiak, A. (2009). Curr. Opin. Struct. Biol. 19, 573–584. [DOI] [PMC free article] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr. 26
- Martinez Molina, D., Wetterholm, A., Kohl, A., McCarthy, A. A., Niegowski, D., Ohlson, E., Hammarberg, T., Eshaghi, S., Haeggström, J. Z. & Nordlund, P. (2007). Nature (London), 448, 613–616. [DOI] [PubMed]
- Morth, J. P., Sørensen, T. L. & Nissen, P. (2006). Acta Cryst. D62, 877–882. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Rinaldo-Matthis, A., Wetterholm, A., Martinez Molina, D., Holm, J., Niegowski, D., Ohlson, E., Nordlund, P., Morgenstern, R. & Haeggström, J. Z. (2010). J. Biol. Chem. 285, 40771–40776. [DOI] [PMC free article] [PubMed]
- Saino, H., Ukita, Y., Ago, H., Irikura, D., Nisawa, A., Ueno, G., Yamamoto, M., Kanaoka, Y., Lam, B. K., Austen, K. F. & Miyano, M. (2011). J. Biol. Chem. 286, 16392–16401. [DOI] [PMC free article] [PubMed]
- Sonoda, Y., Newstead, S., Hu, N.-J., Alguel, Y., Nji, E., Beis, K., Yashiro, S., Lee, C., Leung, J., Cameron, A. D., Byrne, B. & Iwata, S. (2011). Structure, 19, 17–25. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. (2000). Acta Cryst. D56, 965–972. [DOI] [PMC free article] [PubMed]
- Walden, H. (2010). Acta Cryst. D66, 352–357. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: LTC4S with SeDDM, 3b29
Structure factors: contains datablock(s) r3b29sf. DOI: 10.1107/S1744309111042345/wd5163sup1.hkl